Abstract
The aberrant expression of microRNAs (miRNAs) has been found in various types of cancer. The present study found miR-20a to be significantly upregulated in prostate cancer compared with normal prostate tissues. The proliferation and colony formation assays revealed that the downregulation of miR-20a by miR-20a inhibitor suppresses the proliferation of MDA-PCa-2b cells in vitro and also inhibits tumor growth in vivo. Furthermore, a gap junction protein, α 1 (CX43), was identified as a direct target gene of miR-20a. The upregulation of CX43 was detected in MDA-PCa-2b cells after treatment with miR-20a inhibitor both in vitro and in vivo. In conclusion, the findings show that miR-20a significantly contributes to the progression of prostate cancer by targeting CX43.
Keywords: :
Introduction
MicroRNAs (miRNAs) are a class of small RNA molecules, about 21 nucleotides in length that regulate mRNA stability and translation in a post-transcriptional level. They function as negative regulators by binding to the 3′UTR of target mRNAs through base pairing, which resulting target mRNAs cleavage or translation inhibition.Citation1 A group of miRNAs have been reported to exhibit typical expression profiles and play critical roles in cell proliferation, differentiation and apoptosis. And the dysregulation of miRNAs and their targets will result to various diseases including cancers.Citation2,Citation3
Prostate cancer is the most common cancer in males. The incidence of prostate cancer is highest in Western countries and is among the leading causes of cancer-related deaths in men.Citation4 Several treatments are currently available for prostate cancer, including radiotherapy, hormonal therapy and chemotherapy.Citation5,Citation6 However, these methods are still limited in terms of improving the life quality of patients, as well as in the increasing cost of treatment.Citation7 Therefore, the development of innovative and reliable diagnostic or prognostic biomarkers is important in the treatment of prostate cancer.
Recently, miRNAs have been shown to participate in the development, prognosis and chemo-resistance of prostate cancer. MiR-1 has been reported to be a tumor suppressor and can be used as a prognostic marker of prostate cancer.Citation8 MiR-21 is involved in mediating chemo-resistance to docetaxel.Citation9 Furthermore, miR-100,Citation10 miR-205,Citation11 miR-221 and miR-222 are the regulators of prostate cancinogenesis.Citation12 However, reports about miR-20a in prostate cancer is still limited.
Connexins are widely-expressed transmembrane proteins that are known to form gap junction channels. They play critical roles in cell growth,Citation13 tissue regeneration and carcinogenesis.Citation14 As a member of gap junction channels, CX43 also was found to be disregulated in a multiple type of cancer and participates in tumor progression, including gastric,Citation15,Citation16 cervical,Citation17 colorectalCitation18 and prostate cancers.Citation19,Citation20 In breast cancer, the downregulation of CX43 significantly promotes tumor progression,Citation21 while the expression of CX43 could suppress the cancer phenotype of human mammary carcinoma cells and restore differentiation potential.Citation22 All the above these suggested that CX43 can function as a tumor suppressor.
In the present study, miR-20a was found to be overexpressed in prostate cancer. The downregulation of miR-20a in vitro inhibited the proliferation of prostate cancer cell by targeting CX43, α 1. Furthermore, inhibition of miR-20a could suppress the growth of prostate cancer cells in vivo. These results highlighting the role of miR-20a in prostate cancer will be helpful in achieving a deeper understanding of the anti-cancer mechanism ofmiRNA.
Results
miR-20a is highly expressed and inversely correlated with CX43 expression in prostate cancer tissues
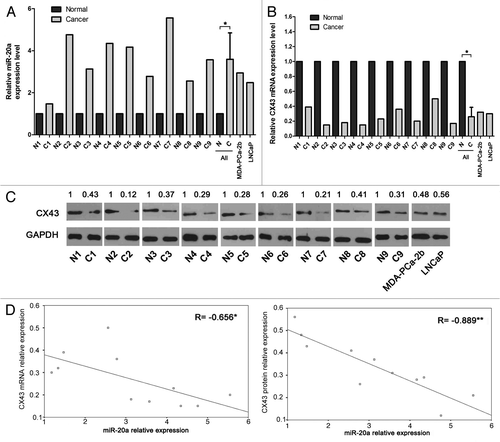
To analyze the expression of miR-20a and CX43 in normal prostate tissues and those with prostate cancer, real-time qRT-PCR was employed to quantify the level of miR-20a and CX43 in nine pairs of matched human prostate cancer tissues/surrounding tissues and two prostate cancer cell lines, MDA-PCa-2b and LNCaP. As shown in , the expression of miR-20a is significantly higher, whereas the expression of CX43 mRNA is lower in prostate cancer specimens and cell lines than that in normal prostate tissues. Furthermore, we investigated the protein expression of CX43 in prostate specimens and cell lines by western blot analysis. As shown in , the protein expression displayed the similar results with the mRNA level, showing the good correlation between the mRNA and protein level of CX43. Moreover, we found the expression of miR-20a was inversely correlated with mRNA and protein expression of CX43 in prostate tissues ().
