Abstract
This study was designed to detect global gene expressions of primary advanced colorectal cancer (ACC) patients who have undergone FOLFOX4 chemotherapy and screen valuable biomarkers to predict the effects of chemotherapy. Samples from primary ACC patients who have undergone FOLFOX4 chemotherapy were collected. Their chemotherapy effects were evaluated and divided into chemotherapy sensitive group (experimental group) and non-sensitive group (control group). Cancerous tissue gene expression profiles were detected by chip technology. Two groups with differentially expressed genes were screened by cluster analysis and significance analysis of microarrays (SAM). Valuable biomarkers were screened by bioinformatics analysis. Immunohistochemical analysis was performed to characterize the pattern of Nkx2–3 and TGFB1I1 expression. Nkx2–3 and TGFB1I1 signal log ratio were used Receiver Operating Characteristic (ROC) analyses to calculate its own predicting accuracy. Thirty cases were divided into experimental group (13 cases) and control group (17 cases). There was evident difference in the tumor cell biology states of the two groups; that is, 25 ESTs (21 genes) were upregulated and 5 ESTs (5 genes) were downregulated. Nkx2–3 protein was observed on the nucleus of the cancer cells and TGFB1I1 protein was observed on the nucleus and cytoplasm of the cancer cells in experimental group. Their prediction accuracies were 85.3% and 76.7% respectively. Nkx2–3 and TGFB1I1 expressions in control group are very low, but highly expressed in the experimental group; Nkx2–3 and TGFB1I1 may be classified as valuable biomarkers, as these can predict the effects of primary ACC patients who will undergo FOLFOX4.
Introduction
Colorectal cancer (CRC) is known as the third highest incidence rate of cancer and is the fourth leading cause of death due to cancer worldwide.Citation1 The World Health Organization (WHO) estimates that 945,000 new cases are diagnosed yearly, resulting in 492,000 deaths.Citation1 Early CRC patients may be cured by surgical operation, but 30–40% of them are confirmed only while in the advanced metastatic stage and hence cannot be cured by surgery alone.Citation2 In addition, more than half of the patients who are assumed initially as curable suffer from recurrence and eventually die of the disease.Citation3 In the past 40 y, fluorouracil (FU)-based chemotherapy has been the main treatment for CRC metastasis. Recently, significant progress with oxaliplatin and other new cytotoxic drugs and monoclonal antibody targeted therapies have been introduced into chemotherapy.Citation4-Citation7 Some of the most commonly combined chemotherapy, such as FOLFOX4 (fluorouracil, leucovorin, and oxaliplatin), can obtain nearly 50% objective remission rates.Citation8 However, about half of the patients on this new combination chemotherapy have remained non-sensitive; the majority eventually appeared as drug-resistant. From among different CRC patients who have received the same treatment of chemotherapy regimens and drug doses, there have been notable individual differences in curative effects resulting in tolerability.Citation9,Citation10 Hence, there is an urgent need to identify some valuable biomarkers based on their expressions in order to predict the effects of chemotherapy, as well as to carry out tailored therapies. At present, many have attempted to explore various chemotherapy reaction predictors.Citation11-Citation21 However, two recent systematic review analyses have shown that most prediction effects are somewhat controversial,Citation22,Citation23 and conducting research on the basic fields of chemotherapy predictors remain a difficult task.
Gene chip technology is important in functional gene detection. It has many advantages, such as high throughput, high efficiency, and low consumption in the analysis of biological information processing and large-scale access to relevant information; it can also screen gene-related cancer occurrence and development, as well as its transfer mechanism and chemotherapy drugs reaction.Citation24-Citation26 It contributes to the research of tumor to the genome level in a systemic and holistic manner.
Herein, by using microarray analysis of gene expression profile, we compared the global gene expression patterns of primary ACC patients who have undergone FOLFOX4 chemotherapy from the experimental group with the control group. The objective of this study was designed to detect gene expressions and screen valuable biomarkers according to their expressions in order to predict the effects of chemotherapy, as well as to implement tailored therapies.
