Abstract
Background: To improve the prognostic evaluation of colorectal cancer requires new molecular markers. Cancerous inhibitor of protein phosphatase 2A (CIP2A) serves as an oncoprotein by targeting PP2A-mediated inhibition of c-Myc. A prognostic role for CIP2A has been demonstrated in gastric, lung, and tongue cancers. Methods: 863 consecutive colorectal cancer patients treated at Helsinki University Central Hospital in 1983-2001 were collected with 752 scored successfully for CIP2A immunohistochemical expression from tumor tissue microarrays. Associations with clinicopathologic variables and molecular markers were explored by the chi-square test, and the Kaplan-Meier method served for survival analysis. Results: CIP2A was overexpressed in 661 (87.9%) specimens. CIP2A overexpression was associated with tumor differentiation grade (p = 0.014), p53 immunopositivity (p = 0.042), EGFR immunopositivity (p = 0.007), and c-Myc nuclear immunopositivity (p = 0.018). In survival analysis, CIP2A failed to show any prognostic significance (p = 0.270, log-rank test). Conclusions: Overexpression of CIP2A in colorectal cancer patients may be an important step in colorectal carcinogenesis. Based on our findings, CIP2A shows no association with patient prognosis in colorectal cancer, but is associated with nuclear c-Myc.
Keywords: :
Introduction
Colorectal cancer is one of the most common cancers in the world. Its incidence in Western countries is high, and is increasing even in geographical areas where it used to be low. Colorectal cancer-related mortality is rising, especially in economically transitioning countries, whereas in many Western countries it has remained constant.Citation1,Citation2 Common risk factors are obesity, smoking, and high lifetime alcohol intake.Citation1,Citation3 Modern treatment involves radical surgery together with postoperative chemotherapy in stage III and also often in stage II colon cancer patients, and for rectal cancer patients usually preoperative radio- or chemoradiotherapy, followed by postoperative chemotherapy in certain cases. Oncological treatments have severe side-effects, so ideally only those patients who benefit should receive adjuvant treatment.
Few prognostic or predictive biomarkers are available for colorectal cancer to aid clinicians in their treatment decisions. Colorectal cancer arises through an adenoma-dysplasia-carcinoma sequence, in which the APC and KRAS gene mutations occur early, TP53 being a late mutation.Citation4,Citation5 Randomized trials have demonstrated, that only patients with wild-type K-RAS benefit from EGFR-targeted therapy in metastatic colorectal cancer.Citation5
Cancerous inhibitor of PP2A (CIP2A) is an oncoprotein that inhibits protein phosphatase 2A (PP2A) -mediated dephosphorylation of c-Myc.Citation6 Overexpression of CIP2A is evident in acute myeloid leukemia, in epithelial carcinomas, and in adenocarcinomas;Citation6-Citation11 though little is known of its possible role in human cancers. A prognostic role for CIP2A has been demonstrated in certain subgroups of gastric cancer.Citation12 In lung cancer, CIP2A is an independent prognostic marker,Citation13 but in colorectal cancer its impact on prognosis remains unknown. The aim of this study was to investigate the association of CIP2A protein expression with clinicopathologic variables and molecular markers such as c-Myc, as well as to study the prognostic role of CIP2A in colorectal cancer.
Results
Immunohistochemistry
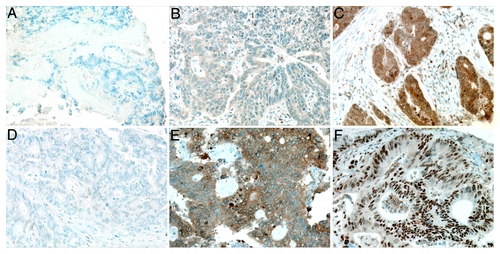
CIP2A cytoplasmic immunoreactivity of the 754 specimens was scored as 0 and regarded as negative (score 0) in 4 (0.7%), weakly positive (score 1) in 52 (9.6%), moderately positive (score 2) in 283 (52.4%), and strongly positive (score 3) in 201 (37.2%). The distribution was similar for the validation series (1 of 212, 0.5% negative; 34, 15.5% weakly positive; 97, 44.1% moderately positive; 80, 36.4% strongly positive). Representative immunostainings are shown in . Cytoplasmic c-Myc negativity was found in 54 of 205 cases (24.5%), weak positivity in 70 (31.8%), moderate positivity in 52 (23.6%), and strong positivity in 29 (13.2%; ). Nuclear c-Myc was expressed to 0–10% in 63 specimens (30.7%), 11–30% in 30 (13.6%), 31–50% in 37 (16.8%), 51–80% in 47 (21.4%), and expressed in over 80% of the nuclei in 28 specimens (12.7%; ).