Figure 1. MiR-20a is highly expressed and inversely correlated with CX43 in prostate cancer tissues and cell lines. (A-C) The expression level of miR-20a (A), CX43 mRNA (B) and CX43 protein level (C) were measured in nine pairs of prostate tissues, MDA-PCa-2b and LNCaP cell lines by real-time qRT-PCR and western blot with U6 snRNA, β-actin or GAPDH as the control. The expression of miR-20a and CX43 mRNA were determined by triplicate measurements for each sample and were quantified using the 2-ΔΔCt method. Representative western photos were shown. The expression level in normal tissues is set to 1. Student′s t test were performed to analyze the significance of differences between sample means. *, p < 0.05. (D-E) The correlation of CX43 mRNA and miR-20a levels (D, r = -0.656) and CX43 protein and miR-20a levels (E, r = -0.889) are shown. *, p < 0.05, **, p < 0.01.
Inhibition of prostate cancer cell viability and proliferation by the downregulation of miR-20a
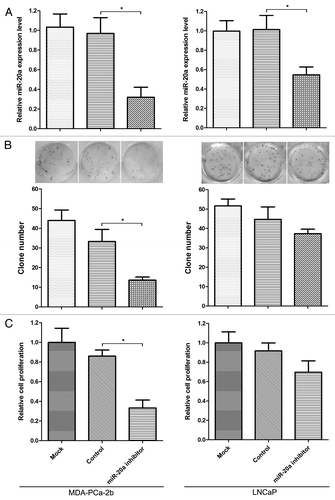
An aberrant expression of miR-20a suggested that miR-20a may regulate metabolism and signal transduction in prostate cancer development. Therefore, we further investigate the effect of altered miR-20a on cell viability and proliferation through CKK-8 and cell colony formation assays. As shown in , miR-20a inhibitor can significantly inhibit the expression level of mature miR-20a in MDA-PCa-2b and LNCaP cells. Results of CKK-8 and colony formation assays showed that miR-20a inhibitor can significantly reduce cell viability and proliferation in MDA-PCa-2b cells in comparison to control group (), whereas there is no stastically difference in cellular proliferation of LNCaP cells between the miR-20a inhibitor transfection group and the control group ().
Figure 2. Downregulation of miR-20a significantly inhibits the proliferation of the MDA-PCa-2b cell line. (A) The expression level of miR-20a following transfection of miR-20a inhibitor in MDA-PCa-2b and LNCaP cells. The control vector and mock transfection were used as controls. 48 h after transfection, cell were harvested and the expression of miR-20a was examined by real-time qRT-PCR. Transfection of miR-20a inhibitor decreased the expression of miR-20a in MDA-PCa-2b and LNCaP cell lines significantly. (B) MDA-PCa-2b and LNCaP cells were transfected with miR-20a inhibitor, and control vector and mock transfection were used as controls. Twenty-four hours after transfection, cells were seeded into 96-well plate for CCK-8 assays (B) or into 12-well plate for cell colony assays (C). The colony formation and CCK-8 assay showed that the knockdown of miR-20a in MDA-PCa-2b cells inhibited cell proliferation more significantly than that in LNCaP cells. *, p < 0.05.
miR-20a directly inhibits CX43 expression
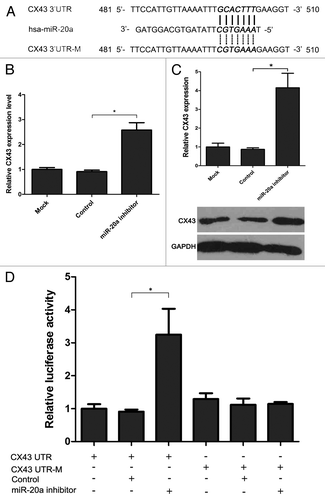
On the basis of the inverse correlation between miR-20a and CX43 expression, we hypothesized that miR-20a may exert its function through inhibiting CX43 expression. To further support the hypothesis, TargetScan database (Version 5.1) was used to predict the target gene of miR-20a. As shown in , the 3′UTR of CX43 was found to contain a predicted miR-20a binding site (481–510 of the 3′UTR, ). To test whether miR-20a could repress the expression of CX43, we investigated the endogenous CX43 protein expression in MDA-PCa-2b cells with downregulated miR-20a by western blot analysis. As a result, inhibition of miR-20a resulted in about 4 fold increase in CX43 protein level (). Furthermore, we constructed luciferase reporter vectors carrying the 3′UTR or mutant 3′UTR of CX43 (CX43 UTR or CX43 UTR-M) and performed the luciferase reporter assay to verify if miR-20a can directly bind the 3′UTR of CX43. As shown in , miR-20a inhbitor can increase luciferase activity with wild-type CX43 3′UTR. However, miR-20a inhbitor does not affect the expression of luciferase with mutated miR-20a binding elements (). From the above results, CX43 is one of the target genes of miR-20a and is repressed by miR-20a.