Results
Patient evaluation of the study sample
The study start enrolled 34 patients; 3 patients could not tolerate chemotherapy, and hence failed to fulfill the FOLFOX4 chemotherapy and exited the research midway. One of these patients was unable to finish R0 resection of the primary tumor after chemotherapy and was also eliminated from the study. Finally, 30 cases completed the required treatment; all were included in the analysis. Among these patients (), none was observed with complete response (CR); 13 cases were with partial response (PR) and were included into the experimental group; and 6 cases were with stable disease (SD) while 11 cases were with progressive disease (PD), and these were included into the control group.
Table 1. Baseline Patient Characteristics (intent-to-treat population)
Microarray data and bioinformatics analysis
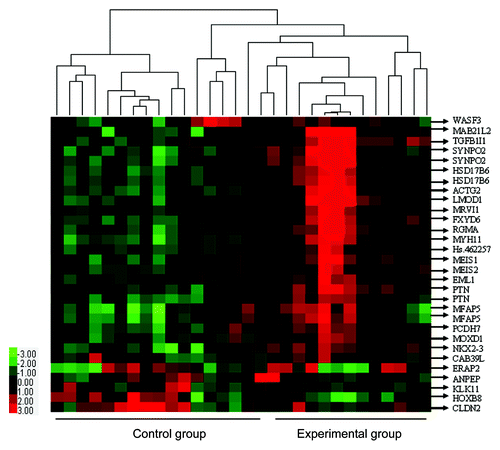
After SAM analysis of microarray data for experimental vs. control group, there were differentially expressed genes or ESTs, upregulated genes or ESTs (9,359), and downregulated genes or ESTs (1,1104). Thirty ESTs were certified as significantly differentially expressed between the two groups, with EST |Score(d)| ≥ 2, with Fold Change ≥ 2 or Fold Change ≤ 0.5. Meanwhile, 25 ESTs (21 genes) were upregulated whereas 5 ESTs (5 genes) were downregulated ( and ). The list of 26 differentially expressed genes was used for unsupervised hierarchical clustering, and the results were analyzed and visualized by a TreeView program ().
Table 2. Upregulated gene (n = 21) for experimental group vs. control group
Table 3. Downregulated gene (n = 5) for experimental group vs. control group
Figure 1. Clustering display of microarray data and functional classifications of differentially expressed genes for experimental group vs. control group. Comparisons of 26 differentially expressed genes (30 ESTs) between the two groups were performed using the SAM software. Upon hierarchical clustering, they have been visualized with TreeView tools. Gene symbols are labeled on the right. Expression levels are represented by a color tag; red represents the highest while green is for the lowest levels of expression.
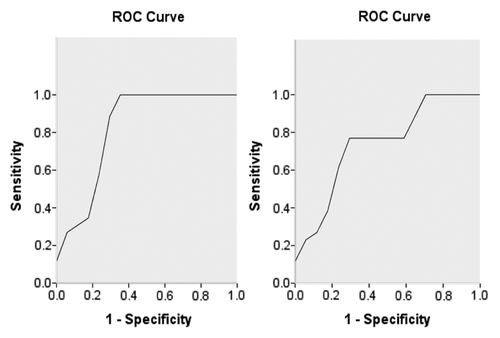
Nkx2–3 and TGFB1I1 prediction accuracies analysis
AUCs of Nkx2–3 and TGFB1I1 were 0.824 and 0.742(), cutoff values were 2.392 and 3.290, sensitivity were 1.000 and 0.769, specificities were 0.706 and 0.765, Youden indexes were 0.706 and 0.534, accuracies were 85.3% and 76.7%, p values were 0.003 and 0.025 respectively.
Immunohistochemistry
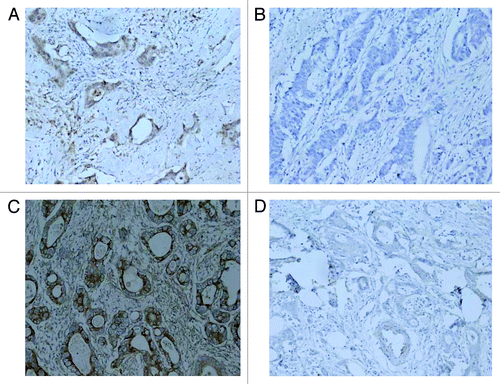
Immunohistochemical analysis revealed that Nkx2–3 protein was observed on the nucleus of the cancer cells in 13 cancer tissues in experimental group, lower or no expression in 17 cancer tissues in control group. There was a statistically significant difference in expression levels between experimental group and control group (p < 0.01, ). The result indicated that higher expression of Nkx2–3 was associated with the sensitivity of FOLFOX4 chemotherapy. TGFB1I1 protein was observed on the nucleus and cytoplasm of the cancer cells in 13 cancer tissues in experimental group, lower or no expression in 17 cancer tissues in control group. There was a statistically significant difference in expression levels between experimental group and control group (p < 0.01, ). The result also indicated that higher expression of TGFB1I1 was associated with the sensitivity of FOLFOX4 chemotherapy.
