Abstract
Epidermal growth factor receptors (EGFR) are overexpressed in a wide range of malignancies including head and neck, colon, and breast cancers. It has been identified that carcinomas with high expression levels of EGFR are more resistant to radiotherapy. Therefore, inhibiting nuclear translocation of EGFR to increase the radiosensitivity of malignant cells expressing EGFR offers the potential for increasing the therapeutic index of radiotherapy. The purpose of the present study was to quantify and to compare the radiosensitizing properties of the well-known anti-EGFR antibodies, cetuximab and nimotuzumab in human epidermoid A431 overexpressing EGFR cells. Cells were treated with two concentrations of the antibodies and then irradiated with a single dose of 4 Gy. The results indicated that the two antibodies induced radiosensitization increasing the percentage of dead/dying cells and the yield of γ-H2AX foci 24 h after irradiation. Whereas cetuximab exhibited a significant increase in radiosensitization at the highest concentration, the effects of nimotuzumab were more modest. A correlation between γ-H2AX foci signals and dead/dying cells was observed. The disparity in modulation of radiation-induced DNA damage by the two antibodies could be associated with the level of their respective intrinsic cytotoxic properties. Overall, the findings highlight the potential therapeutic benefit of combination therapy with anti-EGFR antibodies and radiotherapy for relevant carcinomas.
Introduction
Ionizing radiation (IR) damages DNA directly by energy deposition and indirectly by the generation of reactive oxygen species. IR induces several types of DNA lesions including, base damage (BD), and single- and double-strand breaks (SSB, DSB). To respond to these potentially genotoxic lesions, cells have evolved complex signal transduction, cell-cycle checkpoints and repair pathways. BD and SSB are repaired by different processes such as the base excision repair, nucleotide incision repair, and single strand break repair pathways.Citation1,Citation2
The homologous recombination (HR) and DNA non-homologous end-joining (NHEJ) are the two major repair mechanisms of DSB.Citation3,Citation4
The epidermal growth factor receptor (EGFR) is linked with the repair mechanism of radiation-induced DBS. IR induces the nuclear translocation of EGFR,Citation5 which results in stimulation of the kinase activity of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs).Citation6-Citation8 Small molecules or monoclonal antibodies (mAbs) can inhibit the role of EGFR in the DSBs repair by blocking nuclear translocation, and hence increasing the sensitivity of cells to ionizing radiation.Citation9 This is of particular interest in those carcinomas exhibiting elevated expression levels of EGFR that correlates with radioresistance, as in glioblastomas and some head and neck cancers.Citation10-Citation12 At present blockage of EGFR with mAbs is a new molecular intervention strategy for increasing the radiosensitivity of radioresistant tumors.Citation5
Two of the most promising anti-EGFR antibodies are cetuximab and nimotuzumab.Citation13-Citation16 Preclinical and clinical studies have shown that cetuximab and nimotuzumab enhance the anti-tumor effects of radiotherapy.Citation13-Citation16 However, various assays have shown that cetuximab enhances skin reactions induced by ionizing radiation (IR).Citation13 On the contrary, no increase of radiation induced toxicity on skin have been described for nimotuzumab.Citation14-Citation16
Although it is well known that cetuximab influences the cellular response to radiation increasing the residual DNA damage in tumoral cells,Citation5 the effect of nimotuzumab on radiation-induced residual DNA damage remains unexplored. Here, we directly compared the residual DNA damage following combinations of ionizing radiation and the two mAbs (cetuximab and nimotuzumab) in A431 cells, using γ-H2AX as a molecular DSBs marker.Citation5,Citation9
Results
The number of foci per nuclei in the non-irradiated cells incubated for 24 h with mAbs was similar to those observed in cells non-treated with mAbs. The number of foci per nuclei after incubation with cetuximab was 1.6, while after incubation with nimotuzumab, and in cells non-treated with the mAb the number of foci per nuclei was 1.4 in both situations.
