Abstract
KRAS mutations are proved to confer dramatic resistance to EGFR target therapy of cancer patients. The aim of this study was to establish a convenient and accurate method to screen plasma KRAS mutations of cancer patients since tumor specimens were not always available in clinical practice. A modified PNA-PCR method was established and evaluated in plasma of 19 pancreatic cancer patients. Our results showed that the modified PNA-PCR assay was a sensitive (87.5%, 14/16) and accurate (92.9%, 13/14) method to screen plasma KRAS mutations of pancreatic cancer patients and there was a high consistency of KRAS mutation status between plasma samples and tumor specimens. The modified protocol could not only screen plasma KRAS mutations rapidly and accurately but also had potential to quantify KRAS mutant DNA to predict treatment response of cancer patients and monitor disease progression. It’s should be indicated that this modified assay was only confirmed in the pancreatic cancer patients in this study and need to be verified in other cancer patients.
Introduction
KRAS is a downstream molecule in the EGFR-signaling pathway and its mutation is one of the most important somatic mutations in malignant tumors. KRAS mutations are found in about 80% of pancreatic cancer and 40% of colorectal cancer,Citation1,Citation2 and most of them occur in codon 12 and 13.Citation3 Several phase II and III clinical trials have demonstrated that KRAS mutations confer dramatic resistance to EGFR antibody, such as cetuximab and panitumumab.Citation4-Citation6 The American Society of Clinical Oncology (ASCO), the Food and Drug Administration and the European Medicine Agency have restricted anti-EGFR drugs to treat the set of patients carrying wild-type KRAS tumors in 2009.Citation7 Therefore, it’s of great importance to define subpopulation of patients through KRAS mutations analysis.
However, recent studies suggested that it was not enough to roughly distinguish patients with wild-type KRAS from those with mutations. In 2010, de Roock et al. found that KRAS codon 13 (G13D) may not be absolutely predictive of non-response and patients with G13D mutation in fact could possibly benefit from anti-EGFR therapy.Citation8 Similarly, S. Tejpar et al. also observed that mCRC (metastatic colorectal cancer) patients with KRAS G13D mutation could benefit from the addition of cetuximab to first-line chemotherapy and reported this finding in 2011 ASCO annual meeting. Therefore, it’s necessary to detect the exact pattern of KRAS mutations when testing KRAS mutations status.
Although KRAS mutations can be detected in tumor tissues, obtaining such tissues is not always available in clinical practice, particularly for patients with recurrent and refractory cancer. In this situation, it’s a good alternative to use plasma samples to screen KRAS mutations since patients’ plasma may contain circulating tumor mutant DNA. More importantly, circulating DNA could provide a unique opportunity to repeatedly monitor patients during treatment.Citation9 The circulating DNA fragment might originate from apoptotic or necrotic cells of primary tumor.Citation9 Recently, some researchers found that cancer patients circulating DNA may have close relationship of circulating tumor cells (CTC) and proved that both CTC and circulating DNA could be valuable biomarkers for cancer patients.Citation10 Recently, numerous publications have proved that circulating DNA contained the same genetic alternation as corresponding tumor tissue and could be surrogate tumor tissues for screening genetic alternation.Citation11-Citation14
Although there are many other methods developed to enrich plasma mutant KRAS DNA,Citation13,Citation15,Citation16 most of them need multiple procedures and are not convenient and suitable for clinical use. PNA-PCR (peptide nucleic acids-PCR) method is a relatively convenient and timesaving method.Citation17,Citation18 However, there are still two challenges that need to be solved. One is how to analyze KRAS mutation pattern since almost none of PNA-PCR assay could ascertain KRAS mutation type. The other is whether the plasma screening could replace the tumor tissue’s detection because no study to date has estimated the consistency of plasma and tumor tissue using PNA-PCR method.
In this study, we devised a convenient and accurate method, PNA-PCR plus pyrosequencing assay, to screen plasma KRAS mutations of pancreatic cancer patients and ascertain the mutation pattern simultaneously. In addition, we compared KRAS mutations status between patients’ plasma and corresponding tumor tissue to ensure the consistency of two results.