Figure 3. MiR-20a inhibits the expression of CX43 by binding to 3′-UTR. (A) Sequence alignment of miR-20a with the wild-type and mutant 3′UTR of CX43. (B-C) The expression of CX43 in MDA-PCa-2b cells transfected by miR-20a inhibitor was tested by real-time qRT-PCR detection (B) and western blot (C) and the relative expression was shown. Normalization was performed with β-actin and GAPDH. The expression of CX43 in mock transfection group was set as 1. (D) CX43 UTR or CX43 UTR-M was transfected alone or cotransfected with miR-20a inhibitor or control vectors into MDA-PCa-2b cells. Forty-eight hours later, cells were lyzed and analyzed. The intensity of firefly luciferase in cells cotransfected with CX43 UTR and miR-20a inhibitor was increased after 48 h (p < 0.05). However, miR-20a inhibitor had no effect on the intensity of firefly luciferase when cells were transfected with the 3′UTR mutant vector (p > 0.05). Relative fluorescence intensity of the value with transfection of CX43 UTR alone was set as 1. Values represent means ± SEM from three independent experiments. One-way ANOVA test were performed to analyze the significance of differences. *, p < 0.05.
Restoration of CX43 in MDA-PCa-2b cells can significantly inhibit cell proliferation
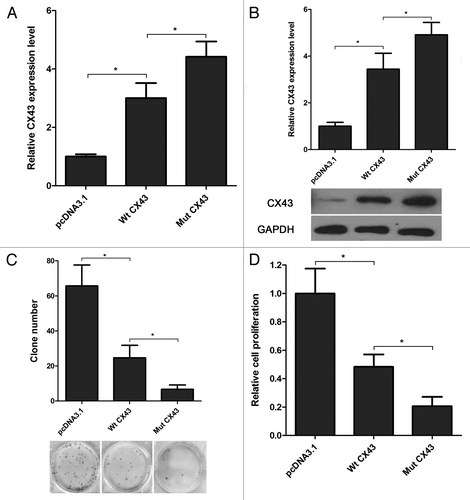
We have proofed that miR-20a is upregulated, whereas CX43 is downregulated in MDA-PCa-2b cells () and miR-20a can inbhit the expression of CX43 directly. So we speculate that miR-20a regulate prostate cancer cell progression through CX43. To support this hypothesis, we construced two different CX43 CDS expression plasmid with wild-type 3′UTR or the mutated 3′UTR that lacked the miR-20a binding site (Wt CX43 or Mut CX43) to reverse the negative effects on CX43 protein level caused by miR-20a in MDA-PCa-2b cells. As shown in , Wt CX43 and Mut CX43 can both enhance the protein level of CX43 in comparision to it control group, pcDNA3.1 transfected cells. And CX43 with mutated 3′UTR (Mut CX43) transfection can increase the CX43 protein more significantly than Wt CX43 transfection because the Mut CX43 is out of control by endogenous miR-20a. Furthermore, we evaluated the effects of Wt CX43 and Mut CX43 on cell viability and proliferation by CKK-8 and cell colony formation asssays. The results showed that Mut CX43 can affected cell viability and proliferation more significantly than Wt CX43 in MDA-PCa-2b cells ().
Figure 4. Overexpression of CX43 reverses miR-20a function in MDA-PCa-2b cells. (A-B) Transfection of CX43 expression plasmid increased the mRNA (A) and protein (B) expression of CX43 in MDA-PCa-2b cells. CX43 expression plasmid with mutant 3′UTR (Mut CX43) can significantly increase the mRNA and protein level of CX43 than Wt CX43. (C-D) The effects of Wt CX43 and Mut CX43 on cell viability and proliferation were evaluated by CKK-8 and cell colony formation assays. Overexpression of CX43 with mutated miR-20a target site in MDA-PCa-2b cells more significantly decreased the proliferation compared with CX43 with wild-type miR-20a target site. One-way ANOVA test were performed to analyze the significance of differences from three independent experiments. *, p < 0.05.
