Abstract
Cullin-3 is a component of the Cullin-Ring ubiquitin ligase (CRL) family that plays an important role in mediating protein degradation. Deregulation of Cullin-3 expression has been observed in human cancers; however, a role for Cullin-3 in tumor progression has not been previously recognized. Using the MCF10DCIS.com human breast cancer xenograft model, we show that Cullin-3 is increasingly expressed during progression from comedo ductal carcinoma in situ (DCIS) to invasive carcinomas. Cullin-3 protein is not detected in early lesions but is noticeably increased in DCIS tumors and significantly overexpressed in invasive cancers. In experimental metastasis assays, high expression of Cullin-3 was observed in the lung site. Importantly, Cullin-3 staining is detected in human breast cancer tissues, not in normal breast tissues and its expression level positively correlates with tumor stage. These data suggest that Cullin-3 may play an important role in tumor progression from DCIS to invasive cancer and may serve as a biomarker for the diagnosis of aggressive breast cancer.
Introduction
Breast cancer is the most commonly diagnosed type of cancer and second leading cause of cancer death for women in the United States. In 2012, it is predicted that 226,870 new cases of breast cancer will be diagnosed and that 39,920 women will die from their disease.Citation1 Even if a woman’s disease is diagnosed at an early stage, there is always the possibility that the tumor will progress. Ductal carcinoma in situ (DCIS) is an intermediate phase of breast cancer that is not immediately life threatening, but it predisposes the patient to developing invasive breast cancer (IBC).Citation2 DCIS is thought to account for approximately 20% of breast cancers diagnosed in the United StatesCitation3 and it is estimated that between 14 and 50% of DCIS tumors will progress to IBC if not treated.Citation4 Therefore, it is critically important to find biomarkers that can be used to diagnose breast cancers as well as assess the likelihood that they will progress to an invasive phenotype.
Ubiquitin-mediated protein degradation plays a critical role in regulating many diverse cellular processes. This regulatory system uses a series of enzymes to transfer ubiquitin molecules to their substrates in order to target them for destruction. The three classes of enzymes involved in this process are the E1 ubiquitin activating enzymes, E2 ubiquitin conjugating enzymes and E3 ubiquitin ligases.Citation5 The E3 ligases determine substrate specificity and contain two major groups, the HECT (homology to E6-associated protein carboxyl terminus) family and the RING (Really Interesting New Gene) family. Within these families, the Cullin-RING ubiquitin ligases (CRLs) are the largest class of E3 ligases.Citation6-Citation8 There are seven types of CRLs, all of which contain a cullin protein, a RING finger protein and a substrate recognition subunit.Citation9-Citation11 Among these seven family members, Cullin-3 and Cullin-4 have been directly linked to breast cancer.Citation12-Citation15 In a study done by Chen et al., Cullin-4A was shown to be amplified and overexpressed in 600PE, MDA-MB-157 and SKBR3 cells. Cullin-4A mRNA expression was detected in 14 of 30 (47%) primary breast tumors analyzed, but not in the adjacent normal breast tissues, indicating that Cullin-4A might function as an oncogene.Citation12,Citation13 Cullin-3, which is the focus of this paper, has also been shown to be overexpressed in breast cancer cell lines and breast tissue samples. This overexpression was associated with decreased Nrf2 protein expression.Citation14 Nrf2 is known to regulate the expression of several detoxification and anti-oxidant enzymes.Citation15 Silencing of Cullin-3, which would stabilize Nrf2, resulted in increased resistance to oxidative stress, as well as to benzo(a)pyrene, doxorubicin and paclitaxel treatments.Citation14 It has also been shown that a cysteine to tyrosine mutation at position 23 in Keap1 (Keap1C23Y), which functions in Cullin-3 substrate recognition, impairs the ability of Keap1 to ubiquitylate Nrf2 and repress its activity via proteasomal degradation.Citation15 This mutation has been found in breast cancers and suggests that the resulting stabilization of Nrf2 could contribute to tumorigenesis by protecting transformed cells from oxidative stress.Citation15 Additionally, RhoBTB2, a candidate tumor suppressor, has been shown to bind specifically to Cullin-3 and was subsequently identified as a substrate of a Cullin-3 based ubiquitin ligase complex.Citation13,Citation16 These results suggest that Cullin-3 plays distinct roles in regulating chemosensitivity and tumorigenesis. It is unknown what role Cullin-3 plays in breast tumor progression. In this paper, we demonstrate that Cullin-3 expression increases with progression of MCF10DCIS.com xenografts and is detectable in experimental lung metastases. Additionally, we show that Cullin-3 is overexpressed in human breast tumors and Cullin-3 expression levels correlate with breast tumor stage.
