Abstract
Purpose: Expression of the PGE2 receptor, EP4, is up-regulated during colorectal carcinogenesis. However the mechanism leading to deregulation of the EP4 receptor is not known. The present study was conducted to investigate the regulation of EP4 receptor by miRNAs.
Experimental Design: We analyzed 26 colon cancers (i.e. 15 adenocarcinomas and 9 adenomas) and 16 normal colon specimens for EP4 receptor expression by immunohistochemistry. A bioinformatics approached identified putative microRNA binding sites with the 3'-UTR of the EP4 receptor. Both colon cancer cell lines and tumor specimens were analyzed for miR-101 and EP4 expression by qRT-PCR and Western analysis respectively and simultaneously in situ hybridizations was used to confirm our results. In vitro and in vivo assays were used to confirm our clinical findings.
Results: We observed an inverse correlation between the levels of miR-101 and EP4 receptor protein. Transfection of LS174T cells with miR-101 significantly suppressed a luciferase reporter containing the EP4 receptor-3'-UTR. In contrast, a mutant EP4 receptor-3'-UTR construct was unaffected. Ectopic expression of miR-101 markedly reduced cell proliferation and motility. Co-transfection of EP4 receptor could rescue colon cancer cells from the tumor suppressive effects of miR-101. Moreover, the pharmacologic inhibition of EP4 receptor signaling or silencing of EP4 receptor phenocopied the effect of miR-101. This is the first study to show that the EP4 receptor is negatively regulated by miR-101.
Conclusions: These data provide new insights in the modulation of EP-4 receptor expression at the post-transcriptional level by miR-101 and suggests therapeutic strategies against miR-101 targets may be warranted.
Introduction
Colorectal cancer (CRC) is the second most common cause of cancer mortality in Western countries and claims > 50,000 lives a year in the United States alone.Citation1 Approximately 70% of patients that are affected by colorectal cancer undergo surgical resection and 30–40% of these patients develop recurrent disease.Citation2 The liver is the most common site of metastatic CRC and complete resection of hepatic metastases is the only curative option; however, surgery can only be performed on approximately 20% of patients at the time of diagnosis and 5-y survival rates average 25–40% despite adjuvant chemotherapy.Citation3 Among patients with metastatic CRC receiving 5-flourouracil (5-FU) as first line chemotherapy and new medications such as Irinotecan, Xeloda, Oxaliplatin, Erbitux, and Avastin, the median time to progression is 8–9 mo and their mean survival is 15–20.5 mo.Citation3–Citation5 Thus it is mandatory to elucidate molecular mechanisms of the metastasis process to improve diagnosis and develop appropriate treatment modalities.
Several lines of evidence demonstrate the functional importance of overproduction of cyclooxygenase (COX)-2 and one of its major metabolites, PGE2 in many human cancers including colorectal cancers.Citation6–Citation8 Preclinical studies implicate both COX-2 and PGE2 in many hallmark characteristics of malignant disease including metastasis (reviewed in refs. Citation7 and Citation8). For example, in vitro studies show that PGE2 stimulates CRC cell proliferation and invasion as well as resistance to apoptosis.Citation9–Citation11 In addition, in vivo studies demonstrate that PGE2 can drive intestinal tumorigenesis in animal models of colon cancer (reviewed in ref. Citation12).
PGE2 signals are transduced via four G-protein coupled cell surface receptors, termed as EP1, EP2, EP3, and EP4 receptors. There is a growing appreciation that the EP4 receptor is an important transducer of PGE2 signaling leading to cell invasion and motility during tumorigenesis. Interestingly, constitutive expression of EP4 promotes proliferation and anchorage-independent growth,Citation13 demonstrating that the EP4 receptor may also be a key regulator of tumor progression. Also EP4 receptor signaling appears to be important for cell movement and motility during development. Studies in zebrafish show that the EP4 receptor transduces PGE2 signaling to regulate appropriate speed of cell migration during gastrulation, demonstrating that regulation of cell motility by EP4 receptor signaling is evolutionarily conserved.Citation14 PGE2 stimulates the proliferation and motility of LS174T adenocarcinoma cells through the EP4 dependent activation of phosphatidylinositol 3-kinase/AKT signaling.Citation15 Whereas PGE2 inhibits apoptosis in human Caco-2 adenocarcinoma cells through a EP4 dependent pathway.Citation16 Furthermore, premalignant aberrant crypt foci formation in EP4 deficient mice following azoxymethane treatment is suppressed compared with the EP4 wild type mice.Citation17 This study also showed a reduction in colon adenomatous polyp formation in mice wild-type for the EP4 receptor treated with the EP4 receptor antagonist ONO-AE2-227.Citation17 Treatment with another EP4 antagonist, ONO-AE3-208, decreased liver metastases after intrasplenic injection of MC26 CRC cells.Citation18 In addition, in vitro studies by our group and others indicate that PGE2/EP4 receptor signaling via ERK activation promotes tumorigenic behavior of CRC cells.Citation9,Citation19 Finally, enforced expression of EP4 receptors promotes the tumorigenic behaviors of HT-29 CRC cancer cells.Citation10 Also there is a limited amount of evidence that suggests that increased EP4 expression occurs in human colon cancers compared with normal colorectal mucosa.Citation11 However, exactly how this occurs remains to be clarified.