Figure 3. Immunohistochemistry analysis of Nkx2–3 and TGFB1I1. (A) Nkx2–3 high expression in cancer tissues in experimental group (200 size); (B) Nkx2–3 no expression in cancer tissues in control group (200 size); (C) TGFB1I1 high expression in cancer tissues in experimental group (200 size); (D) TGFB1I1 no expression in cancer tissues in control group (200 size).
Discussion
It has been widely agreed that personalized treatment is the ideal approach to tumors. A necessary premise of personalized treatment is to screen its valuable biomarkers. FOLFOX4 chemotherapy has been used extensively in ACC patients, but the lack of prediction biomarks of the regime therapeutic effect prohibits ACC patients to achieve ideal personalized treatments. This was the first time that primary ACC patients who have undergone FOLFOX4 chemotherapy had their global gene expression analyzed. The 26 genes identified as differentially expressed almost involved various biological function. Some of these interesting genes may be related to CRC development, the body’s immunes, and drug chemotherapy.
There were two tumor suppressor genes (Nkx2–3 and TGFB1I1), who were the strongest prediction abilities in the upregulated genes.
Nkx2–3 was grouped primarily within the following two functional categories: (1) immune and inflammatory response and (2) cell proliferation, growth and oncogenesis. Ingenuity pathway analysis indicated that the affected top pathway included genes directly involved in Wnt signaling. Nkx2–3 could contribute to the pathogenesis of Inflammatory Bowel Disease (IBD)-associated CRC and sporadic CRC by regulating the Wnt signaling pathway.Citation28 Nkx2–3 might be a new tumor suppressor gene related to sporadic CRC.Citation29 However there is little research in this field now.
TGFB1I1 encodes a coactivator of the androgen receptor, a transcription factor which is activated by androgen and has a key role in male sexual differentiation.Citation30-Citation32 The encoded protein is thought to regulate androgen receptor activity and adjust the epithelial cells of the growth process.Citation33-Citation36 Multiple transcript variants encoding different isoforms have been found for this gene. TGFB1I1 different subtypes have different functions in platelet gathered, adhesion and movement and muscle cells generating process,Citation37-Citation40 these effects perform mainly through integrin signaling pathways,Citation41 serotonin, protein kinase C and TGFB1I1-associated redistribution of the platelet serotonin transporter.Citation42 TGFB1I1 tyrosine phosphorylation functions to regulate signaling associated with lamellipodia formation, a process fundamental to cell motility.Citation43 TGFB1I1 was related to prostate cancer cell differentiation, and decrease of its expression can cause the tumor.Citation44,Citation45 Long-term stimulation by tumor necrosis factor-α could increase TGFB1I1 expression in prostate cancer cells. The different expression of coactivators may contribute to the different response of prostate cancer to androgenic stimulation or endocrine therapy.Citation46,Citation47 The encoded protein may have a role to play in the treatment of prostate cancer.Citation48 In addition, in colorectal cancer TGFB1I1 can reduce colorectal tumor cell growth rate and induce tumor cell apoptosis.Citation49,Citation50 However there is little research on its mechanism of the action now.
Results from the microarray data and immunohistochemistry were not certified by qRT-PCR, and hence might be somewhat flawed. However, the study was based on the reliability of the Affymetrix chip technology combined with comprehensive bioinformatics analysis immunohistochemistry. Therefore, we believe the results are consistent. Clearly, more work is needed to provide molecular biology index for CRC treatment and prognosis based on differentially expressed genes, as well as in establishing a forecasting model for FOLFOX4 chemotherapy.
In summary, Nkx2–3 and TGFB1I1 expression in the control group are very low, but they are highly expressed in the experimental group. Nkx2–3 and TGFB1I1 may be classified as valuable biomarkers, as these can predict the effects of primary ACC patients who will undergo FOLFOX4. Future studies are required to address relationships between their signaling pathways and drug response.