Results and Discussion
Cullin-3 protein expression in breast cell lines
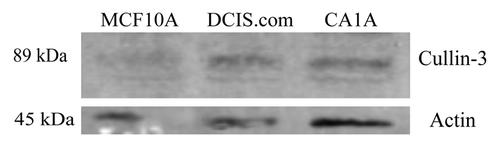
Prior to examining the role of Cullin-3 in tumor progression, it was necessary to establish that Cullin-3 protein is expressed in breast cancer cell lines. Whole cell lysates of MCF10DCIS.com and MCF10CA1A breast cancer cell lines, as well as non-transformed MCF10A cells were analyzed by western blot (). Cullin-3 protein was detected in all three cell lines. These results demonstrate that MCF10DCIS.com and MCF10CA1A cells express Cullin-3 protein.
Increased expression of Cullin-3 protein during MCF10DCIS.com tumor progression
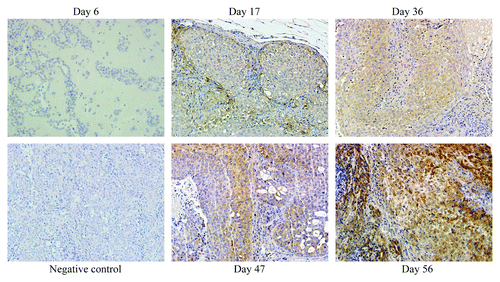
To determine if changes in Cullin-3 protein expression are associated with breast cancer progression, we subcutaneously injected MCF10DCIS.com cells into nude mice. The MCF10DCIS.com cells provide an appropriate model to study progression because these xenografts produce tumors resembling clinical comedo DCIS that progress to invasive cancer.Citation17-Citation19 The cells develop into lesions at day 6, that progress to DCIS (day 17), advanced comedo DCIS (day 36) and invasive cancer (day 56).Citation19 In our study, tumors were excised from the mice at day 6, 17, 36, 47 and 56. Cullin-3 protein expression was then determined by immunohistochemistry. Although western blot analysis showed the comparable levels of Cullin-3 protein among MCF10A, MCF10DCIS.com and MCF10CA1A cells (), in vivo, Cullin-3 was detectable at day 17 and continued to increase in expression at days 36, 47 and 56, () revealing Cullin-3 protein correlation with tumor progression.
Figure 2. The expression of Cullin-3 during breast cancer progression. MCF10DCIS.com cells were sc injected into nude mice. Tumors were excised at day 6, 17, 36, 47 and 56. Cullin-3 expression was determined by IHC (x20). Tumor obtained at day 56 was used as a negative control (without Cullin-3 antibody).
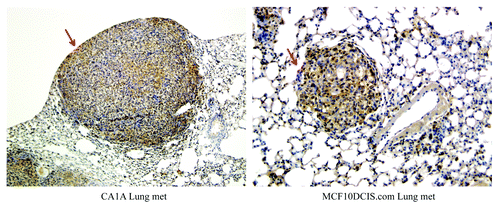
Cullin-3 is overexpressed in lung foci of systemically implanted invasive breast cancers
One of the most common sites of breast tumor metastasis is the lung. To investigate whether or not Cullin-3 is expressed in lung colonization of breast tumors, MCF10CA1A and MCF10DCIS.com cells were injected via tail vein of nude mice and tumors were allowed to form in the lungs.Citation20 Lung sections were stained for Cullin-3 expression. As shown in , these metastatic lung nodules expressed high levels of Cullin-3 protein.
Cullin-3 is overexpressed in human breast tumors
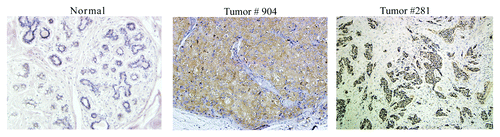
To determine if Cullin-3 has the potential to serve as a biomarker in the clinic, it was necessary to determine if it is expressed in human breast tumors. Two human breast tumors and a normal breast tissue sample were stained for Cullin-3 expression. As shown in , compared with normal breast tissue, which showed no Cullin-3 expression, both tumor number 904 and tumor number 281 showed high levels of Cullin-3 protein. These data are consistent with our data from the MCF10DCIS.com model and suggest that overexpression of Cullin-3 may be clinically relevant.