MicroRNAs (miRNAs) have emerged as important regulators of gene expression and implicated in many human cancers.Citation20,Citation21 The pairing of miRNAs with the mRNAs of their target genes carrying a partially complementary sequence in the 3′-UTR results in translation repression and/or degradation of mRNA and thus silencing of correlated genes.Citation22 Deregulation of the expression of miRNAs that regulated genes involved in cellular proliferation, differentiation or apoptosis, have been observed in different human cancers, including colon cancers.Citation23 MicroRNAs 199 and 101a can regulate the expression of COX-2 during embryo transplantation.Citation24 Also, miR-101 is downregulated in endomentrial, hepatocellular carcinomas, and prostate cancers.Citation25–Citation27 In addition, decreased miR-101 has been found to be involved in COX-2 overexpression in human colon cancers.Citation28 In present study, we assess whether EP4 expression could be regulated by one or more miRNAs in human CRC cells and whether elevated EP4 expression in human CRC could be the result of an aberrant expression of miRNAs.
Results
Prostaglandin EP4 receptor expression increases during tumor progression.
We first examined EP4 receptor expression in human colon cancer specimens. There were low levels of EP4 protein in normal mucosa located mainly on membrane surface (). Similar findings were seen in adenomas (). In contrast malignant lesions (i.e., adenocarcinoma and metastatic lesions) stained strongly for EP4 receptor expression (). EP4 receptor expression was elevated in malignant lesions compared with adenoma and normal colonic epithelium (). The lack of staining in the stromal tissue in both normal as well as cancerous lesions indicates specificity of staining to colonic cells. In addition, there was an increased staining in high grade carcinomas (). Interestingly, in one case, malignant colon cancer cells that had penetrated into the intravascular space displayed strong EP4 staining (data not shown). However, consistent with previous reports, we did not observed a significant increase in EP4 mRNA in malignant lesions compared with normal tissues (data not shown).Citation34 Collectively, these data suggests that elevated EP4 receptor protein expression might occur by post-transcriptional mechanism.
An evolutionarily conserved target sequence for miR-101 binding is present in the 3′-UTR of EP4 gene sequence.
The PTGER4 mRNA encodes for the human EP4 protein. To identify miRNA target sites located within the 1.3 kb 3′-UTR of the human PTGER4/EP4 mRNA, predictions from target predictor algorithms were compiled and compared. We searched TargetScan, PicTar, Pita and miRanda-mirSVR for microRNAs capable of binding to the 3′-UTR of the EP4 receptor. A 100% match at position +18 to +27 in the 3′-UTR was observed in multiple database searches (). Comparing the human sequence for interspecies homology, we found that miR-101 target seed sequence at nt 18 to 27 of the PGTER4/EP4 -3′-UTR is highly conserved among several species including chimpanzee, rhesus, cattle and dog (). Because miR-101 has been found to regulate COX-2 in human colon cancer, we focused our studies on miR-101.Citation28
EP4 protein expression correlates inversely with miR-101 levels in colorectal cancer cell lines and resected patient samples.
First, we determined expression levels of miR-101 and EP4 protein in several CRC cell lines. In cell lines with low miR-101, as measured by qRT-PCR, there were high amounts of EP4 protein (). Conversely, in cell lines in which miR-101 was highly expressed there were low levels of EP4 protein. Next, we sought to corroborate our initial findings in CRC cell lines by analyzing resected tumor and corresponding normal tissues of 12 patients with CRC for EP4 protein (western) and miR-101 (qRT-PCR) levels. Representative examples are shown in . As reported for other tumor entities (for example Strillaci et al.Citation28 and Su et al.Citation26), we observed that miR-101 showed a higher expression in normal colonic tissues compared with corresponding colorectal tumor lesions. However, EP4 protein was still observed to be high in colon tumor samples as previously observed by us () and others.Citation11 Moreover, on normal and CRC tissue sections, in situ hybridization revealed expression of miR-101 in normal colonic epithelium and a loss of miR-101 expression in malignant epithelial cells (). The absence of staining with a scrambled probe demonstrates the specificity of the technique (). To clarify the clinical significance of the altered miR-101 expression in CRC tissue, we performed the sensitive fluorescence-based in situ hybridization method together with the routine clinical immunohistochemical assay to visualize the co-expression of miR101 and EP4 protein in the same tissue section. This data identified malignant epithelial cells co-expressing significant EP4 levels with low miR-101 levels and cells expressing high EP4 levels with no detectable miR-101 expression ( parts b, c and d) confirming that in the later cells the absence of miR-101 expression has allowed the full expression of EP4 levels and therefore that miR-101 represses EP4 expression. Taken together our data from both CRC cell lines and CRC tissue support the notion that the EP4 receptor expression is negatively regulated by miR-101 at the post-transcriptional level.
The EP4-3′-UTR is a target for miR-101.
Given the hypothesis that the EP4 receptor might be a target of miR-101, we asked whether the 3′-UTR of EP4 is a functional target of miR-101. We cloned the miR-101 seed sequence of wildtype-EP4-3′-UTR into a luciferase reporter construct at a position immediately downstream to the luciferase gene (). In parallel, we generated a second reporter construct in which the conserved targeting region of miR-101 within the EP4-3′-UTR was specifically mutated and predicted to abolish binding (). The relative luciferase activity of the reporter that contained wild-type 3′-UTR was significantly suppressed when pcDNA3.1-miR101 was co-transfected (). In contrast, the luciferase activity of mutant reporter was unaffected by simultaneous transfection of pcDNA3.1-miR-101 (). Similar findings were observed with a EP4-3′-UTR in which seed sequence was specifically deleted (data not shown). Furthermore, we examined the effect of miR-101 on the endogenous expression of EP4 by transfecting pcDNA3.1-miR-101 plasmid for long periods of time and demonstrated a time-dependent decrease in the endogenous EP4 receptor protein levels in LS174T colon cancer cells (). These data indicate that miR-101 may regulate the expression of the EP4 receptor at the posttranscriptional level by directly targeting the 3′-UTR sequence of the PTGER4/EP4 mRNA.
miR-101 suppresses colony formation and motility in vitro.
