Abstract
Human promyeloblastic leukemia KG-1a cells exhibit many characteristics similar to leukemia stem cells, which are resistant to chemotherapeutic drugs and hyposensitive to cytotoxic cells. Resveratrol (RES), as a member of plant polyphenols, has gained considerable attention due to its ability to prevent cancer from progressing. In this study, the potential of RES to sensitize KG-1a cells to cytolysis of cytokine-induced killer cells (CIKs) through NKG2D ligands and TNF-related apoptosis-inducing ligand (TRAIL) receptors were investigated. Twenty-five micromolars RES was found to inhibit approximately 50% of KG-1a cell growth and had the least growth-inhibition effect on peripheral blood mononuclear cells (PBMCs) after 24 h. Utilizing cytokines including interleukin-2 (IL-2) and interleukin-15 (IL-15) to activate PBMCs, we obtained substantial CD3+ CD56+ natural killer cell-like T lymphocytes that secreted cytokine interferon-γ (IFN-γ) and expressed NKG2D and TRAIL on their surfaces (i.e., cytokine-induced killer cells, CIKs). RES was shown to render KG-1a cells susceptible to CIK-mediated cytolysis estimated by LDH-release assay. This heightened sensitivity correlated with an increase in cell-surface expression of NKG2D ligands and death receptor 4 (DR4), coupled with a downregulation of cell-surface expression of decoy receptor 1 (DcR1) in KG-1a cells. Blocking NKG2D ligands or TRAIL with monoclonal antibodies could abrogate CIKs-mediated cytolysis. These results demonstrated that increased sensitivity of KG-1a cells, modulated by RES to alloreactive CIKs-mediated cytolysis is a phenomenon attributable to induced expression of NKG2D ligands and activation of TRAIL pathway. Thus, resveratrol combined with alloreactive CIKs merits clinical evaluation as a novel and effective immunotherapy strategy to eliminate residual leukemia stem cells.
Introduction
Leukemia stem cells with self-renewal capacity lead to occurrence of leukemia and may be responsible for leukemia relapse.Citation1 Leukemia stem cells have capacities of drug-resistance and immune-resistance through the impediment of apoptosis pathway and paucity of NKG2D ligands.Citation2,Citation3 It is key to eliminate leukemia stem cells so as to decrease rates of leukemia relapse. A subpopulation of CD34+ CD38- leukemia blasts isolated from acute myeloid leukemia (AML) was the first discovery for leukemia stem cells.Citation4 Human acute myeloid leukemia KG-1a cells possess similar characteristics to AML stem cells, generally serving as a suitable model for studying leukemia stem cells.Citation5
Improved outcome of acute myeloid leukemia after hematopoietic stem cell transplantation (HSCT) highlights the graft-vs.-leukemia (GVL) effect that is the inherent ability to eliminate residual leukemia cells, because immune cells from donor including natural killer cells, natural killer cell-like T cells and mononuclear macrophage cells are all effective in the cytolysis against leukemia cells.Citation6 However, more than 80% of patients with high risk degree of disease were known to relapse after HSCT because residual leukemia stem cells were insensitive to alloreactive immune cell-mediated cytolysis.Citation7 The reversing of the immune-resistance abilities of leukemia stem cells will be pivotal in decreasing relapse after allogeneic HSCT. Utilizing cytokine-induced killer cells (CIKs) may serve as a viable alternative approach for AML-relapsing patients after haplo-identical HSCT.Citation8 However, as to whether leukemia stem cells are sensitive to cytolysis mediated by alloreactive CIKs remains unkown.
Resveratrol (RES), one of naturally occurring dietary compounds, has recently gained considerable attention as cancer-preventive agents and also exhibit anti-tumor activity.Citation9 RES suppresses many cancers involving in arresting G0/G1-S phase cell-cycle progression,Citation10 inhibiting proliferation through the downstream Akt, MAPK and Wnt signaling followed by the activation of caspase signaling,Citation11 and inducing apoptosis via Fas-, P53-, and P21WAF/CIP1-mediated pathways.Citation12 Importantly, RES exhibited inhibitory effects on cancer stem cells and prolonged the survival time of spontaneous tumors in transgenic mice.Citation13,Citation14 On the other hand, RES at low concentrations could enhance cell-mediate immune response through T lymphocytes, B lymphocytes and NK cells, displaying its potential for combining with cancer immunotherapy.Citation15-Citation18 Although leukemia stem cells are resistant to immune cells,Citation20 if RES can reverse leukemia stem cells resistant to alloreactive CIKs-mediated cytolysis, RES would be designed along these lines to eliminate leukemia stem cells so as to reduce leukemia recurrence after HSCT.
Herein, we exploited the potential of resveratrol to sensitize leukemia stem cell-like KG-1a cells to alloreactive CIKs-mediated cytolysis in order to provide a novel and alternative therapeutic regimen. This regimen inclusive of other relevant chemical drugs and immunotherapy could revolutionalise the current approach for the effective removal of leukemia stem cells and ultimate eradication of leukemia.
Results
Resveratrol at low concentration exhibited a growth-inhibitory effect on KG-1a cells, but not on PBMCs
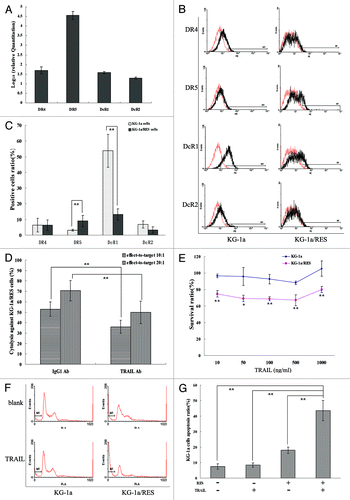
RES inhibited the growth of KG-1a cells in a dose-dependent manner, but 25–100 μM RES had minor inhibitory effects on PBMCs growth. When approximately 50% inhibition concentration (IC50), 25 μM RES was added to KG-1a cells, growths at 24 h and 48 h (both at 25 μM) didn’t either exhibit a notable inhibitory effect on PBMCs (). Under inverted microscope, KG-1a cells were bright, round and smooth with clear edges. In addition there was almost no sign of debris (), while KG-1a cells after treated with 25 μM resveratrol for 24 h (KG-1a/RES cells) had irregular shapes with many granules in the cytoplasm, and there was a little debris (). Under fluorescent microscope, KG-1a cells nucleus stained with DAPI possessed homogenous, regular shaped bodies with complete edges (). On the other hand, the nucleus of KG-1a/RES cells was markedly different with non-homogenous, irregularly shaped, incomplete edges. Some even presented with cauliflower-like, piecemeal necrosis ().