Contribution of miR-20a to the growth of prostate cancer in vivo
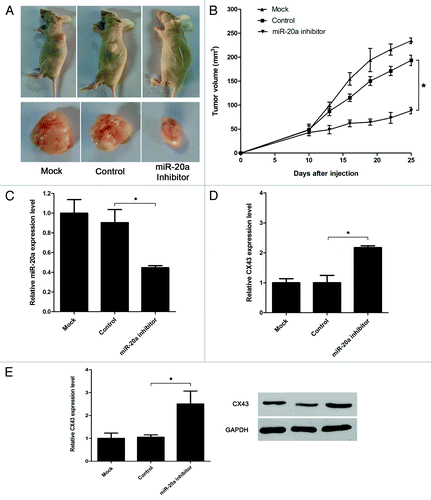
To further investigate the role of miR-20a in vivo, miR-20a inhibitor and its control vector were transfected, and 24 h later, cells were trypsinized and injected into mice subcutaneously. At 25 d post-implantation, the mice were sacrificed, and the tumors were removed. As shown in , the tumor volume of the miR-20a inhibitory and CX43 overexpression group were significantly smaller than that in the control groups. Meanwhile, we detect the expression of miR-20a and CX43 in the tumor tissues by real-time qRT-PCR and western blot. As shown in , the exression of miR-20a was significantly decreased, whereas the expression of CX43 was obviously increased when miR-20a inhibitor was transfected.
Figure 5. In vivo anti-prostate cancer effect of miR-20a. (A) MDA-PCa-2b cells transfected with miR-20a inhibitor or control vector were injected subcutaneously in the nude mice. n = 8 mice per group. Mice were sacrificed 25 d later for evaluation. Gross morphology of tumor was shown. (B) miR-20a inhibitor significantly decreased the volume of tumors compared with the control group. (C-D) Analysis of miR-20a expression level (C) and CX43 mRNA (D) and protein (E) expression level in solid tumors by real-time qRT-PCR and western blot. U6 snRNA, β-actin or GAPDH as the control. One-way ANOVA test were used and mock transfecion group were set as 1. *, p < 0.05.
Discussion
miRNAs are closely related to prostate cancer. The miR-34c, miR-205, miR-34a and miR-15a-miR-16–1 clusters were downregulated and acted as tumor suppressor genes in prostate cancer.Citation23-Citation26Otherwise, miR-21, miR-let7c, miR-100 and miR-218 were found to be upregulated in prostate cancer.Citation27,Citation28 In the present study, miR-20a levels were compared between normal tissues and prostate cancer tissues, and the results indicated that the level of miR-20a was higher in the prostate cancer tissues. Early studies have been shown miR-20a to be highly expressed in prostate cancer, ovarian cancer and osteosarcoma.Citation29-Citation31 The overexpression of miR-20a contributed to chemotherapeutic resistance in colorectal adenocarcinoma cells.Citation32 In breast cancer, however, miR-20a was found to be downregulated, thus inhibiting cell proliferation.Citation33,Citation34 Hence, in the present work, the downregulation of miR-20a was shown to inhibite prostate cancer phenotypes.
In the current study, CX43 was demonstrated to be a new target of miR-20a. CX43 is a member of the Connexin gene family, which is a component of gap junctions. Connexins not only allow direct intercellular communication, but also play critical roles in cell proliferation, differentiation and migration. For example, the activity of TGF-β has been shown to be mediated by CX43.Citation35 The overexpression of CX43 in human glioblastoma cell lines U251 and T98G reduced cell proliferation.Citation36 In human breast cancer, CX43 is important in maintaining cell differentiation and angiogenesis.Citation21 In the present study, expression of CX43 at a low level was verified in prostate cancer compared with normal prostate tissue. In addition, overexpressed CX43 could inhibit prostate cancer cell viability and proliferation. Furthermore, overexpression of CX43 significantly inhibited prostate cancer cell proliferation compared with that of CX43 containing a miR-20a binding site. Because of the endogenous miR-20a cannot bind the mutant 3′UTR of Mut CX43, the expression of CX43 in cells transfected with Mut CX43 is higher and cell viability and proliferation were inhibited more obviously than that of Wt CX43 transfection. Moreover, CX43 expression in prostate cancer has been reported to be significantly associated with established features indicative of worse prognosis, and decreased CX43 expression is a significant predictor of biochemical recurrence free-survival.Citation37 Therefore, miR-20a will be a potential therapeutic drug to target CX43 for the treatment of prostate cancer. This therapeutic effect has also been demonstrated in vivo in the present study.
In summary, the present study showed how miR-20a downregulated CX43 in prostate cancer. The current work also exhibited the positive role of miR-20a in prostate cancer cell proliferation and carcinogenesis through the downregulation of the expression of CX43. This newly identified miR-20a/CX43 link may be useful in understanding prostate cancer bio-mechanisms and also provides a new, potential therapeutic target for treating prostate cancer.