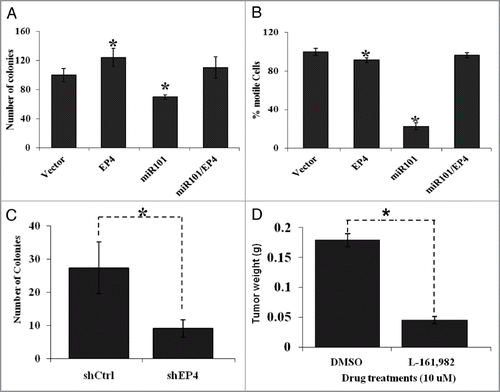
The significant reduction of miR-101 expression in CRC samples and the observations from our luciferase assays prompted us to explore the possible functional significance of miR-101. As an initial step, the capacity of colony formation was evaluated in LS174T colon cancer cells, followed by analysis of motility in transwell motility assays. First, we observed that ectopic expression of EP4 receptor caused an increase in the number of colony forming units. Next, we also saw an enhancement in the percentage of motile cells. In contrast to these observations, we demonstrated that enhanced miR-101 expression suppressed colony formation ability as well as tumor cell motility of CRC cell lines. Interestingly and more importantly, co-expression of EP4 receptor effectively and significantly restored tumorigenic and motile behavior of high miR-101 expressing CRC cell lines, that otherwise remained anti-tumorigenic ( and B). Moreover, genetic silencing of the EP4 receptor (by RNA interference) or pharmacological inhibition of the EP4 receptor signaling (by using EP4 specific antagonist, L-161,982), recapitulated the antitumorigenic effects of miR-101 ( and D). Thus, these functional studies support the notion that EP4 receptor is negatively regulated by miR-101 at the post-transcriptional level.
Discussion
This is the first study to show that the pro-tumorigenic activity of the prostanoid EP4 receptor is negatively regulated by miR-101 at the post-transcriptional level, via a specific target site within the 3′-UTR. It is also the first study to demonstrate that miR-101 inhibits cell proliferation and migration of colon cancer cells in vitro. Finally, we also show for the first time that there is an inverse correlation between the levels of miR-101 and expression of EP4 receptor protein in human colon cancers. We show data strongly suggesting that EP4 receptor expression increases as a result of loss of miR-101.
We observed an increase in EP4 receptor expression in malignant CRC (i.e., primary CRC and metastasis) compared with normal colorectal mucosa and adenomas. This suggests a predominant role for PGE2-EP4 receptor signaling at later stages of colorectal carcinogenesis. Moreover, PGE2-EP4 receptor signaling promotes tumorigenic behavior (proliferation, resistance to apoptosis, motility, and invasion).Citation12 Previous studies report that miR-101 promotes apoptosis, inhibits cell proliferation, as well as cell motility and invasion.Citation27 Based on our present study showing that tumor suppressive effects of miR-101 can be overcome by concomitant ectopic expression of EP4, it is interesting to speculate that loss of miR-101 contributes to colorectal cancer progression at least in part by increased EP4 expression. However, since miR-101 has multiple targets that play role in cancer (i.e., fos, EZH2, Cox-2, N-Myc, Mcl-1), and it is likely that other post-trancriptional targets of miR-101 also play a role in colorectal carcinogenesis.
Inhibition of PGE2 biosynthesis by NSAIDs and COX-2 inhibitors (Coxibs) represents an effective therapy to treat colorectal cancers and other diseases.Citation35 Unfortunately, their clinical utility is limited by their potential to cause either gastro-intestinal toxicity (by NSAIDs)Citation36 or cardiovascular (CV) side effects (by both NSAIDs and Coxib).Citation37 Therefore, there is a vast unmet medical need to discover alternatives for treating cancer and other chronic inflammatory conditions.
The adverse CV effects associated with NSAIDs and Coxibs are not completely understood however it is postulated that the pro-thrombotic and hypertensive effects are caused by inhibition of prostacyclin biosynthesis (for a recent review see ref. Citation38). It is plausible that a selective EP4 antagonist may be efficacious without the potential CV side effects observed with NSAIDs and COX-2 inhibitors since they should not interfere with the biosynthesis of other prostanoids such as prostacyclins and thromboxanes. Indeed, several groups have reported the discovery of potent and selective EP4 receptor antagonists.Citation39–Citation42 However, the effectiveness of this new generation of EP4 antagonists against malignant lesions warrants further investigation.
In conclusion, our present study indicates that the protumorigenic EP4 prostanoid receptor is regulated at the post-transcriptional level by miR-101 via a specific target motif at nucleotides +18 to +27 of the EP4-3′-UTR. Furthermore, miR101 inhibits colon cancer cell proliferation and invasion. Our findings in clinically resected colonic tissues from CRC patients together with previously reported studies of miR-101 in several cancers support the notion that inhibitory pharmacological strategies against miR-101 target genes such as COX-2/ EP4 and EZH2Citation43 could have a strong rationale for therapeutic applications in cancer in the future.