Figure 1. RES had an inhibitory effect on the growth of KG-1a cells in a dose-dependent manner. (A) Effects of RES at various concentration on the growth of KG-1a cells and PBMCs; (B) KG-1a cells were bright, round and smooth with clear edges, in addition there was almost no sign of debris (ori, × 200); (C) KG-1a/RES cells had irregular shapes with many granules in the cytoplasm and there was a little debris (blank arrow) (ori, × 200); (D) KG-1a cells nucleus stained with DAPI possessed homogenous, regular shaped bodies with complete edges (ori, × 1000); (E) KG-1a/RES cells nucleus stained with DAPI were non-homogenous, irregularly shaped, incomplete edges. Some even presented with cauliflower-like (white arrow), piecemeal necrosis (red arrow) (ori, × 1000).
Cytokines IL-2 and IL-15 enhanced CIKs-mediated cytolysis
Peripheral blood mononuclear cells (PBMCs) cultured with cytokines IL-2 and IL-15 for 5 d (also called CIKs) were small, round, bright and suspended. Most were shaped in colony-like forms and possessed clear edges (). Cells smeared and stained with Wright-Giemsa had an assortment of particles (small and thick) in the cytoplasm and highly strung chromatin in the nucleoli, which were characteristic of lymphocytes (). After 5 d, the total number of CIKs was estimated to be (2.67 ± 0.55) times more than that of PBMCs. The IFN-γ concentration in the supernatant of the CIKs medium was (2.69 ± 0.59) times than that of PBMCs medium. CD3+CD56+ and CD3+CD16+ percent of CIKs were respectively (32.70 ± 5.19)% and (5.73 ± 1.20)%, while that of PBMCs were respectively (4.35 ± 1.26)% and (1.48 ± 0.65)%. Expression rate of activating receptors NKG2D on the surface of CIKs was (48.10 ± 7.18)% in contrast to that of the PBMCs (10.58 ± 1.37)%. The extrinsic apoptosis pathway, expression rate of TRAIL on the surface of CIKs was (49.34 ± 9.44)% in contrast to that of PBMCs (9.72 ± 2.11)% (). Assessing the effect-to-target ratio 10:1 or 20:1, CIKs-mediated cytolysis against K562 cells were both higher than that of PBMCs with a statistically significance (p < 0.01) (), suggesting that cytotoxic potential of CIKs stimulated by IL-2 and IL-15 against leukemia cells was enhanced.
Figure 2. Characteristics of CIKs stimulated with IL-2 and IL-15 (** p < 0.01). (A) (a) Cells were small, round, bright, suspended. Most were shaped in colony-like forms and possessed clear edges [(A) ori, × 40; (a) ori, × 200]; (B) Cells smeared and stained with Wright-Giemsa had an assortment of particles (small and thick) in the cytoplasm and highly strung chromatin in the nucleoli (ori, × 1000); (C) Expressions of CD3, CD56, CD16, NKG2D and TRAIL on the surface of PBMCs and CIKs were analyzed by flow cytometry; (D) CIKs-mediated or PBMCs-mediated cytolysis against K562 cells was assayed by LDH release.
![Figure 2. Characteristics of CIKs stimulated with IL-2 and IL-15 (** p < 0.01). (A) (a) Cells were small, round, bright, suspended. Most were shaped in colony-like forms and possessed clear edges [(A) ori, × 40; (a) ori, × 200]; (B) Cells smeared and stained with Wright-Giemsa had an assortment of particles (small and thick) in the cytoplasm and highly strung chromatin in the nucleoli (ori, × 1000); (C) Expressions of CD3, CD56, CD16, NKG2D and TRAIL on the surface of PBMCs and CIKs were analyzed by flow cytometry; (D) CIKs-mediated or PBMCs-mediated cytolysis against K562 cells was assayed by LDH release.](/cms/asset/41dc44fc-250a-4304-8adc-6b66a767dae6/kcbt_a_10919601_f0002.gif)
Resveratrol sensitized KG-1a cells to CIKs-mediated cytolysis
KG-1a cells or KG-1a/RES cells were co-cultured with CIKs. Due to their size, the bigger KG-1a cells were surrounded by the smaller CIKs (). At effect-to-target ratios of 10:1 or 20:1, CIKs-mediated cytolysis against KG-1a/RES cells was higher than against KG-1a cells. Statistical differences existed significantly (p < 0.01) and cytolysis levels increased in an effect-to-target ratio dependent manner. However, there was no statistically significant difference (p > 0.05) in PBMCs-mediated cytolysis against KG-1a cells and KG-1a/RES cells at the varied effect-to-target ratio’s of 10:1 or 20:1. Furthermore, at the same effect-to-target ratio, cytotoxic potential of PBMCs remained distinctly lower than that of CIKs against whatever KG-1a cells or KG-1a/RES cells ().
Figure 3. CIKs mediated cytolysis against KG-1a cells and KG-1a/RES cells (** p < 0.01). (A) CIKs were co-cultured with KG-1a/RES cells and the bigger KG-1a cells (black arrow) were surrounded by the smaller CIKs (white arrow) (ori, × 400); (B) Cytolysis against KG-1a cells and KG-1a/RES cells was mediated by PBMCs or CIKs.
Resveratrol upregulated expressions of ULBP1, ULBP2 and ULBP3 on the surface of KG-1a cells and enhanced CIKs-mediated cytolysis
The mRNA expressions of NKG2D ligands for MICA, MICB, ULBP1, ULBP2 and ULBP3 in KG-1a/RES cells were upregulated to some degree in contrast to that of KG-1a cells (). However, expressions of MICA and MICB on the surface of KG-1a/RES cells were not significantly more than that of KG-1a cells (p > 0.05), and expressions of ULBP1, ULBP2 and ULBP3 on the surface of KG-1a/RES cells were all respectively higher than those of KG-1a cells (p < 0.05) (). When expressions of ULBP1, ULBP2 and ULBP3 on the surface of KG-1a/RES cells were blocked by anti-ULBP1, anti-ULBP2 and anti-ULBP3 antibodies respectively, CIKs-mediated cytolysis against KG-1a/RES cells was decreased (p < 0.05). On the other hand, there was no statistically significant difference in CIKs-mediated cytolysis when blocking cell-surface MICA and MICB in KG-1a/RES cells or not (p > 0.05) ().