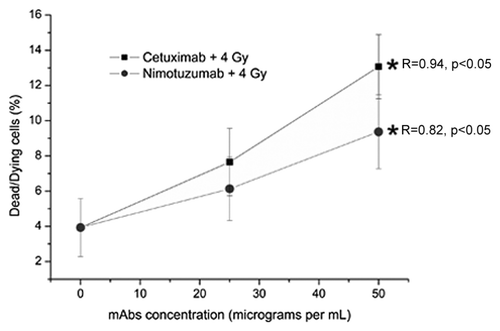
The incubation of A431 epithelial cells with both mAbs 1 h before irradiation results in a significant increase in the percentage of dead/dying cells observed 24 h after irradiation (). However, in spite that the cytotoxic response was 1.7 times greater for cetuximab than for nimotuzumab, there was not a statistical difference between them (p > 0.05).
Figure 1. Values are the mean percentage (%) (± Standard Deviation) of dead/dying A431 cells measured 24 h after irradiation of three independent experiments. The cells were incubated for 1 h with Nimotuzumab (gray circles) and Cetuximab (black squares) mAbs and exposed to ionizing radiation (137Cs, 4 Gy). *ANOVA and Student t-test on slope, p < 0.05. In spite of the cytotoxic response was 1.7 times greater for cetuximab than for nimotuzumab, there was not a statistical difference between then (p > 0.05).
All irradiated cultures, including those were not treated with mAb, showed significantly higher values for DNA damage compared with non-irradiated cultures (data not showed).
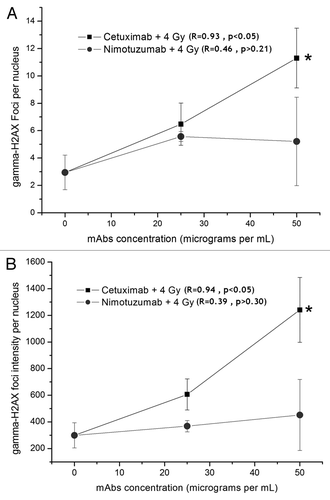
show the residual DNA damage observed after 24 h in cells treated and irradiated with both mAbs. The incubation with mAb increases the foci frequency in the irradiated cultures. When the residual DNA damage was tested 30 min after irradiation, instead of 24 h, there was not an increase the foci frequency. This increase is correlated with the concentration of the cetuximab used. Different behavior was observed after treatment with nimotuzumab, as more γ-H2AX foci were observed in treated cultures but at similar frequency in the two used concentrations.
Figure 2. Detection of residual DNA damage in A431 cells measured 24 h after irradiation (137Cs, 4 Gy). The cells were incubated for 1 h with Nimotuzumab (gray circles) and Cetuximab (black squares) and exposed to ionizing radiation (137Cs, 4 Gy). (A) γ-H2AX foci intensity per nucleus. (B) γ-H2AX foci number per nucleus. Values are means ± standard deviation of three independent experiments. *ANOVA and Student t-test on slope, p < 0.05.