Materials and Methods
Materials, antibodies, cell lines and patient samples.
CRC cell lines (ATCC) were maintained in 1x DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS) and 5 mg/mL penicillin-streptomycin. All transient transfections were performed using LipofectAMINE 2000 (Invitrogen). Formalin fixed archival tissue sections were obtained from the Department of Pathology archives. Frozen tissue specimens (tumor, adjacent normal mucosa) from patients with colorectal cancer were obtained from the Arizona Cancer Center G.I. SPORE tissue core after informed consent and verification by a pathologist.
Construction of 3′-UTR-luciferase plasmid and reporter assays.
We cloned 53 nucleotide sequences of 3′-UTR of the PTGER4 gene (the PTGER4 gene encodes the EP4 mRNA, to avoid confusion we will simply describe our results as the EP4 mRNA) carrying the wild-type miR-101 seed sequence and mutant miR-101 seed sequence into the Xba1 site of pGL3 using the In-fusion Advantage PCR Cloning Kit (Clontech). The orientations of the clones were confirmed using both the PCR and sequencing techniques. The wild-type and mutant clones were named pGL3-EPWt and pGL3-EPMUT (respectively). The pri-miR-101 sequence was cloned into the pcDNA3.1. For reporter assays, LS174T were transiently cotransfected with pGL3-EPWt, pRL-null and pcDNA3.1-miR101 or pGL3-EPMUT, pRL-null and pcDNA3.1-miR-101 using lipofectamine 2000. Reporters assays were performed 48 h posttransfection using the Dual-Luciferase Assay System (Promega), normalized for transfection efficiency by co-transfected Renillaluciferase (pRL-null).
Preparation of cell and tissue lysates and western analysis.
Cells were washed with phosphate-buffered saline and lysed in extraction buffer as previously described.Citation29 Fifty to one hundred milligrams of tissue were homogenized using a sonicator and proteins extracted. Protein concentration was determined by BCA (Pierce). Aliquouts (50 µg) were separated on 10% SDS-PAGE and transferred to PDVF membranes. The membranes were stained with specific antibody followed by horseradish-peroxidase linked immunoglobin G, and visualized by chemiluminescence (ECL).
Real-time PCR-based detection of miR-101.
Small RNA from cells or human normal tissue/matched tumor samples were extracted using mirVana miRNA isolation Kit according to the manufacturer's instructions (Applied Biosystems,). Expression of mature miRNAs was determined by qRT-PCR using SyBR Green and primers specific for miR-101. Signals of miR-101 were normalized to U6-snRNA using the 2-ΔΔCT method. All PCRs were performed in triplicate.
Motility assay.
Cell motility was assessed as previously described by our group.Citation29 Transfected cells were seeded in the upper chamber of an 8 µm pore Falcon transwell insert (24 well format). The bottom chamber was filled with 600 µL of OptiMEM (with antibiotics). Transwell plates containing cells were incubated for 5 d at 37 °C, 5% CO2. Experiment was repeated two times. At the end of the incubation, the medium of the upper and bottom chambers were aspirated and the inserts were placed upside down. The top surface of the inserts was stained with 50 µL of crystal violet stain (0.5% crystal violet in 20% methanol) for 1 min at room temperature. Excess crystal violet solution was removed by plunging the inserts in distilled water several times and then rinsing a second time in distilled water. Non-motile cells inside the insert were removed with a wet cotton swap and the inserts were allowed to dry overnight. Pictures of five fields of each insert were captured under a light microscope and motile cells (cells penetrating through the membrane) were counted as described.Citation30 The numbers of motile cells were expressed as percentage of motile cells over non motile cells. The experiment was repeated three times independently.
Colony formation assay.
Twenty-four hours after transfection, 500 cells were seeded in 10 cm plates and incubating at 37°C for 3 weeks to allow for colonies to form. Media was then removed and colonies were stained in Methylene Blue dye (0.5% Methylene blue and 50% methanol) at room temperature for 10 min. Plates were gently rinsed in water and visible colonies were counted.
Immunohistochemistry.
Immunohistochemistry was performed using a monoclonal EP4 antibody or isotype control antibody. Staining for EP4 receptor protein expression on formalin fixed paraffin embedded colon cancer specimens was performed by methods previously described.Citation31 The intensity of the immunohistochemical signals in each core is graded as (0) negative, (1) weak, (2) moderate, or (3) strong. The proportion of cells staining positively is also evaluated as a percentage. The score is then calculated as the numbers representing intensity times the percentage of cells stained.Citation31 Specimens were scored in a blinded fashion by two investigators (M.N. and A.B.)
In situ hybridization.
Digoxygenin-5′-and 3′-labeled LNA (locked nucleic acid) DNA probe was used to detect miR-101 expression (Exiqon). Hybridization of the LNA probe (40 nM) was performed at 45°C in hybridization chamber. After stringency washes, the sections for the chromogenic in situ were blocked for 1 h and incubated overnight at 4 °C with anti-Digoxygenin-AP (ROCHE) and for the fluorescence in situ, sections were incubated with anti-Digoxygenin-HRP (ROCHE). Chromogenic in situ were reacted with BCIP/NBT to visualized miR101 expression and counterstained with eosin, dehydrated and mounted with Cytoseal XYL mounting media. To visualize miR-101 and EP4 expression in the fluorescence in situ, tyramide-conjugated fluorochromes (NHS-Rhodamine and NHS-Fluorescein) was applied to the slides by performing two tyramide signal amplification reactions. Slides were stained with DAPI and mounted with ProLong Gold anti-fade reagent (Invitrogen). Analysis and photography was done by Deconvolution microscopy.