Materials and Methods
Cell lines and human tissues
Human prostatic carcinoma cell lines LNCaP and MDA-PCa-2b were obtained from the American Type Culture Collection. LNCaP cells were grown in RPMI 1640 containing 10% fetal bovine serum at 37°C in 5% CO2. MDA-PCa-2b cells were maintained in F-12K Medium (GIBCO) with 25 ng/mL cholera toxin, 10 ng/mL epidermal growth factor, 5 μmol/L phosphoethanolamine, 100 pg/mL hydrocortisone, 45 nmol/L selenious acid, 5 μg/mL insulin and 20% fetal bovine serum at 37°C in 5% CO2. Human prostate cancer specimens and its adjacent normal parts that surround cancer cells (n = 9) were obtained from the Southwest Hospital of the Third Military Medical University with documented informed consent in each case.
RNA extraction
RNA from the tissues or cells was isolated according to the instructions of the mirVana™ miRNA Isolation Kit (Ambion). The fraction of large RNA was used to investigate the expression of CX43 genes and the fraction of small RNAs was for the exploration of miRNA expression.
Quantitative real-time reverse transcription polymerase chain reaction (real-time qRT-PCR)
To analyze the expression of miRNAs, real-time qRT-PCR was performed using miRNA-specific primers and the Taqman microRNA assay kit (Applied Biosystems). A mixture of 1 μg the fraction of small RNA together with specific miR-20a-RT or U6-RT primer and water was first prepared in a 13 μl volume. Then the mixture was incubated at 65°C for 10 min and chilled on ice. Finally, the volume was brought up to 20 μl by adding the RT-Buffer, 1 mM dNTPs, 100 U M-MLV Reverse Transcriptase (Promega) and 40U RNase inhibitor (Qiagen). The reaction was incubated at 25°C for 5 min followed by 42°C for 60 min. The enzyme was inactivated by heating at 70°C for 10 min. The cDNAs were used to examine the expression of miR-20a or U6 snRNA. The PCR reaction was performed with specific primer (miR-20a-Fwd or U6-Fwd) and the general primer (Rev), along with the specific TaqMan probes, in 20 μl final volume using 1 μl of cDNA according to the manufacturer's instruction of Taqman microRNA assay kit (Applied Biosystems). The qPCR conditions were the first step at 95°C for 10 min, followed by 40 cycles with 15 sec at 95°C, 10 sec at 58°C, and 20 sec at 72°C. The fluorescence signal was acquired at 72°C and the Ct values were obtained. U6 snRNA was used as internal control. To detect the CX43 level, 2 µg of the fraction of large RNA was reverse transcribed to cDNA using M-MLV reverse transcriptase (Promega) as the above description. For real-time quantitative PCR, CX43 gene (383bp) was amplified from the cDNAs with specific paired primers (UTR-S and UTR-AS), along with the specific TaqMan probes, and β-actin was used as the internal normalizer. PCR cycles were as follows: 94°C for 10 min, followed by 40 cycles of 94°C for 15 sec and 58°C for 30 sec and 72°C for 40 sec. The expression of miR-20a and CX43 gene were determined by triplicate measurements for each sample and were quantified using the 2-ΔΔCt method.
Cell counting kit-8 (CCK-8) assay
Twenty-four hours after transfection, cells were seeded at 3 × 103 cells/well in 96-well plates. After 24 h, 20 μg of CCK-8 (Sigma) in phosphate buffered saline was added to each well. After incubation at 37°C for 4 h, the supernatants were collected and measured using a microculture plate reader (BioTek) at a wavelength of 490 nm. The experiments were repeated three times.
Colony formation assay
Twenty-four hours after transfection, cells were seeded in a six-well plate at a cell density of 1000 cells/well. After two weeks, clones were fixed with methanol and stained with 2% Giemsa solution (Merck, Darmstadt, GERMAN) for 10 min.
Cell invasion assay
Cell invasion assay was performed using Transwell cell culture inserts (Invitrogen). The transfected cells were maintained and allowed to migrate for 24 h. The passed cells were stained with crystal violet solution and photographed under the microscope.