Materials and methods
Eligibility criteria and patient evaluation
From 2008 to 2010, we gathered samples donated by primary ACC patients who had undergone FOLFOX4 chemotherapy. Thereafter, we carried on the primary tumor R0 resection. Clinical data was collected by the Department of Colorectal and Anal Surgery of the Affiliated Union Hospital of Fujian Medical University; this information was then organized as a database. The inclusion criteria were as follows: (1) histologically confirmed metastasis in CRC; (2) ages 18 to 80 y; (3) an Eastern Cooperative Oncology Group (ECOG) performance status of ≤ 1; and (4) patients have not received radiation, chemotherapy or palliative surgery before study registration. The exclusion criteria were as follows: (1) those who have nerves (around sensory neuropathy with dysfunction) and mental illnesses; (2) those with severe renal insufficiency (creatinine clearance is less than 30 mL/min); (3) pregnant and lactating women; and (4) those with another disease and hence cannot tolerate FOLFOX4 chemotherapy. The elimination criteria were as follows: (1) those with incomplete chemotherapy according to FOLFOX4 schedule standards or if they have terminated it halfway and (2) those with unfinished R0 resection of the primary tumor after chemotherapy. A tissue and patient's data usage protocol was approved by the Ethics Committee of the Affiliated Union Hospital of Fujian Medical University. Informed written consent was obtained from each patient. All patients underwent standard FOLFOX4 regimen chemotherapy in four cycles after signing the chemotherapy agreement; then, they were evaluated in accordance with the response evaluation criteria in solid tumors (RECIST).Citation27 Pretreatment evaluations included a complete medical history and physical examination. A baseline radiographic tumor evaluation was required within 4 weeks before study registration. The interim medical history, physical examination, and the laboratory studies listed herein were repeated before the start of each cycle of therapy. Tumor assessment was performed after the fourth cycle of therapy. Patients underwent R0 resection of the primary tumor after chemotherapy within 28 d.
Sample collection
Each sample was collected immediately following resection. Then, each sample was divided in half, one was fixed in formalin and embedded in paraffin, the other floated in ice-cold PBS and finally stored in liquid nitrogen until future total RNA extraction.
RNA extraction
Tissue homogenization and RNA extraction, together with subsequent microarray, was performed at the premises of CapitalBio Corporation. Tissue homogenization and isolation of total RNA were performed according to manufacturer’s instructions using the Macherey-Nagel NucleoSpin RNA II kit (Macherey-Nagel); this RNA isolation kit has significantly reduced the contamination of both genome DNA and proteins. The RNA, extracted with ribosomal 28S and 18S RNA with a ratio of intensities of 1.0–1.5:1, was used in subsequent microarray assay.
Microarray analysis
In comparing the differentially expressed genes of the two groups, an aliquot (2 μg) of total placental RNA was used to synthesize double-stranded cDNA, which was subsequently transcribed into biotin-tagged cRNA using the MessageAmp II aRNA Amplification Kit (Ambion). The cRNA was then fragmented to produce strands of 35–200 bases in length in accordance with protocols (Affymetrix). The fragmented cRNA was hybridized to the Affymetrix GeneChip Human Genome U133 Plus 2.0 Array containing 47,000 transcripts. Microarray hybridization was performed at 45°C with rotation lasting 16 h using the Affymetrix GeneChip Hybridization Oven 640. The arrays were washed and stained (streptavidin-phycoerythrin) by Affymetrix GeneChip Fluidics Station 450 and then scanned on the Affymetrix GeneChip Scanner 3000 in order to analyze the hybridization data. The obtained scanned images were assessed first by visual inspection and then analyzed by the Affymetrix GeneChip Operating Software (GCOS 1.4). To normalize the different arrays, the dChip software was used in global scaling procedures. In the comparative analysis, a two-class unpaired method obtained from the significance analysis of microarrays (SAM; version 3.02, Stanford University) software was used. Significant differentially expressed genes in both the experimental and control groups were compared.
Bioinformatics analysis
Signal log ratio and the gene symbol of differentially expressed genes were inputted in the MAS3.0 software for both gene ontology and pathway analyses.