PNA is a synthetic DNA analog in which the normal phosphodiester backbone is replaced with a 2-aminoethyglycine chain. PNA cannot serve as a primer in PCR and PNA/DNA-hybrids have a higher thermal stability comparing with the corresponding DNA/DNA hybrids. However, a single mismatch could destabilize the PNA-DNA hybrids, causing a Tm shift of 9–16°CCitation17,Citation19,Citation20 Thus, PNA is a good substance to block chain elongation and primer annealing in PCR. Pyrosequencing has been proved to be a fast, convenient and sensitive DNA sequencing method, and has been wildly used in screening KRAS mutations.Citation21-Citation23 In this proof-of-principle study, we selected pancreatic cancer as the target malignancy because most of (~80%) them contain KRAS mutations,Citation1 and it was easy to find more KRAS mutant specimens within limited cancer patients.
Results
Optimization of PNA concentration
To detect all possible KRAS mutations in codon 12 and 13, we employed PNA to suppress the amplification of wild-type KRAS and amplify mutant KRAS DNA exclusively. The concentration of primer was minimized to 0.075 μmol/L because of the competitive relationship of PNA to reverse primer. For complete suppression of wild-type DNA amplification, raising amounts of PNA were used. In the presence of 0.5 μmol/L PNA, no PCR product peak was observed when the template was all wild-type.
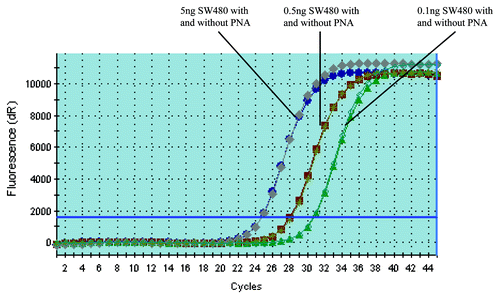
In order to determine if the existence of PNA would affect the amplification of mutant DNA, we compared the amplification status of mutant DNA with and without PNA simultaneously. As shown in , there was no significant difference between these two assays, indicating that the addition of PNA didn’t affect the amplification of KRAS mutant DNA.
Figure 1. PNA doesn’t affect the amplification of KRAS mutant DNA. PCR with and without PNA have almost the same amplification curves when the template is KRAS mutant DNA (The PCR amplification curve with and without PNA are almost totally overlapping each other). The amount of SW480 DNA added is 5, 0.5 and 0.1 ng, respectively (corresponding to the amplification curve from left to right in the figure).
Sensitivity of the assay and quantification of mutant DNA
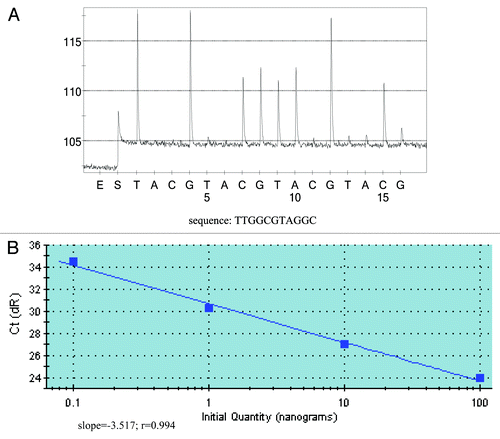
The relative sensitivity of this assay was obtained by using dilutions of SW480 DNA in KRAS wild-type DNA. As shown in , as little as 20 pg of mutant DNA (about six genomes, or 1:5,000 mutant to wild-type ratio) could be detected by this assay. Since the amplification of the wild-type DNA could be completely suppressed (), another favorite advantage of this assay was that it could quantify the mutant DNA simultaneously. After analyzing the standard curve of this PNA-PCR, the regression line was linear (slope = -3.517; r = 0.994) in the range of 0.1 to 100 ng of KRAS mutant DNA ().