Figure 4. Resveratrol upregulated expression of ULBP1, ULBP2 and ULBP3 on the surface of KG-1a cells to enhance CIKs-mediated cytolysis (*p < 0.05, **p < 0.01). (A) mRNA expression of NKG2D ligands in KG-1a/RES cells relative to KG-1a cells by real-time RT-PCR assay; (B and C) Cell-surface expression for NKG2D ligands in KG-1a cells and KG-1a/RES cells by flow cytometry; (D) CIKs-mediated cytolysis against KG-1a/RES cells after blocking expression of MICA/MICB or ULBP1–3 on the surface of KG-1a/RES cells.
Resveratrol upregulated expression of DR4 and decreased expression of DcR1 on the surface of KG-1a cells and enhanced CIKs-mediated cytolysis
The mRNA expressions of TRAIL receptors for DR4, DR5, DcR1 and DcR2 in KG-1a/RES cells were all upregulated to some degree, distinctively DR5 (). On the surface of KG-1a/RES cells, expression of DR5 increased (p < 0.01) and expression of DcR1 decreased (p < 0.01) in contrast to that of KG-1a cells. However, expressions of DR4 and DcR2 on the surface of KG-1a/RES cells were not significantly different from those of KG-1a cells in statistics (p > 0.05) (). When expression of TRAIL on the surface of KG-1a cells was blocked by anti-TRAIL antibody, CIKs-mediated cytolysis against KG-1a/RES cells decreased at the effect-to-target ratio of 10:1 and 20:1 (p < 0.01) (). 10–1000 ng/mL rhTRAIL protein had an inhibitory effect on the proliferation of KG-1a/RES cells in a dose-independent manner, but not on KG-1a cells (). Statistically, there was no difference in the apoptosis rates of KG-1a cells, whether the cells were induced by 10 ng/ml TRAIL or not (p > 0.05). However, there was a marked increase in 10 ng/mL TRAIL-induced apoptosis of KG-1a/RES cells in contrast to that of KG-1a cells (p < 0.01) ().
Figure 5. Resveratrol upregulated expression of DR4 and downregulated expression of DcR1 on the surface of KG-1a cells to enhance CIKs-mediated cytolysis (*p < 0.05, **p < 0.01). A, mRNA expression of TRAIL receptors in KG-1a/RES cells relative to KG-1a cells by real-time RT-PCR assay; (B and C) Cell-surface expression for TRAIL receptors in KG-1a cells and KG-1a/RES cells by flow cytometry; (D) CIKs-mediated cytolysis against KG-1a/RES cells after blocking expression of TRAIL on the surface of CIKs; (E) TRAIL exhibited an inhibitory effect on KG-1a/RES cells, but not on KG-1a cells; (F and G) Apoptosis of KG-1a cells and KG-1a/RES cells was induced with or without 10 ng/ml TRAIL.
Discussion
Human immature promyeloblastic leukemia KG-1a cells express CD34 and lack expression of CD38, which have the ability to self-renew and to initiate human AML in NOD/SCID mice. Like other types of cancer stem cells, KG-1a cells exhibit variable mechanisms designed to resist chemotherapeutic drugs and immune therapy.Citation19 Because a little number of leukemia stem cells isolated from bone marrow, KG-1a cell line served as a substituted model for leukemia stem cells research in our study, similar to Fuchs et al.Citation5
RES has been shown to suppress growth of various leukemia cells such as leukemia U937 cells,Citation20 multiple myeloma cells and T cell leukemia cells,Citation21 acute myeloid leukemia cellsCitation22 and K562 chronic myeloid leukemia cells,Citation23 but not leukemia stem cells. We found that RES had an inhibitory effect on the growth of leukemia stem cell-like KG-1a cells along with changes in cell morphology and nuclear morphology. In other investigations, RES was found to suppress growth of cancer stem-like cells in vitro and vivo,Citation24,Citation25 suggesting that RES will become a novel anti-cancer drug to attack cancer stem cells. IC50 value of RES in KG-1a cells at 24 h was approximately 25 μM lower than those of other leukemia cells, (56.85 ± 2.35) μM of SUP-B15 cells, (65.28 ± 4.89) μM of Kasumi-1 cells and (142.79 ± 2.83) μM of Jurkat cells in the Li et al.’s report,Citation26 suggesting leukemia stem cells were more sensitive to RES, in agreement with some other publications that reported the different sensitivity of leukemia cell lines to RES.Citation27 In our data, 25–100 μM RES has little or no effect on PBMCs, but 25 uM RES can inhibit approximately 50% KG-1a cells proliferation. According to other research, RES could induce a significant enhancement at low concentrations and suppression at high concentrations of both CTL and NK cell cytotoxic activity,Citation15 especially low dose of resveratrol could enhance immune response in vivo,Citation28 therefore 25 μM RES was selected for the following experiments.
Cytokine IL-2 can amplify lymphocytes number in vitro and IL-15 can enhance the cytotoxic potential of lymphocytes by expressing various activated molecule such as NKG2D and TRAIL.Citation8,Citation29,Citation30 Therefore, we gained CIKs by adding IL-2 and IL-15 and found that CIKs could secrete more IFN-γ, express higher levels of NKG2D and TRAIL on the cell-surface, improve cytolytic levels against K562 cells and KG-1a cells relative to PBMCs cytotoxic capabilities. However, in contrast to K562 cells, KG-1a cells were more insensitivate to CIKs-mediated cytolysis, suggesting that leukemia stem cell-like KG-1a cells were resistant to CIKs-mediated cytolysis, as She et al. reported.Citation19 Surprisedly, we found that KG-1a/RES cells rather than KG-1a cells were more susceptible to CIKs-mediated cytolysis, suggesting that RES could enhance sensitivity of KG-1a cells to CIKs-mediated cytolysis. Bae et al. reported that quercetin could enhance susceptibility to NK cell-mediated lysis of K562 leukemia cells,Citation31 but there is still no report about the susceptibility to CIKs-mediated cytolysis of leukemia stem cells.