CAM assay.
The chorioallantoic membrane (CAM) assay was performed as described previously.Citation32,Citation33 Five million HCA-7 cells were implanted on 10-d-old chicken embryo chorioallantoic membrane. After 24 h, tumors were treated daily with 10 µM of EP4 receptor antagonists for 4 d. At the termination of the experiment, tumors were resected, trimmed and weighed
Statistical analysis.
Data were expressed as mean ± standard error of the means of multiple experiments. Paired Students t-test was used to weigh against two groups. Statistical significance was defined as * for p value < 0.05, ** for p value < 0.01 and *** for p value < 0.001.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Figures and Tables
Figure 1 Elevated EP4 receptor expression in human colon cancer specimens. Immunohistochemistry (IHC) staining of EP4 receptor protein was performed in human colon tissue specimens. (A) The staining of normal colonic mucosa shows low EP4 receptor protein expression on the apical membrane. (B) Elevated EP4 staining was observed in adenocarcinomas lesions. (C) Differences between IHC scores for normal, adenomatous, and cancerous lesions are shown as a box and whisker plot. The distribution of pair-wise comparisons of differences in EP4 staining scores shows a significant difference in EP4 receptor staining in malignant colonic lesions compared with normal and adenoma lesions (*p < 0.0001). Each dot represents IHC score (intensity of stain X percentage of cells stained) generated after staining formalin-fixed paraffin embedded tissues for EP4 antibody by IHC.
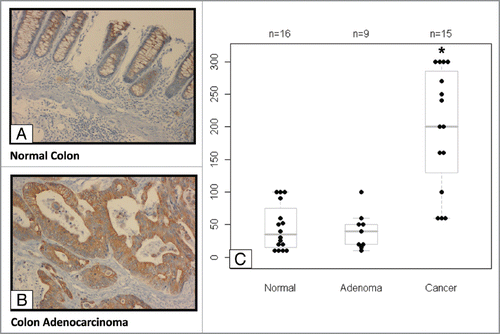
Figure 2 Highly conserved miR-101 seed sequence within the PTGER4 mRNA. (A) Computational prediction of miR-101 seed region on PGTER4 mRNA. The miR-101 target sequences are located on nucleotide 18–24 in 3′-UTR of PTGER4. The target region is denoted with bold letters. (B) The miR-101 seed sequence is highly conserved across five species.
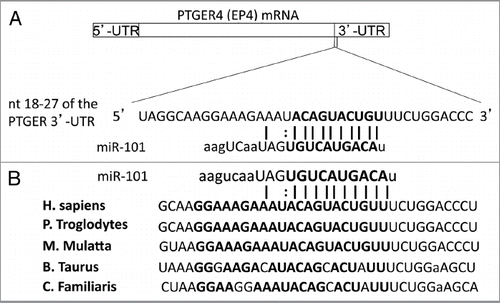
Figure 3 Inverse relationship between EP4 receptor protein and miR-101 expression levels in colon cancer tissues. (A) Western analysis characterizing EP4 receptor protein levels in different colon cancer cells lines. (B) Characterization of miR-101 expression levels in various colon cancer cell lines using qRT-PCR. (C) Western analysis of EP4 receptor protein isolated from paired normal (N) and colon tumor (T) tissue specimens. Tumor colon specimens display higher EP4 receptor protein levels. (D) qRT-PCR analysis showing relative levels of miR-101 in paired normal colon and cancer specimens. The miR-101 expression levels of tumors (T) were normalized against their corresponding levels in normal (N) colon tissues. Note, loss of miR-101 in the tumor specimens is associated with increased EP4 protein expression. **p < 0.01.
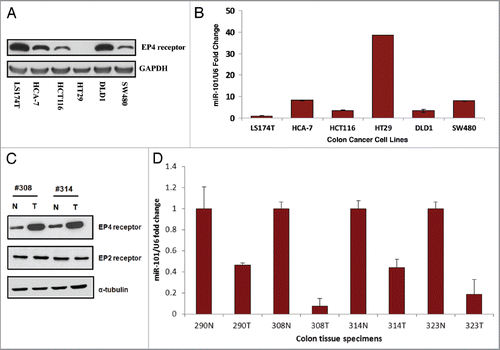
Figure 4 Analysis of miR-101 expression in human colon tissue by in situ hybridization. (A) (a) Paraffin embedded, formalin-fixed non-malignant colon epithelial tissue stained with H&E. (b) Non-malignant colon tissue hybridized with LNA scrambled probe (c) Non-malignant human colon tissue hybridized with LNA miR-101 probe. Note miR-101 expression was observed in normal colon epithelial cells and surrounding tissue. (d) Colon adenocarcinoma tissue stained with H&E. (e) Colon cancer tissue hybridized with LNA scrambled probe. (f) Colon cancer tissue hybridized with LNA miR-101 probe. Malignant adenocarcinoma cells show considerably low miR-101 expression levels or none. Images captured at 20x magnification. (B) (a) DAPI-labeled nuclei. (b) miR-101 expression detected with HRP-DIG-labeled miR-101 probe reacted with tyramide conjugated fluorescein. (c) EP4 expression revealed with HRP-EP4 antibody reacted with tyramide-rhodamine. (d) Co-detection of EP4 and miR-101. Two cell populations are visible in the malignant epithelium, cells co-expressing low miR-101 levels permitting significant EP4 expression (white arrow) and cells that had shut down miR-101 expression allowing high levels of EP4 expression (yellow arrowheads). A third group of cells expressing high miR-101 levels and no EP4 expression is identifiable surrounding the malignant epithelium (white arrowheads). De-convoluted images captured at 40x (water).