Figure 2. Detection sensivitity of PNA-PCR assay and quantification of KRAS mutant DNA. (A) Detection sensitivity of PNA-PCR assay. The sequencing primer used was PF1, and as little as 20 pg of mutant DNA can be detected from 100 ng KRAS wild-type DNA (1:5,000). The sequencing pyrogram shows the absence of wild-type sequence, which indicates that PNA could successfully suppress the amplification of wild-type sequences. (B) Quantification of KRAS mutant DNA. Varying amounts of KRAS mutant DNA were plotted against Ct values (threshold cycle). Slope, r value and regression line are shown.
Genotyping KRAS mutations of pancreatic tissue specimens
Regular PCR plus Sanger sequencing was performed on 19 FFPE (formalin-fixed paraffin-embedded) pancreatic tumor tissues to detect their KRAS mutations and results were shown in . In total, 16 (84.2%) of 19 samples showed KRAS mutation in codon 12 or 13, represented by eight mutations G12D, five mutations G12V, two mutations G13D and one mutation G12R.
Table 1. KRAS mutation status of FFPE tissues and plasma samples
Screening KRAS mutations of plasma samples
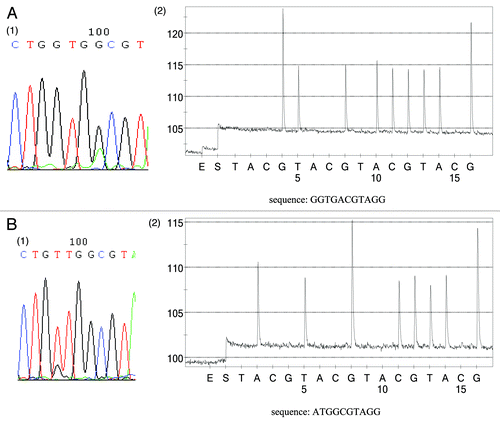
Among the 19 plasma samples, 16 of them were gathered before surgery and the remaining were gathered after operation or recurrence. PCR was first performed in the absence of PNA to prove the successful extraction of cell-free DNA. Then PNA-PCR plus pyroquencing was employed to screen plasma KRAS mutations. As shown in and , KRAS alterations were found in plasma from 14 out of 19 (73.7%) samples (). There were two patients whose FFPE samples were KRAS mutant whereas the corresponding plasma were diagnosed as KRAS wild type. The KRAS mutation types of plasma were identical to those of corresponding tumor tissues in all subjects except for subject Y3 whose plasma carried a GGT to GAT mutation (codon 12) whereas tumor tissue contained a GGT to GTT mutation (). We tested the tumor and plasma samples independently again and the results were the same.
Table 2. Comparison of KRAS mutation rate between plasma and tumor tissue
Figure 3. Comparison of KRAS status between FFPE tumor tissues and corresponding plasma samples. (A) Subject (15) has the same KRAS alternation (G13D) between tumor tissue and plasma sample. A1 KRAS status of FFPE tumor tissue. A2 KRAS status of plasma sample using sequencing primer PF2. (B) Inconsistency between plasma sample and corresponding tumor tissue (subject Y3). Tumor tissue contains a G12V mutation (1) whereas plasma sample carries a G12D mutation (sequencing primer PF1) (2). It should be noted that the grams of pyrosequencing only reveal the KRAS mutant sequence, while no wild-type sequence was observed, which also proved that only KRAS mutant alleles could be amplified in this assay.
In summary, as shown in , there was no significant difference of KRAS mutation rate between plasma and tumor tissues. However, there was a tendency of higher consistency between plasma and tumor tissue in pre-surgery patients than that in post-operation group, although the difference did not reach statistical significance (p = 0.054).
Discussion
In this study we combined modified PNA-PCR method and pyrosequencing to screen and ascertain plasma KRAS mutations of pancreatic cancer patients, and as far as we know, it was the first time to compare KRAS mutation results between plasma samples and corresponding tumor tissues using PNA-PCR method. Our results showed that modified PNA-PCR protocol was a sensitive (87.5%, 14/16) and accurate (92.9%, 13/14) method to screen plasma KRAS mutations and there was a high consistency of KRAS mutations status between plasma samples and tumor specimens.