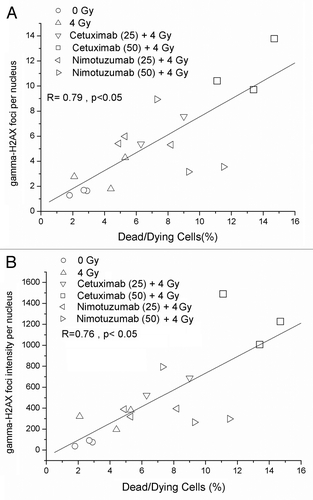
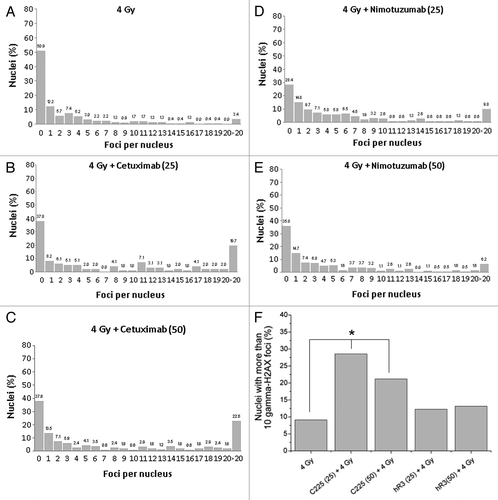
shows the distribution of γ-H2AX foci per cell 24 h after irradiation. For both mAb treatments there was a significant deviation from Poisson distribution (u values greater than 1.96) indicating an overdispersion. Moreover comparing distributions, statistical differences between cetuximab () and nimotuzumab () treatments were observed. After treatment with 25 µg mL−1, both mAbs showed similar values of residual damage. However, the proportion of highly damage cells (> 10 foci per nuclei) was higher for cetuximab treatment (47%) compared with nimotuzumab treatment (20%). Similarly, the proportion of highly damaged cells between the two concentrations increases only for cetuximab treatment ().
Figure 3. (A) Distribution of number of foci per nuclei after irradiation (137Cs, 4 Gy). (B) Nimotuzumab (25 µg/mL) pretreatment plus 4 Gy. (C) Nimotuzumab (50 µg/mL) pretreatment plus 4 Gy. (D) Cetuximab (25 µg/mL) pretreatment plus 4 Gy. (E) Cetuximab (50 µg/mL) pretreatment plus 4 Gy. (F) Percentage of nuclei containing more than 10 foci. *Chi-square trend test, p < 0.05.
The correlation analysis of data show a high degree of association between the percentage of dead/dying cells and the number of γ-H2AX foci per nuclei () and a high degree of correlation was also found between percentage of dead/dying cells and γ-H2AX foci intensity per nuclei ().
Discussion
The incubation of anti-EGFR monoclonal antibodies alone confirms the non-genotoxic effect per se already reported for cetuximab,Citation5 and demonstrates a similar non-genotoxicity behavior for nimotuzumab. The use of non-anti-EGFR antibodies as controls have been tested previously in several experiments with cetuximab and nimotuzumab without any unspecific observed effect.Citation5,Citation15,Citation17
Analyzing the gamma-H2AX response, the radiosensitiviy effect of cetuximab has been tested in several cell lines with different levels of EGFR expression and no significant differences in the radiosensitization induced were observed among them.Citation5,Citation17 Using only one concentration of cetuximab at 60 nM (9 µg mL−1), in A549 and MDA MB 231 tumor cells an increase in the residual DNA damage compared with untreated samples have been observed after X-ray irradiation at doses between 2 and 6 Gy.Citation17
Our results indicate that with cetuximab the number of residual foci observed after 24 h may depend on the used concentrations.
It should be noted that based on cytome criteria, in situ exclusion of cells with apparent characteristics of dead/dying cells is possible at the same time in the population of cells in which DNA damage is measured.Citation11,Citation12 This is an important advantage, as the use of the cytome criteria allow characterization of the cell survival response after mAb treatment without the necessity of clonogenic survival determination several days after irradiation. This is an important advantage of γ-H2AX foci assay compared with other DNA damage detection techniques like comet assay and it is particularly useful in circumstances when cytotoxicity could be a significant confounding factor like in this case.