Cloning of expression constructs
For the construction of luciferase reporter plasmid, the fragment of CX43 3′UTR containing the putative target site of miR-20a (383bp) was amplified by PCR using specific primers (UTR-S and UTR-AS). Another fragment of CX43 3′UTR with mutated 3′UTR (383bp) was also amplified by the PCR side-directed mutagenesis assay. The two additional primers (UTR-MS and UTR-MA) were used in the mutation assay. They were all inserted into pmirGLO dual- luciferase reporter vector (Promega) by PmeI and XbaI separately and verified by DNA sequencing. The resulted plasmids were named pmirGLO/CX43 3′UTR (CX43 UTR) or pmirGLO/CX43 3′UTR-M (CX43 UTR-M). The fragment (1838bp) containing the intact CDS and part of 3′UTR of CX43 gene was amplified using paired primers (CDS-S and CDS-AS) and inserted into the pcDNA3.1 expression plasmid by BamHI and XhoI. The resulting plasmid was the CX43 expression plasmids with wild-type 3′UTR (Wt CX43). The CX43 expression plasmid with mutated 3′UTR (Mut CX43), in which the binding sites of miR-20a were mutated, was also constructed using the PCR side-directed mutagenesis assay. Another two primers (UTR-MS and UTR-MA) were also used. MiR-20a inhibitor (LentimiRa-Off-hsa-miR-20a Vector, mh30322) which expressing antisense oligonucleotides of miR-20a and its control vector (LentimiRa-Off) were purchased from Abmgood (CA, Canada). All the primers that listed in Table S1 were purchased from AuGCT Inc. (Beijing, China)
Luciferase Reporter Assays
The luciferase reporter plasmids (0.3 μg, CX43 3′UTR or CX43 3′UTR-M) were transfected separately or cotransfected with 0.5 μg miR-20a inhibitor or its control vector (Abmgood) using Lipofectamine 2000 (Invitrogen) into MDA-PCa-2b cells cultured in 24-well plates. After 48 h, firefly and renilla luciferase activities were measured by using the Dual-Glo® Luciferase Assay System (Promega) and Varioskan Flash microplate reader (Thermo) according to the manufacturer’s protocol. All the transfections were performed three times. Firefly luciferase was normalized to Renilla luciferase activity.
Western blot
Cells were lysed with 0.4 ml of lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 10 mM EDTA, 1% NP40, 20 mM NaF, 1 mM orthovanadate and protease inhibitor cocktail). Cell lysates were resolved by SDS/PAGE and transferred onto polyvinylidene difluoride membranes (Millipore). Membranes were blocked and incubated with rabbit polyclonal antibody to CX43 (ab11370, Abcam) and rabbit polyclonal antibody to GAPDH (ab9485, abcam). Then they were incubated with goat anti-rabbit IgG-HRP secondary antibody (sc-2004). GAPDH was used as loading control. The expression of protein was assessed by enhanced chemiluminescence and exposure to the chemiluminescent film.
Murine xenograft model
Male BABL/c nude mice (6 weeks old) purchased from the Animal Center of the Chinese Academy of Science (Shanghai, China) were randomly assigned to one of three groups (8 mice per group). The mice used in the experiments were handled in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals. Stable cell lines with low expression of miR-20a were established by transfecting MDA-PCa-2b cells with miR-20a inhibitor and followed by selection for 40 d. Cells were resuspended at a final concentration of 1 × 107cells/0.1ml in phosphate-buffered saline mixed with Matrigel (Becton Dickinson and Company) and injected into the back of nude mice. MDA-PCa-2b cells transfected without DNA were used as the mock transfection group. The mice were monitored, and tumor sizes were measured daily. The mice were killed 25 d after injection.
Statistical analysis
All data in the study were evaluated using SPSS 11.5 (SPSS Inc.). Student’s t test or one-way ANOVA test were performed to analyze the significance of differences between sample means obtained from three independent experiments. Differences were considered significant at values of p < 0.05.
Additional material
Download Zip (91 KB)Acknowledgments
This study was supported by Natural Science Foundation Project of CQ CSTC (CSTC, 2010BB5170) and State Key Laboratory Open Project on Trauma, Burn and Combined Injury (SKLKF201007)
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Material
Supplemental material may be downloaded here: http://www.landesbioscience.com/journals/cbt/article/20841/
References