Our results were consistent with other published literatures and supported that circulating DNA could be used as surrogate tumor tissues for detecting genetic alternations.Citation11-Citation14 Our modified assay could screen 14 plasma KRAS mutations from 16 cases patients whose tumor tissues were confirmed to carry KRAS mutations. These results concurred with the findings of a recent study.Citation13 Although other PNA-PCR studies were also demonstrated to be sensitive to screen circulating KRAS mutations, but in contrast with the current study, the previous study didn’t have matched tumor and plasma samples in all cases.Citation17,Citation18
Another advantage of this PNA-PCR method was that it had potential ability to quantify KRAS mutant DNA. Recently, more and more researches have focused on quantification of mutant circulating DNA. Some research has found that decreased concentration of circulating mutant DNA could be observed in patients with partial or complete clinical remission, whereas persistence of circulating mutant DNA was observed in patients with cancer progression.Citation24,Citation25 These findings indicated that quantitative circulating mutant DNA could be regarded as a useful measure of tumor dynamics to predict treatment response and monitor disease progression. In this study, we found that our assay could suppress the primer annealing to KRAS wild-type alleles, and didn’t interfere the amplification of mutant alleles, which made quantification of mutant KRAS become feasible. Further study concerning whether quantification of circulating KRAS mutant DNA could be a biomarker to predict treatment response of cancer patients was under consideration.
Although this assay showed high accuracy in our study, there was one inconsistency between plasma sample and corresponding tumor tissue. This subject (Y3) contained one type mutation in tumor tissue (GGT→GTT) whereas carries another type mutation in plasma (GGT→GAT). A potential explanation is that during tumor progression the KRAS status changes. Recently, some articles have reported the heterogeneity of KRAS mutations between primary tumor and corresponding metastases.Citation26-Citation28 We should notice that the specimens (tumor tissue and plasma) of this patient were not obtained at the same time. The tissue sample was collected during the time of initial diagnosis, whereas plasma sample was gained after operation and successive chemotherapy. This discordance might hint instable gene status of tumor cells, especially in those suffering from chemotherapeutics stress.
In summary, the current study has proved that the modified PNA-PCR assay is a sensitive, convenient and accurate method to screen plasma KRAS mutations of pancreatic cancer patients and has potential ability to quantify circulating KRAS mutant DNA. Although the modified method got a satisfactory result in this study, it still needs further verification in other cancer patients. Future studies are underway to find out the association between concentration change of mutant DNA and disease progression.
Materials and methods
Cell lines and patients
Cells of cell lines SW480 (colon carcinoma) harboring a homozygous KRAS codon 12 mutation and SW116 (colon carcinoma) carrying wild-type KRAS were used in this study. A total of 19 pancreatic patients’ plasma samples (15 pre-operations and 4 post-operations) and paired FFPE specimens were obtained from the Drum Tower Hospital from November 2008 to December 2009. Among these 19 plasma samples, 15 of them were obtained on the morning before operation and the remaining 4 samples were gained after resection of primary tumor and a serial of chemotherapy. The study was approved by the Institutional Ethic Committee of the hospital and informed consents were obtained from all patients.
DNA extraction from the plasma
Blood samples (2 ml) were withdrawn from peripheral vein and placed in tubes containing EDTA. Plasma samples were centrifuged at 3,000 g for 10 min to remove any residual cells and then stored at -80°C until further use. DNA was extracted from plasma with QIAamp spin columns (catalog no. 51106, QIAamp DNA Mini Kit). The standard protocol was modified to include the addition of polydT DNA carrier facilitating better adsorption of DNA to the Qiagen column. DNA was eluted in a final volume of 35 μl. Negative control (without sample) was performed to exclude the possibility of contamination during extraction.