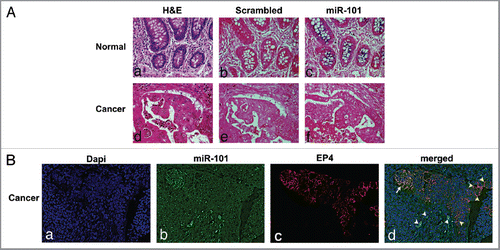
Figure 5 The EP4 receptor is a direct target of miR-101. (A) Predicted miR-101 binding sequence in the human PTGER4 3′-UTR, encoding EP4 receptor. The PTGER4 wildtype and mutant constructs were cloned into the pGL3 plasmid and downstream of the Luciferase gene at the Xba1 restriction site. The location of the wild-type constructs in the plasmid is denoted with yellow and mutant with red. Ninety percent of the seed sequence is mutated in the mutant construct. (B) The LS174T colon cells were co-transfected with pGL3carrying the constructs (wild-type or mutant EP4-3′-UTR and either a miR-101 expressing plasmid or empty vector, and Renilla. Co-transfection of the wildtype construct and miR-101 suppressed EP4 Luciferase expression. The relative luciferase activities were measured and normalized against their control. (C) Western analysis of endogenous EP4 receptor protein expression levels after ectopic miR-101 expression in LS174T cells. The levels of endogenous EP4 receptor protein decreases in a time-dependent manner
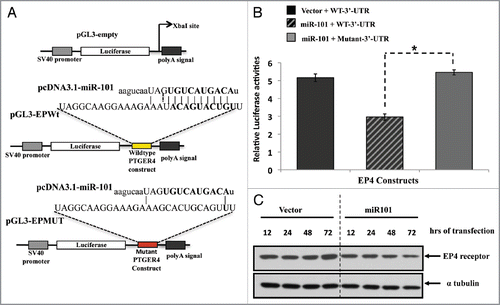
Figure 6 The biological effects of miR-101 and EP4 receptor inhibition in vitro and in vivo. (A) Ectopic expression of miR-101 in LS174T cells suppressed EP4 expression colon tumor cell motility, whereas co-expression of EP4 could overcome miR-10 suppression of motility. (B) Enforced expression of miR-101 attenuated LS174T cell motility, whereas co-expression of EP4 rescued cells from the inhibitory effects of miR-101. (C) RNAi-based silencing of EP4 receptor expression using shRNA (shEP4) reduced colony formation relative to cell containing vector scrambled sequence (shCtrl). (D) Administration of the EP4 receptor antagonist LS161982 inhibited the growth of colon cancer cells (LS174T) grown in the chorioallantoic membrane (CAM) assay. **p < 0.01.
Acknowledgments
We thank Jennifer M. Davenport, Reneé Shañas, and Jenae M. Owens for histology assistance. This work was supported by CA95060, T32CA009213, and P30CA023074.
Reference
- Rothenberg ML, Oza AM, Bigelow RH, Berlin JD, Marshall JL, Ramanathan RK, et al. Superiority of oxaliplatin and fluorouracil-leucovorin compared with either therapy alone in patients with progressive colorectal cancer after irinotecan and fluorouracilleucovorin: interim results of a phase III trial. J Clin Oncol 2003; 21:2059 - 2069; PMID: 12775730; http://dx.doi.org/10.1200/JCO.2003.11.126
- Elias D, Liberale G, Vernerey D, Pocard M, Ducreux M, Boige V, et al. Hepatic and extrahepatic colorectal metastases: when resectable, their localization does not matter, but their total number has a prognostic effect. Ann Surg Oncol 2005; 12:900 - 909; PMID: 16184442; http://dx.doi.org/10.1245/ASO.2005.01.010
- Meyerhardt JA, Mayer RJ. Systemic therapy for colorectal cancer. N Engl J Med 2005; 352:476 - 487; PMID: 15689586; http://dx.doi.org/10.1056/NEJMra040958
- Rougier P, Van Cutsem E, Bajetta E, Niderele N, Possinger K, Labianca R, et al. Randomised trial of irinotecan versus fluorouracil by continuous infusion after fluorouracil failure in patients with metastatic colorectal cancer. Lancet 1998; 352:1407 - 1412; PMID: 9807986; http://dx.doi.org/10.1016/S0140-6736(98)03085-2
- Rougier P, Paillot B, LaPlanche A, Morvan F, Seitz JF, Rekacewicz C, et al. 5-Fluorouracil (5-FU) continuous intravenous infusion compared with bolus administration. Final results of a randomised trial in metastatic colorectal cancer Eur J Cancer 1997; 33:1789 - 1793; PMID: 9470834; http://dx.doi.org/10.1016/S0959-8049(97)00175-5
- Brown JR, DuBois RN. COX-2: a molecular target for colorectal cancer prevention. J Clin Oncol 2005; 23:2840 - 2855; PMID: 15837998; http://dx.doi.org/10.1200/JCO.2005.09.051
- Cha YI, DuBois RN. NSAIDs and cancer prevention: targets downstream of COX-2. Annu Rev Med 2007; 58:239 - 252; PMID: 17100552; http://dx.doi.org/10.1146/annurev.med.57.121304.131253
- Chell S, Kaidi A, Williams AC, Paraskeva C. Mediators of PGE2 synthesis and signalling downstream of COX-2 represent potential targets for the prevention/treatment of colorectal cancer. Biochim Biophys Acta 2006; 1766:104 - 119; PMID: 16859832