The higher values of dead/dying cells and the concentration-dependence of γ-H2AX foci observed after the treatment with cetuximab most likely reflects the differences in the affinity to EGFR between the two mAbs. A difference of one order (10−9 to 10−8 M) between cetuximab and nimotuzumab has been described.Citation18 Another factor that would explain these results is that unlike cetuximab, nimotuzumab requires bivalent binding (two arms) for a stable attachment, which leads to it selectively binding to cells expressing moderate to high EGFR levels.Citation18
The poisson over-dispersion of foci counts is not unexpected if we consider γ-H2AX foci formation as a compound Poisson process that involve the energy deposition and the induction of DNA damage and its repair.Citation19 The interaction between the EGFR and the DNA repair can occur not only by a direct interaction between EGFR and DNA-PK, but also through another pathway in which the EGFR does not need to be translocated to the nucleus, by phosphatidylionitol 3-kinase (PI3K)-Akt or MAPK.Citation11 The different binding affinity of the two mAbs could be an important determinant in the alternative translocation pathway. The significance and persistence of cells with elevated γ-H2AX foci remains unclear. If one assumes that the residual foci are unsolved DSBs, a problem should occur if this damage is able to pass through cell division and hence to promote genomic instability. However, cells have different checkpoints to prevent the progression of DSB, and it has been observed that the G2/M checkpoint has a threshold of DSB, and that cells with more than 10 DSB typically do not progress to M phase.Citation20,Citation21
Finally, the high correlation between the percentage of dead/dying cells and the number of γ-H2AX foci per nuclei and foci intensity per nucleus in non-dead/dying cells probably reflects that γ-H2AX assay mainly measures the induced DSBs,Citation22-Citation24 and that DSBs are critical lesions on DNA with an elevated potential of lethality.
Our results show that in vitro at the tested concentrations, cetuximab has higher radiosensitizing properties than nimotuzumab nevertheless; less adverse effects and similar therapeutic properties have been reported for this anti-EGFR mAb. Cetuximab elicits severe cutaneous toxicity during concomitant radiotherapy which in many cases need modifications of prescribed radiotherapy and/or cetuximab regimes, and might compromise efficacy.13On the contrary, nimotuzumab can be administered concomitant with radiotherapy, including pediatric patients, without serious toxicities.Citation25
Materials and Methods
Cell culture, treatment and irradiation
A431 cells were grown in DMEM (GIBCO) culture medium with 10% fetal bovine serum (GIBCO), 15 mM HEPES (Sigma Chemical Co.) and 1% of Penicillin-Streptomycin liquid (Sigma). The cells were cultured at 37°C in a humidified incubator with 5% of CO2.
Before irradiation, cell cultures were incubated for 1 h with cetuximab (Merck) or nimotuzumab (CIMAB SA) using two concentrations 25 and 50 µg mL−1. These concentrations were chosen because they are in the therapeutic serum levels.Citation14,Citation26,Citation27 Cultures were then irradiated at 4 Gy using a 137Cs source at a dose rate of 5.4 Gy min−1.
Non-irradiated cells and irradiated cells without mAbs treatment were used as controls in irradiated cultures, while non-irradiated cells incubated for 1 h with cetuximab or nimotuzumab at 50 µg mL were used to evaluate the genotoxic effect per se of both mAbs. Twenty-four hours after irradiation the cells were sampled to determine the dead/dying cells percentage and DNA damage.
Cell selection criteria for focus analysis. Detection of dead/dying cells
The criteria for choosing cells for analysis was based on nuclear morphology following the principles developed for the cytome assay.Citation28,Citation29 Cells were considered as normal when they showed a uniformly stained nucleus which is usually oval or round in shape. No other DNA containing structures apart from the nucleus are observed in the normal cells, the analysis of γ-H2AX foci was done in these cells. Cells with abnormal nuclear chromatin include cells with condensed chromatin, karyolytic cells, karyorrhectic cells and pyknotic cells. These types of cells were considered dead/dying and not analyzable for foci.Citation28 The percentage of dead/dying cells from a total of 1,000 cells was considered as a measure of cytotoxicity.