Isolation of DNA from FFPE tissues, SW480 and SW116 cells
After manual macrodissection, genomic DNA was isolated from FFPE samples with RecoverAll total nucleic acid isolation kit (catalog no. AM1975, Ambion) according to the manufacturer’s instructions. Genomic DNA of SW480 and SW116 were extracted using the same isolation kit except for the step of deparaffinization. DNA concentrations were determined by UV (260 nm) spectrophotometer. DNA from cell lines SW480 was serially diluted into 100 ng KRAS wild-type DNA (SW116) to the following ratios: 1:200, 1:500, 1:1,000, 1:2,000, 1:5,000, and stored at -20°C.
PNA-PCR conditions
We used one single PNA clamp PCR assay based on SYBR-Green I to detect different KRAS mutations.Citation29 The PCR primers and PNA of real-time PCR assay were adapted from Gilje et al.Citation29 The sequences of the PCR primers were 5′-GCCTGCTGAAAATGACTGAATATAA-3′ (forward) and 5′-biotin-CGTCAAGGCACTCTTGCCTAC-3′ (reverse). The PNA sequence was 5′-CCTACGCCACCAGCTCC-3′.
Phusion (catalog no. F-530F, Finnzymes) HS-based PNA-PCR was performed in a final volume of 20 μl containing 1 × Phusion HF buffer, 0.2 mmol/L d NTP, 0.075 μmol/L forward and reverse primer, 1 μl 1:500 SYBR Green I, a certain amount of PNA, 1 μl DMSO, 0.02 U/μl Phusion HS DNA polymerase and 100 ng cell line DNA or 4 μl plasma DNA. Thermocycling was performed in an Mx3000P (Stratagene) real-time PCR instrument and the cycling conditions were as follows: 98°C, 30 s for initial denaturation and enzyme activation, 45 cycles of 98°C, 10 s for denaturation, 72°C, 10 s for PNA annealing, 62°C, 20 s for primer annealing, 72°C, 20 s for elongation. The PCR products were monitored by melting curve analysis. Negative control (without sample) was performed to exclude the possibility of contamination. Positive control (KRAS mutant DNA) was performed to make sure successful amplification of PNA-PCR.
Sensitivity of PNA-PCR plus pyrosequencing
Dilutions of KRAS mutant SW480 DNA (1:200, 1:500, 1:1,000, 1:2,000 and 1:5,000, wild type to mutant type ratio) were used to assess the sensitivity of PNA-PCR plus pyrosequencing. Pyrosequencing was performed in Pyrosequencing PSQ96 HS System (Biotage AB). In order to increase detection sensitivity of most KRAS mutations on pyrosequencing, we adopted two slightly different sequencing primers to analyze plasma samples: PF1 (5′-TGTGGTAGTTGGAGCTG-3′) and PF2 (5′-TGTGGTAGTTGGAGCT-3′).Citation18 Nucleotide dispensation order was cyclic (TACG from 5′ to 3′).
Patients’ samples
For plasma samples, 4 μl of plasma DNA was added to PNA-PCR and the PCR products were analyzed on Pyrosequencing with the primers that were identical to those above. For tumor tissue samples, we employed conventional PCR and Sanger sequencing to screen KRAS mutations (primer sequences and conditions are available on request).
Statistical analysis
Chi-square test was performed to compare the qualitative data. A p-value of < 0.05 was considered statistically significant.