- Buchan JR, Parker R. Molecular biology. The two faces of miRNA. Science 2007; 318:1877 - 8; http://dx.doi.org/10.1126/science.1152623; PMID: 18096794
- Foshay KM, Gallicano GI. miR-17 family miRNAs are expressed during early mammalian development and regulate stem cell differentiation. Dev Biol 2009; 326:431 - 43; http://dx.doi.org/10.1016/j.ydbio.2008.11.016; PMID: 19073166
- Mizuno Y, Yagi K, Tokuzawa Y, Kanesaki-Yatsuka Y, Suda T, Katagiri T, et al. miR-125b inhibits osteoblastic differentiation by down-regulation of cell proliferation. Biochem Biophys Res Commun 2008; 368:267 - 72; http://dx.doi.org/10.1016/j.bbrc.2008.01.073; PMID: 18230348
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61:69 - 90; http://dx.doi.org/10.3322/caac.20107; PMID: 21296855
- Crawford ED, Eisenberger MA, McLeod DG, Spaulding JT, Benson R, Dorr FA, et al. A controlled trial of leuprolide with and without flutamide in prostatic carcinoma. N Engl J Med 1989; 321:419 - 24; http://dx.doi.org/10.1056/NEJM198908173210702; PMID: 2503724
- Tannock IF, de Wit R, Berry WR, Horti J, Pluzanska A, Chi KN, et al, TAX 327 Investigators. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med 2004; 351:1502 - 12; http://dx.doi.org/10.1056/NEJMoa040720; PMID: 15470213
- Wilson LS, Tesoro R, Elkin EP, Sadetsky N, Broering JM, Latini DM, et al. Cumulative cost pattern comparison of prostate cancer treatments. Cancer 2007; 109:518 - 27; http://dx.doi.org/10.1002/cncr.22433; PMID: 17186528
- Hudson RS, Yi M, Esposito D, Watkins SK, Hurwitz AA, Yfantis HG, et al. MicroRNA-1 is a candidate tumor suppressor and prognostic marker in human prostate cancer. Nucleic Acids Res 2012; 40:3689 - 703; http://dx.doi.org/10.1093/nar/gkr1222; PMID: 22210864
- Shi GH, Ye DW, Yao XD, Zhang SL, Dai B, Zhang HL, et al. Involvement of microRNA-21 in mediating chemo-resistance to docetaxel in androgen-independent prostate cancer PC3 cells. Acta Pharmacol Sin 2010; 31:867 - 73; http://dx.doi.org/10.1038/aps.2010.48; PMID: 20581857
- Leite KR, Tomiyama A, Reis ST, Sousa-Canavez JM, Sañudo A, Dall’Oglio MF, et al. MicroRNA-100 expression is independently related to biochemical recurrence of prostate cancer. J Urol 2011; 185:1118 - 22; http://dx.doi.org/10.1016/j.juro.2010.10.035; PMID: 21255804
- Majid S, Dar AA, Saini S, Yamamura S, Hirata H, Tanaka Y, et al. MicroRNA-205-directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer 2010; 116:5637 - 49; http://dx.doi.org/10.1002/cncr.25488; PMID: 20737563
- Sun T, Wang Q, Balk S, Brown M, Lee GS, Kantoff P. The role of microRNA-221 and microRNA-222 in androgen-independent prostate cancer cell lines. Cancer Res 2009; 69:3356 - 63; http://dx.doi.org/10.1158/0008-5472.CAN-08-4112; PMID: 19351832
- Loewenstein WR, Rose B. The cell-cell channel in the control of growth. Semin Cell Biol 1992; 3:59 - 79; http://dx.doi.org/10.1016/S1043-4682(10)80008-X; PMID: 1623203
- Trosko JE, Chang CC, Wilson MR, Upham B, Hayashi T, Wade M. Gap junctions and the regulation of cellular functions of stem cells during development and differentiation. Methods 2000; 20:245 - 64; http://dx.doi.org/10.1006/meth.1999.0941; PMID: 10671317
- Tang B, Peng ZH, Yu PW, Yu G, Qian F. Expression and significance of Cx43 and E-cadherin in gastric cancer and metastatic lymph nodes. Med Oncol 2011; 28:502 - 8; http://dx.doi.org/10.1007/s12032-010-9492-5; PMID: 20373058
- Xu CX, Jia Y, Yang WB, Wang F, Shen SR. [Relationship between Helicobacter pylori infection and expression of connexin (Cx) 32 and Cx43 genes in gastric cancer and gastric precancerous lesions]. Zhonghua Yi Xue Za Zhi 2008; 88:1523 - 7; PMID: 18956631
- Li J, Cheng L, Wang LJ, Liu HC, Li L, Wang XL, et al. Cell surface sialic acid inhibits Cx43 gap junction functions in constructed Hela cancer cells involving in sialylated N-cadherin. Mol Cell Biochem 2010; 344:241 - 51; http://dx.doi.org/10.1007/s11010-010-0548-9; PMID: 20803237
- Kanczuga-Koda L, Koda M, Sulkowski S, Wincewicz A, Zalewski B, Sulkowska M. Gradual loss of functional gap junction within progression of colorectal cancer -- a shift from membranous CX32 and CX43 expression to cytoplasmic pattern during colorectal carcinogenesis. In Vivo 2010; 24:101 - 7; PMID: 20133984