Nkx2–3 and TGFB1I1 prediction accuracies analysis
Nkx2–3 and TGFB1I1 signal probe values transformed into signal log ratio using Biweight algorithm and ROC analysis to calculate Area Under The Curve (AUC), cutoff value, sensitivity, specificity, Youden index, judged prediction ability and calculated its own accuracy respectively.
Immunohistochemistry
Immunohistochemical studies of Nkx2–3 and TGFB1I1 were performed on surgical specimens of colorectal cancer, and tissue arrays using the avidin–biotin-peroxidase methods (DakoCytomation) on formalin-fixed, paraffin-embedded tissues. All sections were counterstained with hematoxylin. The primary mouse polyclonal antibody against Nkx2–3 (Santa Cruz Biotechnology, Inc.) and TGFB1I1 (Santa Cruz Biotechnology, Inc.) were used at dilutions of 1:100 and 1:200 respectively.
| Abbreviations: | ||
| BSA | = | body surface area |
| CR | = | complete response |
| MD | = | moderately differentiated |
| PD | = | progressive disease |
| PDi | = | poorly differentiated |
| PR | = | partial response |
| SD | = | stable disease |
| TNM | = | tumor, nodes and metastases |
| WD | = | well-differentiated |
Acknowledgments
The authors are grateful to the insightful work provided the leaders of Affiliated Union Hospital of Fujian Medical University, all of the fellows in Department of Colorectal and Anal Surgery and students that contributed with some of the studies discussed in this article. The source: National Natural Science Foundation of China. Project Approval number: 30872479; Number of grants: 300,000 RMB. Gainer: Pan Chi.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Weitz J, Koch M, Debus J, Höhler T, Galle PR, Büchler MW. Colorectal cancer. Lancet 2005; 365:153 - 65; http://dx.doi.org/10.1016/S0140-6736(05)17706-X; PMID: 15639298
- Arnold CN, Goel A, Blum HE, Boland CR. Molecular pathogenesis of colorectal cancer: implications for molecular diagnosis. Cancer 2005; 104:2035 - 47; http://dx.doi.org/10.1002/cncr.21462; PMID: 16206296
- Lorenz M, Staib-Sebler E, Hochmuth K, Heinrich S, Gog C, Vetter G, et al. Surgical resection of liver metastases of colorectal carcinoma: short and long-term results. Semin Oncol 2000; 27:Suppl 10 112 - 9; PMID: 11049042
- Goldberg RM, Sargent DJ, Morton RF, Fuchs CS, Ramanathan RK, Williamson SK, et al. A randomized controlled trial of fluorouracil plus leucovorin, irinotecan, and oxaliplatin combinations in patients with previously untreated metastatic colorectal cancer. J Clin Oncol 2004; 22:23 - 30; http://dx.doi.org/10.1200/JCO.2004.09.046; PMID: 14665611
- Grothey A, Sargent D. Overall survival of patients with advanced colorectal cancer correlates with availability of fluorouracil, irinotecan, and oxaliplatin regardless of whether doublet or single-agent therapy is used first line. J Clin Oncol 2005; 23:9441 - 2; http://dx.doi.org/10.1200/JCO.2005.04.4792; PMID: 16361649
- Hicklin DJ, Ellis LM. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J Clin Oncol 2005; 23:1011 - 27; http://dx.doi.org/10.1200/JCO.2005.06.081; PMID: 15585754
- Jain RK, Duda DG, Clark JW, Loeffler JS. Lessons from phase III clinical trials on anti-VEGF therapy for cancer. Nat Clin Pract Oncol 2006; 3:24 - 40; http://dx.doi.org/10.1038/ncponc0403; PMID: 16407877
- Saltz LB, Clarke S, Díaz-Rubio E, Scheithauer W, Figer A, Wong R, et al. Bevacizumab in combination with oxaliplatin-based chemotherapy as first-line therapy in metastatic colorectal cancer: a randomized phase III study. J Clin Oncol 2008; 26:2013 - 9; http://dx.doi.org/10.1200/JCO.2007.14.9930; PMID: 18421054
- Meyer UA. Pharmacogenetics and adverse drug reactions. Lancet 2000; 356:1667 - 71; http://dx.doi.org/10.1016/S0140-6736(00)03167-6; PMID: 11089838
- Pullarkat ST, Stoehlmacher J, Ghaderi V, Xiong YP, Ingles SA, Sherrod A, et al. Thymidylate synthase gene polymorphism determines response and toxicity of 5-FU chemotherapy. Pharmacogenomics J 2001; 1:65 - 70; http://dx.doi.org/10.1038/sj.tpj.6500012; PMID: 11913730