| Abbreviations: | ||
| FFPE | = | formalin-fixed paraffin-embedded |
| PNA | = | peptide-nucleic-acid |
| KRAS | = | v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog |
| EGFR | = | epidermal growth factor receptor |
| v-erb-b | = | erythroblastic leukemia viral oncogene homolog, avian |
| ASCO | = | The American Society of Clinical Oncology |
| mCRC | = | metastatic colorectal cancer |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Shi C, Hong SM, Lim P, Kamiyama H, Khan M, Anders RA, et al. KRAS2 mutations in human pancreatic acinar-ductal metaplastic lesions are limited to those with PanIN: Implications for the human pancreatic cancer cell of origin. Mol Cancer Res 2009; 7:230 - 6; http://dx.doi.org/10.1158/1541-7786.MCR-08-0206; PMID: 19208745
- Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: Results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol 2010; 28:466 - 74; http://dx.doi.org/10.1200/JCO.2009.23.3452; PMID: 20008640
- Brink M, de Goeij A, Weijenberg MP, Roemen G, Lentjes M, Pachen MMM, et al. K-ras oncogene mutations in sporadic colorectal cancer in The Netherlands Cohort Study. Carcinogenesis 2003; 24:703 - 10; http://dx.doi.org/10.1093/carcin/bgg009; PMID: 12727799
- Linardou H, Dahabreh IJ, Kanaloupiti D, Siannis F, Bafaloukos D, Kosmidis P, et al. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol 2008; 9:962 - 72; http://dx.doi.org/10.1016/S1470-2045(08)70206-7; PMID: 18804418
- Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 2008; 359:1757 - 65; http://dx.doi.org/10.1056/NEJMoa0804385; PMID: 18946061
- Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 2008; 26:1626 - 34; http://dx.doi.org/10.1200/JCO.2007.14.7116; PMID: 18316791
- Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, et al. American Society of Clinical Oncology Provisional Clinical Opinion: Testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol 2009; 27:2091 - 6; http://dx.doi.org/10.1200/JCO.2009.21.9170; PMID: 19188670
- De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu DS, Siena S, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA 2010; 304:1812 - 20; http://dx.doi.org/10.1001/jama.2010.1535; PMID: 20978259
- Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarker in cancer patients. Nat Rev Cancer 2011; 11:426 - 37; http://dx.doi.org/10.1038/nrc3066; PMID: 21562580
- Schwarzenbach H, Alix-Panabières C, Müller I, Letang N, Vendrell JP, Rebillard X, et al. Cell-free tumor DNA in blood plasma as a marker for circulating tumor cells in prostate cancer. Clin Cancer Res 2009; 15:1032 - 8; http://dx.doi.org/10.1158/1078-0432.CCR-08-1910; PMID: 19188176
- Schwarzenbach H, Felix KHC, Isbarn H, Huland H, Pantel K. Genomic profling of cell-free DNA in blood and bone marrow of prostate cancer patients. J Cancer Res Clin Oncol 2011; 137:811 - 9; http://dx.doi.org/10.1007/s00432-010-0941-5; PMID: 20683729
- Bai H, Mao L, Wang HS, Zhao J, Yang L, An TT, et al. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in chinese patients with stages IIIB to IV non-small-cell lung cancer. J Clin Oncol 2009; 27:2653 - 9; http://dx.doi.org/10.1200/JCO.2008.17.3930; PMID: 19414683
- Wang S, An T, Wang J, Zhao J, Wang Z, Zhuo M, et al. Potential clinical significance of a plasma-based KRAS mutation analysis in patients with advanced non-small cell lung cancer. Clin Cancer Res 2010; 16:1324 - 30; http://dx.doi.org/10.1158/1078-0432.CCR-09-2672; PMID: 20145159
- Rosell R, Moran T, Queralt C, Porta R, Cardenal F, Camps C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 2009; 361:958 - 67; http://dx.doi.org/10.1056/NEJMoa0904554; PMID: 19692684