Figure 1. Representative images of CIP2A and c-Myc expression in colorectal cancer. (A) CIP2A negative, (B) weakly positive, and (C) strongly positive immunoreactivity. (D) Negative c-Myc cytoplasmic and nuclear immunoreactivity. (E) Strongly positive cytoplasmic c-Myc immunoreactivity, and (F) over 80% positive nuclear c-Myc immunoreactivity. Original magnification was 200x.
Associations with clinicopathologic variables
When the association between CIP2A expression and clinicopathologic variables was explored, in the test series, stronger CIP2A expression was more evident in adenocarcinomas than in mucinous carcinomas (p < 0.0001, chi-square test, ). Strong CIP2A expression was also associated with WHO grade (p = 0.014, chi-square test). CIP2A did not associate with age, gender, Dukes classification, or tumor location (colon vs. rectum). In the validation series, we found associations between CIP2A positivity and histological tumor grade (p = 0.018), pT- (p = 0.024), and pM-classifications (p = 0.038, chi-square test, ). An association appeared between CIP2A and nuclear c-Myc immunoreactivity (p = 0.018, ), but not with cytoplasmic c-Myc immunoreactivity (p = 0.455, data not shown). CIP2A positivity was in the test series associated with p53-immunopositivity (p = 0.042) and EGFR-positivity (p = 0.007), but not with proliferation index (p = 0.488, chi-square test, ).
Table 1. Association of CIP2A with clinicopathologic variables in a test series with 540 colorectal cancer patients
Table 2. Association of CIP2A with clinicopathologic variables in a validation series with 212 colorectal cancer patients
Table 3. Association of CIP2A with molecular biomarkers in a test series with 540 colorectal cancer patients
In the validation series, we found an association between cytoplasmic c-Myc immunoreactivity and female gender (p = 0.048, Table S1). Nuclear c-Myc was associated with WHO grade (p = 0.018, Table S2).
Survival analysis
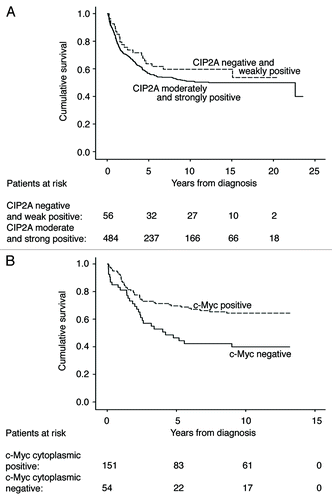
The 5-y disease-specific overall survival rate for patients with moderately and strongly positive CIP2A tumors was 56.8% (95% CI 52.1–61.5) in the test series compared with 63.7% (95% CI 50.6–76.8) for those with negative and weakly positive CIP2A tumors (p = 0.270, log-rank test, ). In the validation series, results were similar, with a 5 y survival rate for moderately and strongly positive CIP2A tumors of 64.2% (95% CI 56.8–71.6) compared with 61.3% (95% CI 44.1–78.5) for those with negative and weakly positive CIP2A tumors (p = 0.378, log-rank test, data not shown).
Figure 2. Survival analysis for CIP2A and c-Myc expression in colorectal cancer patients. (A) Disease-specific overall survival analysis according to the Kaplan-Meier method for cytoplasmic CIP2A immunoreactivity in the test series (p = 0.270, log-rank test). (B) Disease-specific overall survival analysis according to the Kaplan-Meier method for cytoplasmic c-Myc immunoreactivity in the validation series (p = 0.003, log-rank test).
As the biological background for colon and for rectal tumors is considered to differ, we stratified our survival analysis for colon and rectal tumors. For colon cancer patients, we found no difference in survival between patients with moderate or strong CIP2A compared with those patients with CIP2A-negative or weak CIP2A expression (p = 0.874 in the test series; p = 0.650 in the validation series, log-rank test). Results were similar for rectal tumors (p = 0.071 in the test series; p = 0.317 in the validation series, log-rank test).