To exploiting the mechanisms that RES can enhance sensitivity of KG-1a cells to CIKs-mediated cytolysis, we found that RES could significantly upregulate the expressions of the UL16 binding protein (ULBP) family ULBP1–3 on the surface of KG-1a cells, but not the MHC class I chain related proteins (MIC) A and MICB. ULBP1–3 and MICA/MICB can trigger an activating signal in the activated lymphocytes through recognizing Natural Killer group 2D (NKG2D).Citation32 In general, the MICA/MICB are frequently found on epithelial tumors derived from various tissues, while ULBP1–3 have been detected on leukemic cells from patients.Citation33 Studies in vitro had demonstrated that the expression of an NKG2D ligand is sufficient to trigger cytolysis by lymphocytes expressing NKG2D.Citation34 Sodium butyrate, a potent repressor of histone deacetylase (HDAC), could upregulate expression of NKG2D ligand MICA/MICB in HeLa and HepG2 cell lines and increase their susceptibility to NK lysis.Citation35 Low-dose Bortezomib mediated a specific dual antitumor effect in hepatocellular carcinoma by inhibiting tumor cell proliferation and increasing MICA/B protein expression levels on the surface of hepatoma cells, thus stimulating cytotoxicity of cocultured NK cells,Citation36 similarly to our findings that RES could improve CIKs-mediated cytolysis by upregulating the cell-surface expression of NKG2D ligands ULBP1–3 in KG-1a cells because after expressions of cell-surface ULBP1–3 were all blocked, cytolysis of CIKs against KG-1a/RES cells decreased. Additionally, our data showed that RES could significantly upregulate the mRNA of MICA/MICB and ULBP1–3 in the KG-1a cells, different from the protein distributions on cell surface. Relatively little is known about the molecular mechanisms that trigger NKG2D ligands gene expression, chiefly involving in DNA damage response pathway,Citation37 heat shock transcriptional elements,Citation38 nuclear factor (NF)-κB signalingCitation39 and PI3K/AKT/mTOR pathway.Citation40 Further investigations for the molecular mechanism triggering NKG2D ligands by RES are the critical need for offering a better understanding of the role of RES in cancer immunotherapy.
On the other hand, in our study KG-1a cells were found to lack DR4 and DR5, yet express DcR1 and DcR2, consequently resulting in KG-1a cells resistance to TRAIL-induced growth-inhibition and apoptosis, consistent with Zauli et al.’s report that primary leukemias had shown poor susceptibility to TRAIL-mediated cytotoxicity.Citation41 TRAIL, TNF-related apoptosis-inducing ligand, is highly expressed on immune cells after being stimulated by IL-2 or IL-15,Citation42 including two agonistic receptors DR4 and DR5 and the other two antagonistic receptors DcR1 and DcR2.Citation43 Upon numerous investigations it has been determined that RES is capable of sensitizing various cancer cells to TRAIL induced apoptosis through upregulation of DR5 expression,Citation44,Citation45 including neuroblastoma, medulloblastoma, glioblastoma, melanoma, T cell leukemia, as well as pancreatic, breast, and colon carcinomas,Citation46 similarly to our findings that RES not only upregulated the expression of DR5, but also downregulated the expression of DcR1 on the surface of KG-1a cells, as a result to sensitize KG-1a cells to TRAIL-induced growth-inhibition and apoptosis. Moreover, when expression of cell-surface TRAIL on CIKs was blocked, CIKs-mediated cytolysis against KG-1a/RES cells decreased. This maybe was the other possible explanation for the RES ability to improve KG-1a cells susceptibility to CIKs-mediated cytolysis by regulating the expression of TRAIL receptors. Unfortunately, little is known about the molecular mechanisms to regulation of TRAIL receptors by RES, albeit in inconsistence with mRNA expression and protein contribution of TRAIL receptors regulated by RES in our study. Future investigations directed at dissecting the mechanisms controlling the regulation of TRAIL receptors should offer a better understanding of the role of the TRAIL pathway in cancer immunotherapy.
A great deal of research demonstrated the existence of leukemia stem cells, which have become a novel target for cancer therapy.Citation47 However, most currently available therapeutic approaches, including chemotherapy, radiotherapy and allogeneic HSCT all lack the ability to effectively kill leukemia stem cells.Citation48,Citation49 Our preliminary studies in vitro have been very promising and encouraging especially with regards to the use of resveratrol and alloreactive CIKs as combined therapeutic protocols to eliminate leukemia stem cells. Thus, further exploration in vivo should allow us to gain much needed insight to the mechanisms for synergy reaction. Ultimately, clinical trials will provide ample evidence as to whether a combinant treatment utilizing RES and alloreactive CIKs can serve as a realistic and effective therapy regimen that will reduce leukemia recurrence.
Materials and Methods
Reagents
RPMI-1640 (Hyclone, SH30809.01B), fetal bovine serum (Gibco, 10099–141), penicillin and streptomycin (PAA, P11–010), resveratrol (RES) (Sigma, R5010), trypan blue stain (Gibco, 15250–061), DAPI stain (Vysis, 30–804931), Ficoll-Paque (1.077 g/ml) solution (TBD, LST1077), recombinant human IL-2 (rhIL-2) (R&D, 1081-IL), recombinant human IL-15 (rhIL-15) (R&D, 247-IL), anti-human CD3-PECy5 antibody (Biolegend, 300309), anti-human CD16- FITC antibody (Biolegend, 302005), anti-human CD56- PE antibody (Biolegend, 318305), PE mouse anti-human TRAIL antibody (Biolegend, 308205), FITC mouse anti-human NKG2D antibody (ebioscience, 11–5878–71), human IFN-γ Cytokine ELISA kit (Enzo, ADI -900–136), LDH-release assay kit (Promega, G1782), mouse anti-human MICA antibody (R&D, MAB1300), mouse anti-human MICB antibody (R&D, MAB1599), mouse IgG2B isotype control antibody (R&D, MAB004), mouse anti-human ULBP1 antibody (R&D, MAB1380), mouse anti-human ULBP2 antibody (R&D, MAB1298), mouse anti-human ULBP3 antibody (R&D, MAB1517), mouse IgG2A isotype control antibody (R&D, MAB003), PE-conjugated goat anti-mouse IgG1 antibody (Santa Cruz Biotechnology, SC-3738), mouse anti-human TRAIL antibody (R&D, MAB375), mouse nonspecific IgG1 antibody (R&D, MAB002), soluble recombinant human TRAIL (rhTRAIL) (R&D, 375-TL/CF), sodium 3-[1-(phenylaminocarbonyl)-3, 4-tetrazolium]-bis(4-methoxy-6-nitro) benzene sulfonic acid hydrate (XTT) reagent (Kengen, KGA314), Cell Cycle and Apoptosis Analysis Kit (Beyotime, C1052), PE-conjugated anti-human DR4 antibody (Biolegend, 307205), anti-human DR5 antibody (Biolegend, 307405), anti-human DcR1 antibody (Biolegend, 307005), anti-human DcR2 antibody (Biolegend, 309005), PE-conjugated nonspecific mouse IgG1 antibody (Biolegend, 400113), TRIzol Reagent (Invitrogen, 15596–026), cDNA Synthesis kit (Fermentas, K1622), Real time PCR Master Mix (SYBR Green) (DBI, DBI-2043).