- Gustavsson B, Kaiser C, Carlsson G, Wettergren Y, Odin E, Lindskog EB, et al. Molecular determinants of efficacy for 5-FU-based treatments in advanced colorectal cancer: mRNA expression for 18 chemotherapy-related genes. Int J Cancer 2009; 124:1220 - 6; http://dx.doi.org/10.1002/ijc.23852; PMID: 19051292
- Kwon HC, Roh MS, Oh SY, Kim SH, Kim MC, Kim JS, et al. Prognostic value of expression of ERCC1, thymidylate synthase, and glutathione S-transferase P1 for 5-fluorouracil/oxaliplatin chemotherapy in advanced gastric cancer. Ann Oncol 2007; 18:504 - 9; http://dx.doi.org/10.1093/annonc/mdl430; PMID: 17322540
- Selvakumaran M, Pisarcik DA, Bao R, Yeung AT, Hamilton TC. Enhanced cisplatin cytotoxicity by disturbing the nucleotide excision repair pathway in ovarian cancer cell lines. Cancer Res 2003; 63:1311 - 6; PMID: 12649192
- Chang PM, Tzeng CH, Chen PM, Lin JK, Lin TC, Chen WS, et al. ERCC1 codon 118 C-T polymorphism associated with ERCC 1 expression and outcome of FOLFOX-4 treatment in Asian patients with metastatic colorectal carcinoma. J. Cancer Sci 2008; 12:278 - 83
- Park DJ, Zhang W, Stoehlmacher J, Tsao-Wei D, Groshen S, Gil J, et al. ERCC1 gene polymorphism as a predictor for clinical outcome in advanced colorectal cancer patients treated with platinum-based chemotherapy. Clin Adv Hematol Oncol 2003; 1:162 - 6; PMID: 16224397
- Ryu JS, Hong YC, Han HS, Lee JE, Kim S, Park YM, et al. Association between polymorphisms of ERCC1 and XPD and survival in non small-cell lung cancer patients treated wide cisplatin combination chemothempy. Lung Cancer 2004; 1:311 - 6; http://dx.doi.org/10.1016/j.lungcan.2003.11.019
- Su D, Ma S, Liu P, Jiang Z, Lv W, Zhang Y, et al. Genetic polymorphisms and treatment response in advanced non-small cell lung cancer. Lung Cancer 2007; 56:281 - 8; http://dx.doi.org/10.1016/j.lungcan.2006.12.002; PMID: 17222938
- Viguier J, Boige V, Miquel C, Pocard M, Giraudeau B, Sabourin JC, et al. ERCC1 codon 118 polymorphism is a predictive factor for the tumor response to oxaliplatin/5-fluorouracil combination chemotherapy in patients with advanced colorectal cancer. Clin Cancer Res 2005; 11:6212 - 7; http://dx.doi.org/10.1158/1078-0432.CCR-04-2216; PMID: 16144923
- Chen J, Xie F, Chen K, Wang D, Jiang H, Li J, et al. ERCC5 promoter polymorphisms at -763 and +25 predict the response to oxaliplatin-based chemotherapy in patients with advanced colorectal cancer. Cancer Biol Ther 2009; 8:1424 - 30; http://dx.doi.org/10.4161/cbt.8.14.8889; PMID: 19458483
- Suh KW, Kim JH, Kim Y, Kim YB, Lee C, Choi S. Which gene is a dominant predictor of response during FOLFOX chemotherapy for the treatment of metastatic colorectal cancer, the MTHFR or XRCC1 gene?. Ann Surg Oncol 2006; 13:1379 - 85; http://dx.doi.org/10.1245/s10434-006-9112-y; PMID: 17009149
- Stoehlmacher J, Park DJ, Zhang W, Groshen S, Tsao-Wei DD, Yu MC, et al. Association between glutathione S-transferase P1, T1, and M1 genetic polymorphism and survival of patients with metastatic colorectal cancer. J Natl Cancer Inst 2002; 94:936 - 42; http://dx.doi.org/10.1093/jnci/94.12.936; PMID: 12072547
- Braun MS, Richman SD, Quirke P, Daly C, Adlard JW, Elliott F, et al. Predictive biomarkers of chemotherapy efficacy in colorectal cancer: results from the UK MRC FOCUS trial. J Clin Oncol 2008; 26:2690 - 8; http://dx.doi.org/10.1200/JCO.2007.15.5580; PMID: 18509181
- Koopman M, Venderbosch S, Nagtegaal ID, van Krieken JH, Punt CJ. A review on the use of molecular markers of cytotoxic therapy for colorectal cancer, what have we learned?. Eur J Cancer 2009; 45:1935 - 49; http://dx.doi.org/10.1016/j.ejca.2009.04.023; PMID: 19473832
- Hou L, Li Y, Jia YH, Wang B, Xin Y, Ling MY, et al. Molecular mechanism about lymphogenous metastasis of hepatocarcinoma cells in mice. World J Gastroenterol 2001; 7:532 - 6; PMID: 11819823
- Duggan DJ, Bittner M, Chen Y, Meltzer P, Trent JM. Expression Profiling using DNA microarrays. Nature Gene ties Supplement 1999; 21:10-14.