In the validation data set, we also studied the role of c-Myc immunoreactivity on survival. The 5 y disease-specific overall survival rate for patients with negative cytoplasmic c-Myc immunoreactivity was 46.5% (95% CI 32.6–60.4), compared with 69.6% (95% CI 61.8–77.4) for those with cytoplasmic immunopositive c-Myc (p = 0.003, log-rank test, ). Nuclear c-Myc immunoreactivity showed no prognostic significance (p = 0.842, log-rank test, data not shown).
In multivariate survival analysis of the test series, the following variables were entered: age, gender, Dukes classification, grade, histological type, tumor location (colon or rectum), p53 immunoreactivity, proliferation index, and CIP2A expression. Only age and Dukes classification remained as independent prognostic factors (data not shown).
Discussion
Here, we report overexpression of CIP2A protein in colorectal cancer patients, with the majority of the samples (484 of 540) having moderate or strong positive immunopositivity for CIP2A. Only four samples were completely negative, and 56 were weakly positive (10%). Nevertheless, CIP2A was associated with very few clinicopathologic variables. The only association was between histological tumor type and CIP2A expression. CIP2A expression was associated with nuclear c-Myc immunoreactivity (p = 0.018). In survival analysis, CIP2A expression failed to add any significant information (p = 0.232). Interestingly, negative cytoplasmic c-Myc immunoreactivity showed a prognostic value in colorectal cancer (p = 0.003), whereas nuclear c-Myc immunoreactivity showed none (p = 0.0842).
Though we failed to show a prognostic value for CIP2A, the strengths of this study are a large, well-characterized colorectal cancer patient cohort with a long follow-up time, and a standard protocol for evaluating CIP2A immunoreactivity.Citation12,Citation14,Citation15 The test series included over 550 patients and the validation series over 240. The clinicopathologic characteristics in both data sets were similar. CIP2A was associated with histologic tumor type in both data sets, but we found no prognostic significance of CIP2A expression in either of them. The limitation of protein expression studies using immunohistochemistry is the subjectivity of sample scoring, although the researchers work independently and blinded to the clinical data. Furthermore, up to 10% of the specimens were lost in the staining of tumor tissue microarrays.
CIP2A has shown prognostic significance in lung, tongue, and ovarian cancer,Citation13-Citation15 as well as in certain subgroups of gastric cancer.Citation12 In colorectal cancer, we failed to identify CIP2A as a prognostic marker. This difference may be due to varying protein expression levels and the different role of CIP2A in various cancers, as well as the different nature of colon cancer, compared with that of other epithelial malignancies. In breast cancer, CIP2A was associated with high grade and high proliferation index, markers of aggressive disease, but not with survival.Citation16 Likewise, in gastric,Citation12 ovarian,Citation15 and tongue cancer,Citation14 CIP2A was associated with markers of an aggressive disease, an association absent from the colorectal cancer patient specimens. Similar to our results, in prostate cancer, Vaarala et al.Citation9 found no association between CIP2A expression and survival. We found an association between CIP2A and c-Myc protein expression in colorectal cancer, in line with results on gastric cancerCitation12 and with those by Dong et al.Citation13 on lung cancer. In gastric cancer, we have earlier demonstrated a positive feedback-regulating mechanism, in which CIP2A downregulates c-Myc protein expression, and vice versa.Citation12 Wang et al.Citation17 have shown that stage II/III colorectal patients with a R497K polymorphism of the EGFR gene may have a better survival outcome and respond more frequently to 5-fluorouracil and oxaliplatin treatment, concluding that the R497 polymorphism, which is associated with c-Myc activation, may be important for controlling tumor recurrence. Furthermore, it has been suggested that gene expression signatures associated with USP22, a novel deubiquitinating enzyme required for Myc-driven transcription, such as of BMI-1, c-Myc, and cyclin D2, may be associated with colorectal cancer disease progression.Citation18
Colon cancer is related to increased Ras signaling and loss of TGF-β signaling.Citation19 Junttila et al.Citation6 demonstrated that CIP2A is a downstream target in the Ras signaling pathway, suggesting that high CIP2A expression in colorectal cancer may be linked to the adenoma-carcinoma sequence and thus may serve as a step in colorectal cancer carcinogenesis. Because we found an association between CIP2A and EGFR expression, together with our earlier findings in gastric cancer,Citation12 and because CIP2A overexpression is dependent on the EGFR-MEK1/2-ETS1 signaling pathway,Citation20 we suggest that CIP2A in colorectal cancer may be a potentially important downstream target of EGFR-therapy.