Cell line and culture
Human acute myeloid leukemia KG-1a cells were cultured in RPMI-1640 with 10% inactivated fetal bovine serum, penicillin and streptomycin at 37°C under 5% CO2, which were kindly presented by Professor Zengxuan Song (Chinese Academy of Medical Sciences and Peking Union of Medical College, China). Human erythromyeloblastoid leukemia K562 cells were cultured in RPMI-1640 with 10% inactivated fetal bovine serum, penicillin and streptomycin at 37°C under 5% CO2.
Cell viability assay for resveratrol treatment
KG-1a cells in logarithmic phase at 5 × 105 cells/ml were treated with 12.5 μM, 25 μM, 50 μM, 100 μM and 200 μM RES for 24 h and 48 h at 37°C under 5% CO2, respectively. Each assay was performed in triplicate. All cells within each assay were collected and living cells in each assay were counted after 0.4% trypan blue staining. Cell viability ratio% = treated group/untreated group × 100%. Aliquote cells in each assay were stained with DAPI for 5 min at room temperature and observed under fluorescence microscope (Leica).
Cytokine-induced killer cells cultured with IL-2 and IL-15
Peripheral blood samples were obtained from five healthy volunteers by informed consent in accordance with the guidelines of the Ethical Committee of Southern Medical University. Peripheral blood mononuclear cells (PBMCs) were purified and cultured in RPMI-1640 with 10% inactivated fetal bovine serum, 30 ng/mL rhIL-2 and 10 ng/mL rhIL-15, penicillin and streptomycin at 37°C under 5% CO2 for 5 d. Half of medium was changed every other day. After 5 d, aliquote cells were checked by flow cytometry analysis with labeled antibodies: CD3-PECy5, CD16-FITC, CD56-PE, TRAIL-PE and NKG2D-FITC. Others were plated in 96-well (1 × 105 / 100 μL) with fresh medium at 37°C under 5% CO2 for 24 h. The supernatants were collected on the 5th day whereby the IFN-γ concentration in each assay was measured using human IFN-γ Cytokine ELISA kit according to manufacturer’s instructions.
Preparation for KG-1a/RES cells
KG-1a cells in logarithmic phase at 5 × 105 cells/ml were treated with 25 μM RES for 24 h at 37°C under 5% CO2. All of cells were collected and washed with PBS in low speed centrifuge to remove RES and dead cells for twice, and cells were collected and resuspended with fresh RPMI-1640 medium with 10% inactivated fetal bovine serum, which were KG-1a/RES cells.
Analysis of CIKs-mediated cytolysis on KG-1a cells by LDH-release
CIKs-mediated cytolysis against KG-1a cells was measured in an LDH-release assay, as described in LDH-release assay kit protocol. Both of target cells and effect cells were washed with PBS and were resuspended with fresh RPMI-1640 with 5% inactivated fetal bovine serum before co-incubation. Briefly, target cells (1 × 104) were incubated with effector cells at effect/target ratio of 10:1 or 20:1 for 4 h. Target cells included KG-1a cells and KG-1a/RES cells for 24 h. The resulting light absorbance was measured in a microplate reader (Bio-Tek, USA) at 490 nm. All tests were performed in triplicate and the amount of LDH released was calculated according to the following formula: % cytotoxicity = [(T – S - E) / (M - S)] × 100%, where t = experimental release of effect/target co-culture cells, S = spontaneous release of target cells, E = spontaneous release of effect cells, M = total release of target cells.
Analysis of mRNA expression by real-time quantitative-PCR (Q-PCR)
Total RNA was isolated from KG-1a/RES cells or KG-1a cells using TRIzol-Reagent according to the manufacturer’s instructions. The purity and concentration were measured by spectrophotometer. Reverse transcription (RT) reactions were performed using cDNA Synthesis kit according to the manufacturer’s instructions. Q-PCR reactions were performed by means of SYBR green staining and reaction systems were referred to the manufacturer’s instructions. Transcript copy number per sample was calculated by normalization to β-actin expression as the following Equation 2-△△CT. The relative level of gene expression for each sample was calculated: gene expression ratio = log10 (KG-1a/RES normalization value / KG-1a normalization value). Primers sequence as following: MICA, sense 5′-GTGCCCCAGTCCTCCAGAGCTCAG-3′, antisense 5′-GTGGCATCCCTGTGGTCACTCGTC-3′, 635bp; MICB sense 5′-GGCGTCAGGATGGGGTATCTTTGA-3′, antisense 5′-GGCAGGAGCAGTCGTGAGTTTGCC-3′, 690bp; ULBP1, sense 5′-CTGCAGGCCAGGATGTCTTGTGAG-3′, antisense 5′-TGAGGGTGGTGGCCATGGCCTTGG-3′, 319bp; ULBP2, sense 5′-CTGCAGGCAAGGATGTCTTGTGAG-3′, antisense 5′-TGAGGGTGGTGGCTGTGGCCCTGA-3′, 327bp; ULBP3, sense 5′-CTGCAGGTCAGGATGTCTTGTGAG-3′, antisense 5′-TGAGGGTGGTGGCTATGGCTTTGG-3′, 321bp; DR4, sense 5′-CTGAGCAACGCAGACTCGCTGTCCAC-3′, antisense 5′-TCCAAGGACACGGCAGAGCCTGTGCCAT-3′, 505bp; DR5, sense 5′-CCTCATGGACAATGAGATAAAGGTGGCT-3′, antisense 5′-CCAAATCTCAAAGTACGCACAAACGG-3′, 501bp; DcR1, sense 5′-GAAGAATTTGGTGCCAATGCCACT-3′, antisense 5′-CTCTTGGACTTGGCTGGGAGATGT-3′, 611bp; DcR2, sense 5′-CTTTTCCGGCGGCGTTCATGTCCT-3′, antisense, 5′-GTTTCTTCCGTTCTGCTTCCCTTTTA-3′, 460bp; β-actin, sense 5′-TGACGTGGACATCCGCAAAG-3′, antisense 5′-CTGGAAGGTGGACAGCGAGG-3′, 200bp.