- Cherukuri DP, Chen XB, Goulet AC, Young RN, Han Y, Heimark RL, et al. The EP4 receptor antagonist, L-161,982, blocks prostaglandin E2-induced signal transduction and cell proliferation in HCA-7 colon cancer cells. Exp Cell Res 2007; 313:2969 - 2979; PMID: 17631291; http://dx.doi.org/10.1016/j.yexcr.2007.06.004
- Hawcroft G, Ko CW, Hull MA. Prostaglandin E2-EP4 receptor signalling promotes tumorigenic behaviour of HT-29 human colorectal cancer cells. Oncogene 2007; 26:3006 - 3019; PMID: 17130837; http://dx.doi.org/10.1038/sj.onc.1210113
- Chell SD, Witherden IR, Dobson RR, Moorghen M, Hermann AA, Qualtrough D, et al. Increased EP4 receptor expression in colorectal cancer progression promotes cell growth and anchorage independence. Cancer Res 2006; 66:3106 - 3113; PMID: 16540660; http://dx.doi.org/10.1158/0008-5472.CAN-05-3702
- Wang D, Dubois RN. Eicosanoids and cancer. Nat Rev Cancer 2010; 10:181 - 193; PMID: 20168319; http://dx.doi.org/10.1038/nrc2809
- Ma X, Kundu N, Rifat S, Walser T, Fulton AM. Prostaglandin E receptor EP4 antagonism inhibits breast cancer metastasis. Cancer Res 2006; 66:2923 - 2927; PMID: 16540639; http://dx.doi.org/10.1158/00085472.CAN-05-4348
- Cha YI, Kim SH, Sepich D, Buchanan FG, Solnica-Krezel L, DuBois RN. Cyclooxygenase-1-derived PGE2 promotes cell motility via the G-protein-coupled EP4 receptor during vertebrate gastrulation. Genes Dev 2006; 20:77 - 86; PMID: 16391234; http://dx.doi.org/10.1101/gad.1374506
- Sheng H, Shao J, Washington MK, DuBois RN. Prostaglandin E2 increases growth and motility of colorectal carcinoma cells. J Biol Chem 2001; 276:18075 - 1881; PMID: 11278548; http://dx.doi.org/10.1074/jbc.M009689200
- Leone V, di Palma A, Ricchi P, Acquaviva F, Giannouli M, Di Prisco AM, et al. PGE2 inhibits apoptosis in human adenocarcinoma Caco-2 cell line through RasPI3K association and cAMP-dependent kinase A activation. Am J Physiol Gastrointest Liver Physiol 2007; 293:G673 - G681; PMID: 17640974; http://dx.doi.org/10.1152/ajpgi.00584.2006
- Mutoh M, Watanabe K, Kitamura T, Shoji Y, Takahashi M, Kawamori T, et al. Involvement of prostaglandin E receptor subtype EP(4) in colon carcinogenesis. Cancer Res 2002; 62:28 - 32; PMID: 11782353
- Yang L, Huang Y, Porta R, Yanagisawa K, Gonzalez A, Segi E, et al. Host and direct antitumor effects and profound reduction in tumor metastasis with selective EP4 receptor antagonism. Cancer Res 2006; 66:9665 - 9672; PMID: 17018624; http://dx.doi.org/10.1158/0008-5472.CAN-06-1271
- Pozzi A, Yan X, Macias-Perez I, Wei S, Hata AN, Breyer RM, et al. Colon carcinoma cell growth is associated with prostaglandin E2/EP4 receptor-evoked ERK activation. J Biol Chem 2004; 279:29797 - 29804; PMID: 15123663; http://dx.doi.org/10.1074/jbc.M313989200
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006; 6:857 - 866; PMID: 17060945; http://dx.doi.org/10.1038/nrc1997
- Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci USA 2004; 101:11755 - 11760; PMID: 15284443; http://dx.doi.org/10.1073/pnas.0404432101
- Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev 2004; 18:504 - 511; PMID: 15014042; http://dx.doi.org/10.1101/gad.1184404
- Valeri N, Gasparini P, Fabbri M, Braconi C, Veronese A, Lovat F, et al. Modulation of mismatch repair and genomic stability by miR-155. Proc Natl Acad Sci USA 2010; 107:6982 - 6987; PMID: 20351277; http://dx.doi.org/10.1073/pnas.1002472107
- Chakrabarty A, Tranguch S, Daikoku T, Jensen K, Furneaux H, Dey SK. MicroRNA regulation of cyclooxygenase-2 during embryo implantation. Proc Natl Acad Sci USA 2007; 104:15144 - 15149; PMID: 17848513; http://dx.doi.org/10.1073/pnas.0705917104
- Hiroki E, Akahira J, Suzuki F, Nagase S, Ito K, Suzuki T, et al. Changes in microRNA expression levels correlate with clinicopathological features and prognoses in endometrial serous adenocarcinomas. Cancer Sci 2010; 101:241 - 249; PMID: 19891660; http://dx.doi.org/10.1111/j.1349-7006.2009.01385.x
- Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, et al. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res 2009; 69:1135 - 1142; PMID: 19155302; http://dx.doi.org/10.1158/0008-5472.CAN-08-2886
- Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 2008; 322:1695 - 1699; PMID: 19008416; http://dx.doi.org/10.1126/science.1165395