In conclusion, CIP2A expression may be an important step in colorectal carcinogenesis. Based on our findings, CIP2A overexpression has no prognostic significance in colorectal cancer but has a strong association with nuclear c-Myc and histological tumor grading. In contrast to an association between CIP2A and prognosis shown for gastric,Citation12 tongueCitation14 and lung cancer,Citation13 its absence in colorectal cancer underscores its distinct characteristics differing those of from many other epithelial malignancies.
Patients and Methods
Patients
At the Department of Surgery, Helsinki University Central Hospital, 643 consecutive colorectal cancer patients treated in 1989–98 comprised the test set of the study population. Their median age was 65. Radical surgery was performed for 196 patients, whereas treatment was considered palliative for 88. Survival data came from the Finnish Cancer Registry and Statistics Finland. Median time at the end of follow-up was 4.7 y (range 0–24.7), with a 5 y disease-specific overall survival rate of 54.4% [95% confidence interval (CI) 50.5–58.3%]. The local ethics committee approved the study.
For validation, data on 220 consecutive colorectal cancer patients treated at the same hospital in 1998–2001 were collected. Median follow-up time was 6.0 y (range 0–13.2), with a 5 y disease-specific overall survival rate of 64.8% (95% CI 58.1–71.5%). The test series comprised 55% of the colon tumors, whereas the figure for the validation series was 36% (p < 0.001, chi-square test). The distribution of age, gender, WHO differentiation grade, and Dukes classification was in both data sets similar (Table S3).
Preparation of tumor tissue micorarrays
Tumor samples were fixed in formalin, embedded in paraffin, and stored in the archives of the Department of Pathology, Helsinki University Central Hospital. Representative cancer areas were marked on H&E stained slides from each tumor by an experienced pathologist. Three punches were taken from every tumor specimen and arranged in recipient paraffin blocks with a tissue microarray instrument (Beecher Instruments) as described.Citation21-Citation23
Immunohistochemistry
Tumor tissue microarray blocks were freshly cut into 4-uM sections. For the detailed immunohistochemistry protocol, see Khanna et al.Citation12 For antigen retrieval, slides were treated in a PreTreatment module (Lab Vision Corp.) in TRIS-HCl (pH 8.5) or Tris-EDTA (pH 9.0) buffer for 20 min at 98°C. Staining of sections was performed in an Autostainer 480 (Lab Vision Corp.) by the Dako REAL EnVision Detection System, Peroxidase/DAB+, Rabbit/Mouse (K4065, Dako). Tissues were incubated in the rabbit polyclonal CIP2A antibody, at a dilution of 1:3000 for one hour at room temperature,Citation24 the mouse monoclonal c-Myc antibody (clone 9E10, Santa Cruz Biotechnology) at 1:200, the mouse monoclonal Ki-67 antibody (clone MIB-1, Dako) at 1:100, the rabbit polyclonal p53 antibody (DO-7, Dako) at 1:50, and the monoclonal EGFR antibody (NCL-EGFR clone EGFR.113, Novocastra, Leica Biosystems Newcastle Ltd) at 1:10.
Scoring of samples
CIP2A expression was scored independently for intensity of cytoplasmic immunoreactivity while blinded to patient information by J.H. and C.B. or J.H. and S.K. Negative cytoplasmic staining was scored as 0, weakly positive as 1, moderately strong positivity or focally strong positivity as 2, and homogeneously strong positivity as 3. C-Myc immunoreactivity was scored separately for cytoplasmic and nuclear expression. Cytoplasmic c-Myc expression was scored for intensity in a similar manner as for CIP2A expression, and nuclear c-Myc expression was scored according to percentage of immunopositive nuclei (0, 1–10, 11–30, 31–50, 51–80 and > 80%). Nuclear p53, nuclear Ki-67, and cytoplasmic EGFR immunopositivity was evaluated by percentage of stained cells (Tables S4 and S5). No immunoreactivity was scored as 0, 1–10% as 1, 11–49% as 2 and more than 50% as 3. Specimens with discordant scores underwent re-evaluation with a multiheaded microscope, and the consensus score served for further analysis. The highest score of the triplicates of each sample was regarded as representative in further analysis. In the test series, 540 (84.0%) were scored successfully for CIP2A, and in the validation series 212 (96.4%). For c-Myc, we managed to score 205 specimens (93%). In the final analysis CIP2A scores 0–1 were grouped to represent negative and weakly positive CIP2A expression, and scores 2–3 to represent moderately and strongly positive CIP2A. Nuclear c-Myc, p53 and Ki-67 expression was analyzed with 0–10% representing negative nuclear immunoreactivity. Cytoplasmic EGFR immunoreactivity was analyzed as ≤ 10 and > 10%.