Analysis of the expression of NKG2D ligands and TRAIL receptors by FACS
Expression of NKG2D ligands on the surface of KG-1a/RES cells or KG-1a cells were determined by flow cytometry after cells were incubated with MICA antibody, MICB antibody, ULBP1 antibody, ULBP2 antibody and ULBP3 antibody respectively and subsequent were stained with the PE-conjugated anti-mouse IgG1 antibody. Nonspecific mouse IgG2B antibody was used as an isotype control for MICA and MICB antibody and nonspecific mouse IgG2A antibody was used as an isotype control for ULBP1, ULBP2 and ULBP3 antibody. Expression of TRAIL receptors on the surface of KG-1a/RES cells or KG-1a cells were determined by staining with the PE-conjugated DR4 antibody, DR5 antibody, DcR1 antibody and DcR2 antibody respectively. PE-conjugated nonspecific mouse IgG1 antibody was used as an isotype control. Flow cytometric analysis was performed. All experiments were independently repeated 3 times.
Analysis of CIKs-mediated cytolysis on KG-1a/RES cells after blocking the expression of NKG2D ligands
Some KG-1a/RES cells were blocked with MICA and MICB antibodies or IgG2B isotype control antibody as target cells, other KG-1a/RES cells were blocked with ULBP1, ULBP2 and ULBP3 antibodies or IgG2A isotype control antibody as target cells. CIKs-mediated cytolysis against KG-1a/RES cells was measured by an LDH-release assay, as described in LDH-release assay kit protocol.
Analysis of CIKs-mediated cytolysis after blocking the expression of TRAIL on CIKs
CIKs were blocked with TRAIL antibody or IgG1 isotype control antibody as effect cells. CIKs-mediated cytolysis against KG-1a/RES cells was measured by an LDH-release assay, as described in LDH-release assay kit protocol.
Measurement of KG-1a/RES cells viability after TRAIL treatment
KG-1a/RES cells (5 × 103 cells / 100 μL) were incubated with various concentration (10 ng/mL, 50 ng/mL, 100 ng/mL, 500 ng/mL, 1000 ng/mL) soluble recombinant human TRAIL (rhTRAIL) for 24 h. 50 μL XTT reagent was added to each well. The assay is based on the cleavage of the yellow tetrazolium salt XTT to form an orange formazan dye by metabolically active cells. The absorbance OD of the formazan product, reflecting cell viability, was measured at 450 nm. Each assay was performed in triplicate. Cell viability ratio % = (treated group OD - medium group OD) / (untreated group OD - medium group OD) × 100%.
Measurement of apoptosis after KG-1a/RES cells were treated with TRAIL
For analysis of cell apoptosis, KG-1a/RES cells at a density of 5 × 105 cells/mL were cultured with 10 ng/mL rhTRAIL for 24 h, then added with 70% cooled alcohol for one night at 4°C. According to the manufacturer’s instructions, cells were stained with Propidium Iodide stain including RNase A for 30 min at 37°C and cells apoptosis was assessed by flow cytometry (BD, USA).
Statistical analysis
The data was analyzed with SPSS 13.0 software package and represented as the mean ± standard deviation ( ± s). Comparisons of the means among groups were performed using one-way analysis of variance (ANOVA). If variances were not homogenous among groups, correctional method of F test by Welch was performed. If variances were homogenous among groups, the LSD method was applied for multiple comparisons, otherwise Dunnett’s T3 method was utilized. Paired comparison was performed using compared samples t-test. p < 0.05 was considered to be statistically significant.
Acknowledgments
The authors thank Amewoke Adamaley at Southern Medical University as undergraduate from USA for revising the manuscript. KG-1a cells were kindly provided by Professor Zengxuan Song (Chinese Academy of Medical Sciences and Peking Union of Medical College, China). This work was supported by the National Natural Science Foundation of China (Grant No. 30973454).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Konopleva MY, Jordan CT. Leukemia stem cells and microenvironment: biology and therapeutic targeting. J Clin Oncol 2011; 29:591 - 9; http://dx.doi.org/10.1200/JCO.2010.31.0904; PMID: 21220598
- Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science 2011; 331:1565 - 70; http://dx.doi.org/10.1126/science.1203486; PMID: 21436444
- O’Sullivan T, Dunn GP, Lacoursiere DY, Schreiber RD, Bui JD. Cancer immunoediting of the NK group 2D ligand H60a. J Immunol 2011; 187:3538 - 45; http://dx.doi.org/10.4049/jimmunol.1100413; PMID: 21876033
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 1997; 3:730 - 7; http://dx.doi.org/10.1038/nm0797-730; PMID: 9212098
- Fuchs D, Daniel V, Sadeghi M, Opelz G, Naujokat C. Salinomycin overcomes ABC transporter-mediated multidrug and apoptosis resistance in human leukemia stem cell-like KG-1a cells. Biochem Biophys Res Commun 2010; 394:1098 - 104; http://dx.doi.org/10.1016/j.bbrc.2010.03.138; PMID: 20350531
- Ringdén O, Karlsson H, Olsson R, Omazic B, Uhlin M. The allogeneic graft-versus-cancer effect. Br J Haematol 2009; 147:614 - 33; http://dx.doi.org/10.1111/j.1365-2141.2009.07886.x; PMID: 19735262
- Linn YC, Niam M, Chu S, Choong A, Yong HX, Heng KK, et al. The anti-tumour activity of allogeneic cytokine-induced killer cells in patients who relapse after allogeneic transplant for haematological malignancies. Bone Marrow Transplant 2011; http://dx.doi.org/10.1038/bmt.2011.202; PMID: 21986635