- Lamiche C, Clarhaut J, Strale PO, Crespin S, Pedretti N, Bernard FX, et al. The gap junction protein Cx43 is involved in the bone-targeted metastatic behaviour of human prostate cancer cells. Clin Exp Metastasis 2012; 29:111 - 22; http://dx.doi.org/10.1007/s10585-011-9434-4; PMID: 22080401
- Hernandez M, Shao Q, Yang XJ, Luh SP, Kandouz M, Batist G, et al. A histone deacetylation-dependent mechanism for transcriptional repression of the gap junction gene cx43 in prostate cancer cells. Prostate 2006; 66:1151 - 61; http://dx.doi.org/10.1002/pros.20451; PMID: 16652385
- Shao Q, Wang H, McLachlan E, Veitch GI, Laird DW. Down-regulation of Cx43 by retroviral delivery of small interfering RNA promotes an aggressive breast cancer cell phenotype. Cancer Res 2005; 65:2705 - 11; http://dx.doi.org/10.1158/0008-5472.CAN-04-2367; PMID: 15805269
- Hirschi KK, Xu CE, Tsukamoto T, Sager R. Gap junction genes Cx26 and Cx43 individually suppress the cancer phenotype of human mammary carcinoma cells and restore differentiation potential. Cell Growth Differ 1996; 7:861 - 70; PMID: 8809403
- Hagman Z, Larne O, Edsjö A, Bjartell A, Ehrnström RA, Ulmert D, et al. miR-34c is downregulated in prostate cancer and exerts tumor suppressive functions. Int J Cancer 2010; 127:2768 - 76; http://dx.doi.org/10.1002/ijc.25269; PMID: 21351256
- Bhatnagar N, Li X, Padi SK, Zhang Q, Tang MS, Guo B. Downregulation of miR-205 and miR-31 confers resistance to chemotherapy-induced apoptosis in prostate cancer cells. Cell Death Dis 2010; 1:e105; http://dx.doi.org/10.1038/cddis.2010.85; PMID: 21368878
- Fujita Y, Kojima K, Hamada N, Ohhashi R, Akao Y, Nozawa Y, et al. Effects of miR-34a on cell growth and chemoresistance in prostate cancer PC3 cells. Biochem Biophys Res Commun 2008; 377:114 - 9; http://dx.doi.org/10.1016/j.bbrc.2008.09.086; PMID: 18834855
- Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med 2008; 14:1271 - 7; http://dx.doi.org/10.1038/nm.1880; PMID: 18931683
- Folini M, Gandellini P, Longoni N, Profumo V, Callari M, Pennati M, et al. miR-21: an oncomir on strike in prostate cancer. Mol Cancer 2010; 9:12; http://dx.doi.org/10.1186/1476-4598-9-12; PMID: 20092645
- Leite KR, Sousa-Canavez JM, Reis ST, Tomiyama AH, Camara-Lopes LH, Sañudo A, et al. Change in expression of miR-let7c, miR-100, and miR-218 from high grade localized prostate cancer to metastasis. Urol Oncol 2011; 29:265 - 9; http://dx.doi.org/10.1016/j.urolonc.2009.02.002; PMID: 19372056
- Fan X, Liu Y, Jiang J, Ma Z, Wu H, Liu T, et al. miR-20a promotes proliferation and invasion by targeting APP in human ovarian cancer cells. Acta Biochim Biophys Sin (Shanghai) 2010; 42:318 - 24; http://dx.doi.org/10.1093/abbs/gmq026; PMID: 20458444
- Pesta M, Klecka J, Kulda V, Topolcan O, Hora M, Eret V, et al. Importance of miR-20a expression in prostate cancer tissue. Anticancer Res 2010; 30:3579 - 83; PMID: 20944140
- Huang G, Nishimoto K, Zhou Z, Hughes D, Kleinerman ES. miR-20a encoded by the miR-17-92 cluster increases the metastatic potential of osteosarcoma cells by regulating Fas expression. Cancer Res 2012; 72:908 - 16; http://dx.doi.org/10.1158/0008-5472.CAN-11-1460; PMID: 22186140
- Chai H, Liu M, Tian R, Li X, Tang H. miR-20a targets BNIP2 and contributes chemotherapeutic resistance in colorectal adenocarcinoma SW480 and SW620 cell lines. Acta Biochim Biophys Sin (Shanghai) 2011; 43:217 - 25; http://dx.doi.org/10.1093/abbs/gmq125; PMID: 21242194
- Yu Z, Wang C, Wang M, Li Z, Casimiro MC, Liu M, et al. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J Cell Biol 2008; 182:509 - 17; http://dx.doi.org/10.1083/jcb.200801079; PMID: 18695042
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006; 103:2257 - 61; http://dx.doi.org/10.1073/pnas.0510565103; PMID: 16461460
- Hirschi KK, Burt JM, Hirschi KD, Dai C. Gap junction communication mediates transforming growth factor-beta activation and endothelial-induced mural cell differentiation. Circ Res 2003; 93:429 - 37; http://dx.doi.org/10.1161/01.RES.0000091259.84556.D5; PMID: 12919949
- Huang RP, Fan Y, Hossain MZ, Peng A, Zeng ZL, Boynton AL. Reversion of the neoplastic phenotype of human glioblastoma cells by connexin 43 (cx43). Cancer Res 1998; 58:5089 - 96; PMID: 9823317
- Benko G, Spajić B, Demirović A, Stimac G, Kru Sbreve Lin B, Tomas D. Prognostic value of connexin43 expression in patients with clinically localized prostate cancer. Prostate Cancer Prostatic Dis 2011; 14:90 - 5; http://dx.doi.org/10.1038/pcan.2010.51; PMID: 21173791