- Strillacci A, Griffoni C, Sansone P, Paterini P, Piazzi G, Lazzarini G, et al. MiR-101 downregulation is involved in cyclooxygenase-2 overexpression in human colon cancer cells. Exp Cell Res 2009; 315:1439 - 1447; PMID: 19133256; http://dx.doi.org/10.1016/j.yexcr.2008.12.010
- Chandramouli A, Mercado-Pimentel ME, Hutchinson A, Gibadulinova A, Olson ER, Dickinson S, et al. The induction of S100p expression by the Prostaglandin E (PGE)/EP4 receptor signaling pathway in colon cancer cells. Cancer Biol Ther 2010; 10:1056 - 1066; PMID: 20890108; http://dx.doi.org/10.4161/cbt.10.10.13373
- Alvarez-Díaz S, Valle N, Garcia JM, Pena C, Freije JM, Quesada V, et al. Cystatin D is a candidate tumor suppressor gene induced by vitamin D in human colon cancer cells. J Clin Invest 2009; 119:2343 - 2358; PMID: 19662683; http://dx.doi.org/10.1172/JCI37205
- Doldan A, Chandramouli A, Shanas R, Bhattacharyya A, Cunningham JT, Nelosn MA, et al. Loss of the eukaryotic initiation factor 3f in pancreatic cancer. Mol Carcinog 2008; 47:235 - 244; PMID: 17918192; http://dx.doi.org/10.1002/mc.20379
- Deryugina EI, Quigley JP. Chapter 2. Chick embryo chorioallantoic membrane models to quantify angiogenesis induced by inflammatory and tumor cells or purified effector molecules. Methods Enzymol 2008; 444:21 - 41; PMID: 19007659; http://dx.doi.org/10.1016/S0076-6879(08)02802-4
- Deryugina EI, Quigley JP. Chick embryo chorioallantoic membrane model systems to study and visualize human tumor cell metastasis. Histochem Cell Biol 2008; 130:1119 - 1130; PMID: 19005674; http://dx.doi.org/10.1007/s00418-008-0536-2
- Gustafsson A, Hansson E, Kressner U, Nordgren S, Andersson M, Wang W, et al. EP1-4 subtype, COX and PPAR gamma receptor expression in colorectal cancer in prediction of disease-specific mortality. Int J Cancer 2007; 121:232 - 240; PMID: 17290397; http://dx.doi.org/10.1002/ijc.22582
- Flower RJ. The development of COX2 inhibitors. Nat Rev Drug Discov 2003; 2:179 - 191; PMID: 12612644; http://dx.doi.org/10.1038/nrd1034
- Bombardier C, Laine L, Reicin A, Shapiro D, Burgos-Vargas R, Davis B, et al. VIGOR Study Group. Comparison of upper gastrointestinal toxicity of rofecoxib and naproxen in patients with rheumatoid arthritis. N Engl J Med 2000; 343:1520 - 1528; PMID: 11087881; http://dx.doi.org/10.1056/NEJM200011233432103
- Solomon SD, McMurray JJ, Pfeffer MA, Wittes J, Finn Fowler R, et al. Cardiovascular risk associated with celecoxib in a clinical trial for colorectal adenoma prevention. N Engl J Med 2005; 352:1071 - 1080; PMID: 15713944; http://dx.doi.org/10.1056/NEJMoa050405
- Funk CD, FitzGerald GA. COX-2 inhibitors and cardiovascular risk. J Cardiovasc Pharmacol 2007; 50:470 - 479; PMID: 18030055; http://dx.doi.org/10.1097/FJC.0b013e318157f72d
- Gauvreau D, Dolman SJ, Hughes G, O'Shea PD, Davies IW. Scalable synthesis of a prostaglandin EP4 receptor antagonist. J Org Chem 2010; 75:4078 - 4085; PMID: 20469914; http://dx.doi.org/10.1021/jo1004197
- Burch JD, Farand J, Colucci J, Sturino C, Ducharme Y, Friesen RW, et al. Naphthalene/quinoline amides and sulfonylureas as potent and selective antagonists of the EP4 receptor. Bioorg Med Chem Lett 2011; 21:1041 - 1046; PMID: 21215624; http://dx.doi.org/10.1016/j.bmcl.2010.12.014
- Li L, Mathieu MC, Denis D, Therien AG, Wang Z. The identification of substituted benzothiophene derivatives as PGE(2) subtype 4 receptor antagonists: From acid to non-acid. Bioorg Med Chem Lett 2011; 21:734 - 737; PMID: 21208803; http://dx.doi.org/10.1016/j.bmcl.2010.11.118
- Chen Q, Muramoto K, Masaaki N, Ding Y, Yang H, Mackey M, et al. A novel antagonist of the prostaglandin E(2) EP(4) receptor inhibits Th1 differentiation and Th17 expansion and is orally active in arthritis models. Br J Pharmacol 2010; 160:292 - 310; PMID: 20423341; http://dx.doi.org/10.1111/j.14765381.2010.00647.x
- Smits M, Mir SE, Nilsson RJ, van der Stoop PM, Niers JM, Marquez VE, Cloos J, et al. Down-regulation of miR-101 in endothelial cells promotes blood vessel formation through reduced repression of EZH2. PLoS ONE 2011; 6:e16282; PMID: 21297974; PMID: 10.1371/journal.pone.0016282