- Rettinger E, Kuçi S, Naumann I, Becker P, Kreyenberg H, Anzaghe M, et al. The cytotoxic potential of interleukin-15-stimulated cytokine-induced killer cells against leukemia cells. Cytotherapy 2012; 14:91 - 103; http://dx.doi.org/10.3109/14653249.2011.613931; PMID: 21973023
- Tan AC, Konczak I, Sze DM, Ramzan I. Molecular pathways for cancer chemoprevention by dietary phytochemicals. Nutr Cancer 2011; 63:495 - 505; http://dx.doi.org/10.1080/01635581.2011.538953; PMID: 21500099
- Vanamala J, Reddivari L, Radhakrishnan S, Tarver C. Resveratrol suppresses IGF-1 induced human colon cancer cell proliferation and elevates apoptosis via suppression of IGF-1R/Wnt and activation of p53 signaling pathways. BMC Cancer 2010; 10:238; http://dx.doi.org/10.1186/1471-2407-10-238; PMID: 20504360
- He X, Wang Y, Zhu J, Orloff M, Eng C. Resveratrol enhances the anti-tumor activity of the mTOR inhibitor rapamycin in multiple breast cancer cell lines mainly by suppressing rapamycin-induced AKT signaling. Cancer Lett 2011; 301:168 - 76; http://dx.doi.org/10.1016/j.canlet.2010.11.012; PMID: 21168265
- Roccaro AM, Leleu X, Sacco A, Moreau AS, Hatjiharissi E, Jia X, et al. Resveratrol exerts antiproliferative activity and induces apoptosis in Waldenström’s macroglobulinemia. Clin Cancer Res 2008; 14:1849 - 58; http://dx.doi.org/10.1158/1078-0432.CCR-07-1750; PMID: 18347188
- Gullett NP, Ruhul Amin AR, Bayraktar S, Pezzuto JM, Shin DM, Khuri FR, et al. Cancer prevention with natural compounds. Semin Oncol 2010; 37:258 - 81; http://dx.doi.org/10.1053/j.seminoncol.2010.06.014; PMID: 20709209
- Provinciali M, Re F, Donnini A, Orlando F, Bartozzi B, Di Stasio G, et al. Effect of resveratrol on the development of spontaneous mammary tumors in HER-2/neu transgenic mice. Int J Cancer 2005; 115:36 - 45; http://dx.doi.org/10.1002/ijc.20874; PMID: 15688416
- Falchetti R, Fuggetta MP, Lanzilli G, Tricarico M, Ravagnan G. Effects of resveratrol on human immune cell function. Life Sci 2001; 70:81 - 96; http://dx.doi.org/10.1016/S0024-3205(01)01367-4; PMID: 11764009
- Zunino SJ, Storms DH. Resveratrol alters proliferative responses and apoptosis in human activated B lymphocytes in vitro. J Nutr 2009; 139:1603 - 8; http://dx.doi.org/10.3945/jn.109.105064; PMID: 19549761
- Lu CC, Chen JK. Resveratrol enhances perforin expression and NK cell cytotoxicity through NKG2D-dependent pathways. J Cell Physiol 2010; 223:343 - 51; PMID: 20082299
- Soto BL, Hank JA, Darjatmoko SR, Polans AS, Yanke EM, Rakhmilevich AL, et al. Anti-tumor and immunomodulatory activity of resveratrol in vitro and its potential for combining with cancer immunotherapy. Int Immunopharmacol 2011; 11:1877 - 86; http://dx.doi.org/10.1016/j.intimp.2011.07.019; PMID: 21854876
- She M, Niu X, Chen X, Li J, Zhou M, He Y, et al. Resistance of leukemic stem-like cells in AML cell line KG1a to natural killer cell-mediated cytotoxicity. Cancer Lett 2011; http://dx.doi.org/10.1016/j.canlet.2011.12.017; PMID: 22198207
- Tang Z, Liu XY, Zou P. Resveratrol inhibits the secretion of vascular endothelial growth factor and subsequent proliferation in human leukemia U937 cells. J Huazhong Univ Sci Technolog Med Sci 2007; 27:508 - 12; http://dx.doi.org/10.1007/s11596-007-0508-0; PMID: 18060622
- Reis-Sobreiro M, Gajate C, Mollinedo F. Involvement of mitochondria and recruitment of Fas/CD95 signaling in lipid rafts in resveratrol-mediated antimyeloma and antileukemia actions. Oncogene 2009; 28:3221 - 34; http://dx.doi.org/10.1038/onc.2009.183; PMID: 19561642
- Cakir Z, Saydam G, Sahin F, Baran Y. The roles of bioactive sphingolipids in resveratrol-induced apoptosis in HL60: acute myeloid leukemia cells. J Cancer Res Clin Oncol 2011; 137:279 - 86; http://dx.doi.org/10.1007/s00432-010-0884-x; PMID: 20401667
- Kartal M, Saydam G, Sahin F, Baran Y. Resveratrol triggers apoptosis through regulating ceramide metabolizing genes in human K562 chronic myeloid leukemia cells. Nutr Cancer 2011; 63:637 - 44; http://dx.doi.org/10.1080/01635581.2011.538485; PMID: 21500096
- Shankar S, Nall D, Tang SN, Meeker D, Passarini J, Sharma J, et al. Resveratrol inhibits pancreatic cancer stem cell characteristics in human and KrasG12D transgenic mice by inhibiting pluripotency maintaining factors and epithelial-mesenchymal transition. PLoS One 2011; 6:e16530; http://dx.doi.org/10.1371/journal.pone.0016530; PMID: 21304978
- Pandey PR, Okuda H, Watabe M, Pai SK, Liu W, Kobayashi A, et al. Resveratrol suppresses growth of cancer stem-like cells by inhibiting fatty acid synthase. Breast Cancer Res Treat 2011; 130:387 - 98; http://dx.doi.org/10.1007/s10549-010-1300-6; PMID: 21188630
- Li T, Wang W, Chen H, Li T, Ye L. Evaluation of anti-leukemia effect of resveratrol by modulating STAT3 signaling. Int Immunopharmacol 2010; 10:18 - 25; http://dx.doi.org/10.1016/j.intimp.2009.09.009; PMID: 19796711
- Luzi C, Brisdelli F, Cinque B, Cifone G, Bozzi A. Differential sensitivity to resveratrol-induced apoptosis of human chronic myeloid (K562) and acute lymphoblastic (HSB-2) leukemia cells. Biochem Pharmacol 2004; 68:2019 - 30; http://dx.doi.org/10.1016/j.bcp.2004.07.002; PMID: 15476673