- Wen WH, Bernstein L, Lescallett J, Beazer-Barclay Y, Sullivan-Halley J, White M, et al. Comparison of TP53 mutations identified by oligonucleotide microarray and conventional DNA sequence analysis. Cancer Res 2000; 60:2716 - 22; PMID: 10825146
- Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst 2000; 92:205 - 16; http://dx.doi.org/10.1093/jnci/92.3.205; PMID: 10655437
- Yu W, Lin Z, Pastor DM, Hegarty JP, Chen X, Kelly AA, et al. Genes regulated by Nkx2-3 in sporadic and inflammatory bowel disease-associated colorectal cancer cell lines. Dig Dis Sci 2010; 55:3171 - 80; http://dx.doi.org/10.1007/s10620-010-1138-0; PMID: 20165982
- Wang X, Zbou C, Qiu G, Fan J, Tang H, Peng Z. Screening of new tumor suppressor genes in sporadic colorectal cancer patients. Hepatogastroenterology 2008; 55:2039 - 44; PMID: 19260473
- Cai G, Huang H, Shapiro E, Zhou H, Yeh S, Melamed J, et al. Expression of androgen receptor associated protein 55 (ARA55) in the developing human fetal prostate. J Urol 2005; 173:2190 - 3; http://dx.doi.org/10.1097/01.ju.0000158119.34126.70; PMID: 15879885
- Yund EE, Hill JA, Keller RS. Hic-5 is required for fetal gene expression and cytoskeletal organization of neonatal cardiac myocytes. J Mol Cell Cardiol 2009; 47:520 - 7; http://dx.doi.org/10.1016/j.yjmcc.2009.06.006; PMID: 19540241
- Heitzer MD, DeFranco DB. Hic-5/ARA55, a LIM domain-containing nuclear receptor coactivator expressed in prostate stromal cells. Cancer Res 2006; 66:7326 - 33; http://dx.doi.org/10.1158/0008-5472.CAN-05-2379; PMID: 16849583
- Dabiri G, Tumbarello DA, Turner CE, Van de Water L. Hic-5 promotes the hypertrophic scar myofibroblast phenotype by regulating the TGF-beta1 autocrine loop. J Invest Dermatol 2008; 128:2518 - 25; http://dx.doi.org/10.1038/jid.2008.90; PMID: 18401422
- Dabiri G, Tumbarello DA, Turner CE, Van de Water L. TGF-beta1 slows the growth of pathogenic myofibroblasts through a mechanism requiring the focal adhesion protein, Hic-5. J Invest Dermatol 2008; 128:280 - 91; PMID: 17671518
- Wang H, Song K, Krebs TL, Yang J, Danielpour D. Smad7 is inactivated through a direct physical interaction with the LIM protein Hic-5/ARA55. Oncogene 2008; 27:6791 - 805; http://dx.doi.org/10.1038/onc.2008.291; PMID: 18762808
- Stavit Drori1, Geoffrey D. Girnun, Liqiang Tou, Jeffrey D. Szwaya, Elisabetta Mueller, Xia Kia, et al. Hic-5 regulates an epithelial program mediated by PPARgamma. Genes Dev 2005; 19(3): 361-375.