Statistical analysis
Evaluation of the associations between CIP2A expression and clinicopathologic variables was by the chi-square test assessed (IBM SPSS Statistics, version 19.0 for Mac; SPSS, Inc., an IBM Company). Disease-specific overall survival was counted from date of surgery to date of death from colorectal cancer, or until end of follow-up. Survival analyses were done according to the Kaplan-Meier method and compared with the log-rank test (IBM SPSS Statistics). The Cox proportional hazard model served for multivariate survival analysis, with Dukes classification and grade entered as categorical variables.
| Abbreviations: | ||
| CIP2A | = | cancerous inhibitor of PP2A |
| PP2A | = | protein phosphatase 2A |
Additional material
Download Zip (206.6 KB)Acknowledgments
The CIP2A antibody was a kind gift from Dr. Edward K. Chan, University of Florida, USA. We thank Päivi Peltokangas, Tuire Koski, and Elina Aspiala for their excellent technical assistance.
Financial support from: Finska Läkaresällskapet (C.B. and C.H.), the Finnish Cancer Society (A.R.), the Finnish Dentists’ Association Apollonia (J.H.), the Dorothea Olivia Foundation (C.H.), Helsinki University Central Hospital Research Funds (A.R. and C.H.), the K. Albin Johansson Foundation (C.B.), the Karl Walter and Jarl Walter Perkléns minne Foundation (C.H.), Medicinska understödsföreningen Liv och Hälsa (C.H.), the Sigrid Jusélius Foundation (A.R. and C.H.), and the Waldemar von Frenckell Foundation (C.B.).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Note
Supplementary materias can be found at: http://www.landesbioscience.com/journals/cbt/article/18922/
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61:69 - 90; http://dx.doi.org/10.3322/caac.20107; PMID: 21296855
- Center MM, Jemal A, Ward E. International trends in colorectal cancer incidence rates. Cancer Epidemiol Biomarkers Prev 2009; 18:1688 - 94; http://dx.doi.org/10.1158/1055-9965.EPI-09-0090; PMID: 19505900
- Ferrari P, Jenab M, Norat T, Moskal A, Slimani N, Olsen A, et al. Lifetime and baseline alcohol intake and risk of colon and rectal cancers in the european prospective investigation into cancer and nutrition (EPIC). Int J Cancer 2007; 121:2065 - 72; http://dx.doi.org/10.1002/ijc.22966; PMID: 17640039
- Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, et al. Genetic alterations during colorectal-tumor development. N Engl J Med 1988; 319:525 - 32; http://dx.doi.org/10.1056/NEJM198809013190901; PMID: 2841597
- Pritchard CC, Grady WM. Colorectal cancer molecular biology moves into clinical practice. Gut 2011; 60:116 - 29; http://dx.doi.org/10.1136/gut.2009.206250; PMID: 20921207
- Junttila MR, Puustinen P, Niemelä M, Ahola R, Arnold H, Bottzauw T, et al. CIP2A inhibits PP2A in human malignancies. Cell 2007; 130:51 - 62; http://dx.doi.org/10.1016/j.cell.2007.04.044; PMID: 17632056
- Li W, Ge Z, Liu C, Liu Z, Bjorkholm M, Jia J, et al. CIP2A is overexpressed in gastric cancer and its depletion leads to impaired clonogenicity, senescence, or differentiation of tumor cells. Clin Cancer Res 2008; 14:3722 - 8; http://dx.doi.org/10.1158/1078-0432.CCR-07-4137; PMID: 18559589
- Katz J, Jakymiw A, Ducksworth MK, Stewart CM, Bhattacharyya I, Cha S, et al. CIP2A expression and localization in oral carcinoma and dysplasia. Cancer Biol Ther 2010; 10:694 - 9; http://dx.doi.org/10.4161/cbt.10.7.12895; PMID: 21068540
- Vaarala MH, Vaisanen MR, Ristimäki A. CIP2A expression is increased in prostate cancer. J Exp Clin Cancer Res 2010; 29:136; http://dx.doi.org/10.1186/1756-9966-29-136; PMID: 20964854