- Feng YH, Zhou WL, Wu QL, Li XY, Zhao WM, Zou JP. Low dose of resveratrol enhanced immune response of mice. Acta Pharmacol Sin 2002; 23:893 - 7; PMID: 12370094
- Buddingh EP, Schilham MW, Ruslan SE, Berghuis D, Szuhai K, Suurmond J, et al. Chemotherapy-resistant osteosarcoma is highly susceptible to IL-15-activated allogeneic and autologous NK cells. Cancer Immunol Immunother 2011; 60:575 - 86; http://dx.doi.org/10.1007/s00262-010-0965-3; PMID: 21240486
- Correia MP, Costa AV, Uhrberg M, Cardoso EM, Arosa FA. IL-15 induces CD8+ T cells to acquire functional NK receptors capable of modulating cytotoxicity and cytokine secretion. Immunobiology 2011; 216:604 - 12; http://dx.doi.org/10.1016/j.imbio.2010.09.012; PMID: 20956026
- Bae JH, Kim JY, Kim MJ, Chang SH, Park YS, Son CH, et al. Quercetin enhances susceptibility to NK cell-mediated lysis of tumor cells through induction of NKG2D ligands and suppression of HSP70. J Immunother 2010; 33:391 - 401; http://dx.doi.org/10.1097/CJI.0b013e3181d32f22; PMID: 20386467
- González S, López-Soto A, Suarez-Alvarez B, López-Vázquez A, López-Larrea C. NKG2D ligands: key targets of the immune response. Trends Immunol 2008; 29:397 - 403; http://dx.doi.org/10.1016/j.it.2008.04.007; PMID: 18602338
- Ogbomo H, Michaelis M, Klassert D, Doerr HW, Cinatl J Jr.. Resistance to cytarabine induces the up-regulation of NKG2D ligands and enhances natural killer cell lysis of leukemic cells. Neoplasia 2008; 10:1402 - 10; PMID: 19048119
- Waldhauer I, Steinle A. NK cells and cancer immunosurveillance. Oncogene 2008; 27:5932 - 43; http://dx.doi.org/10.1038/onc.2008.267; PMID: 18836474
- Zhang C, Wang Y, Zhou Z, Zhang J, Tian Z. Sodium butyrate upregulates expression of NKG2D ligand MICA/B in HeLa and HepG2 cell lines and increases their susceptibility to NK lysis. Cancer Immunol Immunother 2009; 58:1275 - 85; http://dx.doi.org/10.1007/s00262-008-0645-8; PMID: 19139882
- Armeanu S, Krusch M, Baltz KM, Weiss TS, Smirnow I, Steinle A, et al. Direct and natural killer cell-mediated antitumor effects of low-dose bortezomib in hepatocellular carcinoma. Clin Cancer Res 2008; 14:3520 - 8; http://dx.doi.org/10.1158/1078-0432.CCR-07-4744; PMID: 18519785
- Tang KF, Ren H, Cao J, Zeng GL, Xie J, Chen M, et al. Decreased Dicer expression elicits DNA damage and up-regulation of MICA and MICB. J Cell Biol 2008; 182:233 - 9; http://dx.doi.org/10.1083/jcb.200801169; PMID: 18644891
- Qiao Y, Liu B, Li Z. Activation of NK cells by extracellular heat shock protein 70 through induction of NKG2D ligands on dendritic cells. Cancer Immun 2008; 8:12; PMID: 18613644
- Molinero LL, Fuertes MB, Girart MV, Fainboim L, Rabinovich GA, Costas MA, et al. NF-kappa B regulates expression of the MHC class I-related chain A gene in activated T lymphocytes. J Immunol 2004; 173:5583 - 90; PMID: 15494508
- Tokuyama M, Lorin C, Delebecque F, Jung H, Raulet DH, Coscoy L. Expression of the RAE-1 family of stimulatory NK-cell ligands requires activation of the PI3K pathway during viral infection and transformation. PLoS Pathog 2011; 7:e1002265; http://dx.doi.org/10.1371/journal.ppat.1002265; PMID: 21966273
- Zauli G, Bosco R, Secchiero P. Molecular targets for selective killing of TRAIL-resistant leukemic cells. Expert Opin Ther Targets 2011; 15:931 - 42; http://dx.doi.org/10.1517/14728222.2011.580278; PMID: 21548717
- Smyth MJ, Cretney E, Kelly JM, Westwood JA, Street SE, Yagita H, et al. Activation of NK cell cytotoxicity. Mol Immunol 2005; 42:501 - 10; http://dx.doi.org/10.1016/j.molimm.2004.07.034; PMID: 15607806
- Sayers TJ. Targeting the extrinsic apoptosis signaling pathway for cancer therapy. Cancer Immunol Immunother 2011; 60:1173 - 80; http://dx.doi.org/10.1007/s00262-011-1008-4; PMID: 21626033
- Shankar S, Chen Q, Siddiqui I, Sarva K, Srivastava RK. Sensitization of TRAIL-resistant LNCaP cells by resveratrol (3, 4′, 5 tri-hydroxystilbene): molecular mechanisms and therapeutic potential. J Mol Signal 2007; 2:7; http://dx.doi.org/10.1186/1750-2187-2-7; PMID: 17718901
- Rushworth SA, Micheau O. Molecular crosstalk between TRAIL and natural antioxidants in the treatment of cancer. Br J Pharmacol 2009; 157:1186 - 8; http://dx.doi.org/10.1111/j.1476-5381.2009.00266.x; PMID: 19664138
- Jacquemin G, Shirley S, Micheau O. Combining naturally occurring polyphenols with TNF-related apoptosis-inducing ligand: a promising approach to kill resistant cancer cells?. Cell Mol Life Sci 2010; 67:3115 - 30; http://dx.doi.org/10.1007/s00018-010-0407-6; PMID: 20508968
- Naka K, Hoshii T, Tadokoro Y, Hirao A. Molecular pathology of tumor-initiating cells: lessons from Philadelphia chromosome-positive leukemia. Pathol Int 2011; 61:501 - 8; http://dx.doi.org/10.1111/j.1440-1827.2011.02688.x; PMID: 21884299
- Li Y, Wicha MS, Schwartz SJ, Sun D. Implications of cancer stem cell theory for cancer chemoprevention by natural dietary compounds. J Nutr Biochem 2011; 22:799 - 806; http://dx.doi.org/10.1016/j.jnutbio.2010.11.001; PMID: 21295962
- Buss EC, Ho AD. Leukemia stem cells. Int J Cancer 2011; 129:2328 - 36; http://dx.doi.org/10.1002/ijc.26318; PMID: 21796620