- Osada M, Ohmori T, Yatomi Y, Satoh K, Hosogaya S, Ozaki Y. Involvement of Hic-5 in platelet activation: integrin alphaIIbbeta3-dependent tyrosine phosphorylation and association with proline-rich tyrosine kinase 2. Biochem J 2001; 355:691 - 7; PMID: 11311131
- Nishiya N, Tachibana K, Shibanuma M, Mashimo J-I. Kiyoshi Nose1. Hic-5-reduced cell spreading on fibronectin: competitive effects between paxillin and Hic-5 through interaction with focal adhesion kinase. Mol Cell Biol 2001; 21:5331 - 45; http://dx.doi.org/10.1128/MCB.21.16.5332-5345.2001
- Shibanuma M, Iwabuchi Y, Nose K. Possible involvement of hic-5, a focal adhesion protein, in the differentiation of C2C12 myoblasts. Cell Struct Funct 2002; 27:21 - 7; http://dx.doi.org/10.1247/csf.27.21; PMID: 11937715
- Gao ZL, Deblis R, Glenn H, Schwartz LM. Differential roles of HIC-5 isoforms in the regulation of cell death and myotube formation during myogenesis. Exp Cell Res 2007; 313:4000 - 14; http://dx.doi.org/10.1016/j.yexcr.2007.05.023; PMID: 17935713
- Shibanuma M, Kim-Kaneyama JR, Sato S, Nose K. A LIM protein, Hic-5, functions as a potential coactivator for Sp1. J Cell Biochem 2004; 91:633 - 45; http://dx.doi.org/10.1002/jcb.10754; PMID: 14755691
- Carneiro AM, Blakely RD. Serotonin-, protein kinase C-, and Hic-5-associated redistribution of the platelet serotonin transporter. J Biol Chem 2006; 281:24769 - 80; http://dx.doi.org/10.1074/jbc.M603877200; PMID: 16803896
- Hetey SE, Lalonde DP, Turner CE. Tyrosine-phosphorylated Hic-5 inhibits epidermal growth factor-induced lamellipodia formation. Exp Cell Res 2005; 311:147 - 56; http://dx.doi.org/10.1016/j.yexcr.2005.08.011; PMID: 16183059
- Shibanuma M, Mashimo J, Kuroki T, Nose K. Characterization of the TGF beta 1-inducible hic-5 gene that encodes a putative novel zinc finger protein and its possible involvement in cellular senescence. J Biol Chem 1994; 269:26767 - 74; PMID: 7929412
- Heitzer MD, DeFranco DB. Hic-5/ARA55: a prostate stroma-specific AR coactivator. Steroids 2007; 72:218 - 20; http://dx.doi.org/10.1016/j.steroids.2006.11.010; PMID: 17166536
- Fujimoto N, Mizokami A, Harada S, Matsumoto T. Different expression of androgen receptor coactivators in human prostate. Urology 2001; 58:289 - 94; http://dx.doi.org/10.1016/S0090-4295(01)01117-7; PMID: 11489729
- Rahman MM, Miyamoto H, Lardy H, Chang C. Inactivation of androgen receptor coregulator ARA55 inhibits androgen receptor activity and agonist effect of antiandrogens in prostate cancer cells. Proc Natl Acad Sci U S A 2003; 100:5124 - 9; http://dx.doi.org/10.1073/pnas.0530097100; PMID: 12700349
- Mestayer1 C, Blanchère M, Jaubert F, Dufour B, Mowszowicz I. Expression of androgen receptor coactivators in normal and cancer prostate tissues and cultured cell lines. Prostate 2003; 56:191 - 200
- Bertucci F, Salas S, Eysteries S, Nasser V, Finetti P, Ginestier C, et al. Gene expression profiling of colon cancer by DNA microarrays and correlation with histoclinical parameters. Oncogene 2004; 23:1377 - 91; http://dx.doi.org/10.1038/sj.onc.1207262; PMID: 14973550
- Cui W, Wang X, Liu YC, Wan YL, Guo HJ, Zhu J. [Expression of HIC-5/ARA55 in colonrectal cancer and its mechanisms of action]. Beijing Da Xue Xue Bao 2006; 38:280 - 3; PMID: 16778972