- Qu W, Li W, Wei L, Xing L, Wang X, Yu J.. CIP2A is overexpressed in esophageal squamous cell carcinoma. Med Oncol 2010; In press http://dx.doi.org/10.1007/s12032-010-9768-9; PMID: 21140243
- Wang J, Li W, Li L, Yu X, Jia J, Chen C.. CIP2A is over-expressed in acute myeloid leukaemia and associated with HL60 cells proliferation and differentiation. Int J Lab Hematol 2011; 33:290 - 8; http://dx.doi.org/10.1111/j.1751-553X.2010.01288.x; PMID: 21219591
- Khanna A, Böckelman C, Hemmes A, Junttila MR, Wiksten JP, Lundin M, et al. MYC-dependent regulation and prognostic role of CIP2A in gastric cancer. J Natl Cancer Inst 2009; 101:793 - 805; http://dx.doi.org/10.1093/jnci/djp103; PMID: 19470954
- Dong QZ, Wang Y, Dong XJ, Li ZX, Tang ZP, Cui QZ, et al. CIP2A is overexpressed in non-small cell lung cancer and correlates with poor prognosis. Ann Surg Oncol 2011; 18:857 - 65; http://dx.doi.org/10.1245/s10434-010-1313-8; PMID: 20842459
- Böckelman C, Hagström J, Mäkinen LK, Keski-Säntti H, Häyry V, Lundin J, et al. High CIP2A immunoreactivity is an independent prognostic indicator in early-stage tongue cancer. Br J Cancer 2011; 104:1890 - 5; http://dx.doi.org/10.1038/bjc.2011.167; PMID: 21610708
- Böckelman C, Lassus H, Hemmes A, Leminen A, Westermarck J, Haglund C, et al. Prognostic role of CIP2A expression in serous ovarian cancer. Brit J Cancer 2011; 105:989 - 95; http://dx.doi.org/10.1038/bjc.2011.346; PMID: 21897396
- Côme C, Laine A, Chanrion M, Edgren H, Mattila E, Liu X, et al. CIP2A is associated with human breast cancer aggressivity. Clin Cancer Res 2009; 15:5092 - 100; http://dx.doi.org/10.1158/1078-0432.CCR-08-3283; PMID: 19671842
- Wang WS, Chen PM, Chiou TJ, Liu JH, Lin JK, Lin TC, et al. Epidermal growth factor receptor R497K polymorphism is a favorable prognostic factor for patients with colorectal carcinoma. Clin Cancer Res 2007; 13:3597 - 604; http://dx.doi.org/10.1158/1078-0432.CCR-06-2601; PMID: 17575224
- Liu Y, Yang Y, Xu H, Dong X. Implication of USP22 in the regulation of BMI-1, c-myc, p16INK4a, p14ARF, and cyclin D2 expression in primary colorectal carcinomas. Diagn Mol Pathol 2010; 19:194 - 200; http://dx.doi.org/10.1097/PDM.0b013e3181e202f2; PMID: 21052002
- Grady WM, Markowitz SD. Genetic and epigenetic alterations in colon cancer. Annu Rev Genomics Hum Genet 2002; 3:101 - 28; http://dx.doi.org/10.1146/annurev.genom.3.022502.103043; PMID: 12142355
- Khanna A, Okkeri J, Bilgen T, Tiirikka T, Vihinen M, Visakorpi T, et al. ETS1 mediates MEK1/2-dependent overexpression of cancerous inhibitor of protein phosphatase 2A (CIP2A) in human cancer cells. PLoS ONE 2011; 6:e17979; http://dx.doi.org/10.1371/journal.pone.0017979; PMID: 21445343
- Torhorst J, Bucher C, Kononen J, Haas P, Zuber M, Kochli OR, et al. Tissue microarrays for rapid linking of molecular changes to clinical endpoints. Am J Pathol 2001; 159:2249 - 56; http://dx.doi.org/10.1016/S0002-9440(10)63075-1; PMID: 11733374
- Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 1998; 4:844 - 7; http://dx.doi.org/10.1038/nm0798-844; PMID: 9662379
- Kallioniemi OP, Wagner U, Kononen J, Sauter G. Tissue microarray technology for high-throughput molecular profiling of cancer. Hum Mol Genet 2001; 10:657 - 62; http://dx.doi.org/10.1093/hmg/10.7.657; PMID: 11257096
- Soo Hoo L, Zhang JY, Chan EK. Cloning and characterization of a novel 90 kDa 'companion' auto-antigen of p62 overexpressed in cancer. Oncogene 2002; 21:5006 - 15; http://dx.doi.org/10.1038/sj.onc.1205625; PMID: 12118381