Immunofluorescence staining
Thirty minutes and twenty-four hours after irradiation the cultures were trypsinized, collected and spotted onto slides with a cytocentrifuge (Thermo Shandon cytospin). The cells were fixed with 2% paraformaldehyde (PFA) in phosphate-buffered saline (PBS; pH 7.0), without Mg2+ and Ca2+ (Merck) for 10 min at room temperature (RT, 25°C), washed with PBS and treated with PBS plus 0.3% Triton X-100 at RT for 10 min to permeabilize. Cells were fixed with methanol at 0°C for 10 min. The slides with cells were then washed with PBS. After the permeabilization and fixation steps the cells were blocked in PBS with 2% of bovine serum albumin (BSA, Sigma) at RT for 30 min and were incubated for 1 h at 37°C with primary anti-γH2AX monoclonal antibody (1:400; Abcam), washed in PBS and incubated with fluorescein isothiocyanate (FITC) conjugated goat anti-mouse secondary antibody (1:400; Abcam) for 1 h at 37°C. After extensive washing, slides were mounted with coverslips using 4´,6-di-amidino-2-phenylindole (DAPI) Vectashield solution (Vector Laboratories).
Image acquisition and processing
Slides were analyzed using an epifluorescence microscope (Provis AX70, Olympus). The images were acquired using a 100× PlanApo objective and a non-cooled digital (Charge Coupled Device) camera. Fields were randomly selected on the basis of DAPI-counterstained nuclei. After acquisition of the DAPI and FITC images, the nuclear and focal structures were defined automatically using CellProfiler software (www.cellprofiler.org). A CellProfiler pipeline (available upon request) was designed to count nuclei and foci and for the characterization of foci in size and signal intensity. The CellProfiler pipeline was tested against visual detection of foci and a linear response with a slope not different to one and an intercept not different to zero was obtained.
Data and statistical analyses
The γ-H2AX and percentage of dead/dying cells data follows a Gaussian distribution and was represented by the mean and standard deviation of the mean of independent irradiations. The γ-H2AX and percentage of dead/dying cells data were analyzed by analysis of variance (ANOVA) trend analysis and linear regression. Regression results were compared using a Student t-test. For each culture, a sufficient number of nuclei were analyzed to guarantee that at least 100 foci be scored. The distribution of cell by the number of foci was compared by Chi-square test. The fit of foci per cell to the Poisson distribution was tested by the u-test. P < 0.05 was considered as being statistically significant. Correlation between the percentage of dead/dying cells and the number of γ-H2AX foci per nuclei was analyzed by Pearson correlation.
Conclusion
Our results indicate that the two anti-EGFR mAbs are radiosensitizers increasing the percentage of dead/dying cells. However significant differences exist between cetuximab and nimotuzumab in the modulation of the residual DNA damage response measured as the yield and distribution of γ-H2AX foci per nuclei 24 h after irradiation in A431 tumor cells under the experimental conditions. These disparities are most likely associated with their differing levels of intrinsic cytotoxicity.
| Abbreviations: | ||
| EGFR | = | epidermal growth factor receptor |
| TKI | = | tyrosine kinase inhibitor |
| mAb | = | monoclonal antibody |
| SSB | = | single strand breaks |
| DSB | = | double strand breaks |
| DNA-PKcs | = | DNA-dependent protein kinase catalytic subunit |
| FBS | = | fetal bovine serum |
| BSA | = | bovine serum albumin FITC, fluorescein iso-thiocyanate |
| DAPI | = | 4´,6-di-amidino-2-phenylindole |
Acknowledgments
The authors wish to thanks Jessica Martinez from the Autonomous University of Barcelona for her technical assistance. The authors also wish to thanks the advice of Leonard Barrios from the Autonomous University of Barcelona. The critical review of Tom C. Karagiannis is greatly appreciated. Part of this work was supported by IAEA fellowship CUB/10021.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Fortini P, Dogliotti E. Base damage and single-strand break repair: mechanisms and functional significance of shortand long-patch repair subpathways. DNA Repair (Amst) 2007; 6:398 - 409; http://dx.doi.org/10.1016/j.dnarep.2006.10.008; PMID: 17129767