- Dobrzycka B, Terlikowski SJ, Mazurek A, Kowalczuk O, Niklinska W, Chyczewski L, et al. Circulating free DNA, p53 antibody and mutations of KRAS gene in endometrial cancer. Int J Cancer 2010; 127:612 - 21; http://dx.doi.org/10.1002/ijc.25077; PMID: 19960433
- Ryan BM, Lefort F, McManus R, Daly J, Keeling PWN, Weir DG, et al. A prospective study of circulating mutant KRAS2 in the serum of patients with colorectal neoplasia: strong prognostic indicator in postoperative follow up. Gut 2003; 52:101 - 8; http://dx.doi.org/10.1136/gut.52.1.101; PMID: 12477769
- Luo JD, Chan EC, Shih CL, Chen TL, Liang Y, Hwang TL, et al. Detection of rare mutant K-ras DNA in a single-tube reaction using peptide nucleic acid as both PCR clamp and sensor probe. Nucleic Acids Res 2006; 34:e12; http://dx.doi.org/10.1093/nar/gnj008; PMID: 16432256
- Däbritz J, Hänfler J, Preston R, Stieler J, Oettle H. Detection of Ki-ras mutations in tissue and plasma samples of patients with pancreatic cancer using PNA-mediated PCR clamping and hybridisation probes. Br J Cancer 2005; 92:405 - 12; http://dx.doi.org/10.1038/sj.bjc.6602319; PMID: 15655549
- Thiede C. Bayerd rffer E, Blasczyk R, Wittig B, Neubauer A. Simple and sensitive detection of mutations in the ras proto-oncogenes using PNA-mediated PCR clamping. Nucleic Acids Res 1996; 24:983; http://dx.doi.org/10.1093/nar/24.5.983; PMID: 8600471
- Ørum H, Nielsen PE, Egholm M, Berg RH, Buchardt O, Stanley C. Single base pair mutation analysis by PNA directed PCR clamping. Nucleic Acids Res 1993; 21:5332; http://dx.doi.org/10.1093/nar/21.23.5332; PMID: 8265345
- Ogino S, Kawasaki T, Brahmandam M, Yan LY, Cantor M, Namgyal C, et al. Sensitive Sequencing method for KRAS mutation detection by pyrosequencing. J Mol Diagn 2005; 7:413 - 21; http://dx.doi.org/10.1016/S1525-1578(10)60571-5; PMID: 16049314
- Gharizadeh B, Herman ZS, Eason RG, Jejelowo O, Pourmand N. Large-scale pyrosequencing of synthetic DNA: A comparison with results from Sanger dideoxy sequencing. Electrophoresis 2006; 27:3042 - 7; http://dx.doi.org/10.1002/elps.200500834; PMID: 16800029
- Luthra R, Zuo Z. COLD-PCR finds hot application in mutation analysis. Clin Chem 2009; 55:2077; http://dx.doi.org/10.1373/clinchem.2009.136143; PMID: 19833831
- Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008; 14:985 - 90; http://dx.doi.org/10.1038/nm.1789; PMID: 18670422
- Yung TKF, Chan KCA, Mok TSK, Tong J, To KF, Lo YMD. single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer Patients. Clin Cancer Res 2009; 15:2076 - 84; http://dx.doi.org/10.1158/1078-0432.CCR-08-2622; PMID: 19276259
- Monaco SE, Nikiforova MN, Cieply K, Teot LA, Khalbuss WE, Dacic S. A comparison of EGFR and KRAS status in primary lung carcinoma and matched metastases. Hum Pathol 2010; 41:94 - 102; http://dx.doi.org/10.1016/j.humpath.2009.06.019; PMID: 19740513
- Baldus SE, Schaefer KL, Engers R, Hartleb D, Stoecklein NH, Gabbert HE. Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res 2010; 16:790 - 9; http://dx.doi.org/10.1158/1078-0432.CCR-09-2446; PMID: 20103678
- Kalikaki A, Koutsopoulos A, Trypaki M, Souglakos J, Stathopoulos E, Georgoulias V, et al. Comparison of EGFR and K-RAS gene status between primary tumours and corresponding metastases in NSCLC. Br J Cancer 2008; 99:923 - 9; http://dx.doi.org/10.1038/sj.bjc.6604629; PMID: 19238633
- Gilje B, Heikkila R, Oltedal S, Tjensvoll K, Nordgard O. High-fidelity DNA polymerase enhances the sensitivity of a peptide nucleic acid clamp PCR assay for K-ras mutations. J Mol Diagn 2008; 10:325 - 31; http://dx.doi.org/10.2353/jmoldx.2008.070183; PMID: 18556764