- Caldecott KW. Mammalian single-strand break repair: mechanisms and links with chromatin. DNA Repair (Amst) 2007; 6:443 - 53; http://dx.doi.org/10.1016/j.dnarep.2006.10.006; PMID: 17118715
- Wyman C, Kanaar R. Homologous recombination: down to the wire. Curr Biol 2004; 14:R629 - 31; http://dx.doi.org/10.1016/j.cub.2004.07.049; PMID: 15296782
- Hefferin ML, Tomkinson AE. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair (Amst) 2005; 4:639 - 48; http://dx.doi.org/10.1016/j.dnarep.2004.12.005; PMID: 15907771
- Dittmann K, Mayer C, Fehrenbacher B, Schaller M, Raju U, Milas L, et al. Radiation-induced epidermal growth factor receptor nuclear import is linked to activation of DNA-dependent protein kinase. J Biol Chem 2005; 280:31182 - 9; http://dx.doi.org/10.1074/jbc.M506591200; PMID: 16000298
- Bandyopadhyay D, Mandal M, Adam L, Mendelsohn J, Kumar R. Physical interaction between epidermal growth factor receptor and DNAdependent protein kinase in mammalian cells. J Biol Chem 1998; 273:1568 - 73; http://dx.doi.org/10.1074/jbc.273.3.1568; PMID: 9430697
- Burma S, Chen DJ. Role of DNA-PK in the cellular response to DNA double-strand breaks. DNA Repair (Amst) 2004; 3:909 - 18; http://dx.doi.org/10.1016/j.dnarep.2004.03.021; PMID: 15279776
- Burma S, Chen BP, Chen DJ. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair (Amst) 2006; 5:1042 - 8; http://dx.doi.org/10.1016/j.dnarep.2006.05.026; PMID: 16822724
- Balaban N, Moni J, Shannon M, Dang L, Murphy E, Goldkorn T. The effect of ionizing radiation on signal transduction: antibodies to EGF receptor sensitize A431 cells to radiation. Biochim Biophys Acta 1996; 1314:147 - 56; http://dx.doi.org/10.1016/S0167-4889(96)00068-7; PMID: 8972728
- Ang KK, Berkey BA, Tu X, Zhang HZ, Katz R, Hammond EH, et al. Impact of epidermal growth factor receptor expression on survival and pattern of relapse in patients with advanced head and neck carcinoma. Cancer Res 2002; 62:7350 - 6; PMID: 12499279
- Hatanpaa KJ, Burma S, Zhao D, Habib AA. Epidermal growth factor receptor in glioma: signal transduction, neuropathology, imaging, and radioresistance. Neoplasia 2010; 12:675 - 84; PMID: 20824044
- Nyati MK, Morgan MA, Feng FY, Lawrence TS. Integration of EGFR inhibitors with radiochemotherapy. Nat Rev Cancer 2006; 6:876 - 85; http://dx.doi.org/10.1038/nrc1953; PMID: 17036041
- Pryor DI, Porceddu SV, Burmeister BH, Guminski A, Thomson DB, Shepherdson K, et al. Enhanced toxicity with concurrent cetuximab and radiotherapy in head and neck cancer. Radiother Oncol 2009; 90:172 - 6; http://dx.doi.org/10.1016/j.radonc.2008.09.018; PMID: 18976827
- Crombet T, Osorio M, Cruz T, Roca C, del Castillo R, Mon R, et al. Use of the humanized anti-epidermal growth factor receptor monoclonal antibody nimotuzumab in combination with radiotherapy in the treatment of locally advanced head and neck cancer patients. J Clin Oncol 2004; 22:1646 - 54; http://dx.doi.org/10.1200/JCO.2004.03.089; PMID: 15117987
- Akashi Y, Okamoto I, Iwasa T, Yoshida T, Suzuki M, Hatashita E, et al. Enhancement of the antitumor activity of ionising radiation by nimotuzumab, a humanised monoclonal antibody to the epidermal growth factor receptor, in non-small cell lung cancer cell lines of differing epidermal growth factor receptor status. Br J Cancer 2008; 98:749 - 55; http://dx.doi.org/10.1038/sj.bjc.6604222; PMID: 18253126
- Allan DGP. Nimotuzumab: Evidence of clinical benefit without rash. Oncologist 2005; 10:760 - 1; http://dx.doi.org/10.1634/theoncologist.10-9-760; PMID: 16249358
- Dittmann K, Mayer C, Rodemann H-P. Inhibition of radiation-induced EGFR nuclear import by C225 (Cetuximab) suppresses DNA-PK activity. Radiother Oncol 2005; 76:157 - 61; http://dx.doi.org/10.1016/j.radonc.2005.06.022; PMID: 16024112
- Garrido G, Tikhomirov IA, Rabasa A, Yang E, Gracia E, Iznaga N, et al. Bivalent binding by intermediate affinity of nimotuzumab A contribution to explain antibody clinical profile. Cancer Biol Ther 2011; 11:1 - 3; http://dx.doi.org/10.4161/cbt.11.4.14097; PMID: 21189451
- Puig P, Barquinero JF. An application of compound Poisson modeling to biological dosimetry. Proc R Soc A 2010; doi:10.1098/rspa.2010.0384.
- Deckbar D, Birraux J, Krempler A, Tchouandong L, Beucher A, Walker S, et al. Chromosome breakage after G2 checkpoint release. J Cell Biol 2007; 176:749 - 55; http://dx.doi.org/10.1083/jcb.200612047; PMID: 17353355
- Löbrich M, Jeggo PA. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Natl Rev 2007; 7:861 - 9; http://dx.doi.org/10.1038/nrc2248; PMID: 17943134
- Redon CE, Nakamura AJ, Zhang Y-W, Ji J, Bonner WM, Kinders RJ, et al. Histone γH2AX and Poly (ADP-Ribose) as Clinical Pharmacodynamic Biomarkers. Clin Cancer Res 2010; 16:4532 - 42; http://dx.doi.org/10.1158/1078-0432.CCR-10-0523; PMID: 20823146
- Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, et al. γH2AX and cancer. Natl Rev 2008; 8:957 - 67; http://dx.doi.org/10.1038/nrc2523
- Banath JP, Peggy L. Expression of phosphorylated histone H2AX as a surrogate of cell killing by drugs that create DNA double-strand breaks. Cancer Res 2003; 63:4347 - 50
- Massimino M, Bode U, Biassoni V, Fleischhack G. Nimotuzumab for pediatric diffuse intrinsic pontine gliomas. Expert Opin Biol Ther 2011; 11:247 - 56; PMID: 21171927
- Crombet T, Torres L, Neninger E, Catala´ M, Solano M, Perera A, et al. Pharmacological evaluation of humanized anti-epidermal growth factor receptor, monoclonal antibody nimotuzumab, in patients with advanced epithelial-derived cancer. J Immunother 2003; 26:139 - 48; http://dx.doi.org/10.1097/00002371-200303000-00006; PMID: 12616105
- Fracasso P, Burris H, Arquette M, Govindan R, Gao F, Wright L, et al. A phase 1 escalating single-dose and weekly fixed-dose study of cetuximab: Pharmacokinetic and pharmacodynamic rationale for dosing. Clin Cancer Res 2007; 13:986 - 93; http://dx.doi.org/10.1158/1078-0432.CCR-06-1542; PMID: 17289894
- Thomas P, Harvey S, Gruner T, Fenech M. The buccal cytome and micronucleus frequency is substantially altered in Down’s syndrome and normal ageing compared to young healthy controls. Mutat Res 2008; 638:37 - 47; http://dx.doi.org/10.1016/j.mrfmmm.2007.08.012; PMID: 17920640
- González JE, Roch-Lefèvre SH, Mandina T, García O, Roy L. Induction of γ-H2AX foci in human exfoliated buccal cells after in vitro exposure to ionizing radiation. Int J Radiat Biol 2010; 86:752 - 9; http://dx.doi.org/10.3109/09553002.2010.484476; PMID: 20597838