Cullin-3 expression correlates with breast tumor stage
Next we investigated whether Cullin-3 protein expression is associated with breast cancer progression. To this end, we immunostained a breast tissue microarray containing normal tissue, benign tumors and stages 1 through 3 of breast cancer for Cullin-3. As shown in , Cullin-3 expression was not detected in normal tissues. 33% of benign tumors showed Cullin-3 staining. Cullin-3 staining intensity increased from stage 0 to IIIb, with stage II and stage III tumors showing similar levels of Cullin-3 staining (). The Cochran-Armitage testCitation21 was used to evaluate the linear trend of Cullin-3 expression among the samples ordered as normal breast, fibroadenoma, cystosarcoma phyllodes, Stage 0/I, Stage II, and Stage III. Using R, a statistical programming language,Citation22 and scores of 1 through 6, the observed linear trend of Cullin-3 expression is unlikely to be chance alone (p value < 0.0016). These results mirror the changes observed in the MCF10DCIS.com xenograft model. These results indicate that Cullin-3 protein expression correlates with breast tumor stage and that it may play a role in the progression of human breast tumors.
Figure 5. Analysis of Cullin-3 expression in a breast tissue microarray. A breast tissue microarray including 6 normal breast tissues, 6 benign breast tumors and 60 invasive ductal carcinomas was stained for Cullin-3 expression by IHC. Normal breast tissues are indicated in red, fibroadenomas in blue, cystosarcoma phyllodes in aqua, stage 0/I tumors in purple and stage II tumors in orange and stage III tumors in yellow. Tumors were considered positive for Cullin-3 staining if more than 40% of the tissue core showed Cullin-3 staining.
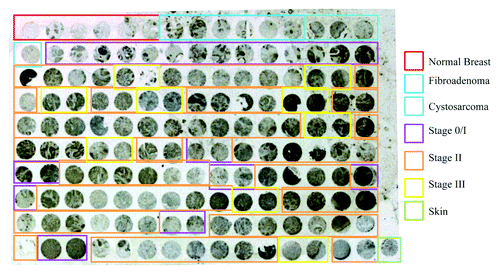
Table 1. Quantification of Cullin-3 expression in a breast tissue microarray
It is well known that breast cancer is a heterogeneous disease. Therefore, it is useful when diagnosing and monitoring disease progression to have biomarkers that can identify which tumors might be particularly aggressive or more likely to develop an invasive phenotype. Our study shows that Cullin-3, a member of the Cullin-RING ubiquitin ligase family, could serve as such a marker. Cullin-3 has been previously associated with breast cancer, but its association with tumor progression was unknown. Here we show that Cullin-3 is increasingly expressed during progression in both the MCF10DCIS.com xenograft model and in clinical samples. Importantly, expression could not be detected in normal tissues. While further investigation is needed to determine the mechanism by which Cullin-3 participates in breast cancer progression from DCIS to an invasive phenotype, we believe that increased expression of Cullin-3 protein promotes degradation of proteins, such as tumor suppressor genes, whose expression is critical for negatively regulating cancer cell growth, proliferation and invasion, leading to cancer progression and metastasis. Previous work by Wilkins et al. that identified RhoBTB2, a candidate tumor suppressor, as a substrate of Cullin-3 E3 ligases,Citation16 suggests that this might be an appropriate model to use to begin to investigate the contributions of Cullin-3 to tumor progression. The mechanism by which Cullin-3 is increasingly expressed during tumor progression is currently under investigation. Additionally, further study of Cullin-3 based E3 ligases, components of the ubiquitin proteasome pathway, may open avenues to explore novel drug targets to limit cancer progression.
Materials and Methods
Western blot analysis
Whole cell lysates were prepared by plating 1 x 106 cells in 60 mm dishes. Cells were allowed to adhere overnight and lysed in 2X Laemmli Sample buffer the following day. Protein expression was measured by western blot. Whole cell lysates were electrophoresed through an 11% polyacrylamide gel. Proteins were transferred to a polyvinylidene diflouride membrane (EMD Millipore) using a semi-dry transfer method. Membranes were blocked for 30 min in 0.1% casein in PBS. Membranes were incubated in primary Cullin-3 antibody (BD Biosciences) overnight at 4°C, washed for ten minutes three times in PBST buffer and then incubated with secondary antibody for 1 h. Bound antibody was detected using the Odyssey Infrared imaging system (Li-Cor Biosciences) according to the manufacturer’s protocol.
Generation of MCF10DCIS.com xenografts
MCF10DCIS.com xenograft was established as described previously.Citation23 Briefly, 1 x 106 MCF10DCIS.com cells in 0.1 ml Matrigel were subcutaneously injected near the nipple of gland of female nude mice (Taconic Farms). Xenografts were removed at day 6, 17, 36, 47 and 56 and then fixed in buffered formalin.
Generation of lung foci of systemic implanted invasive breast cancers
Experimental metastasis (lung colonization) of two highly invasive breast cancer cell lines MCF10CA1A and MCF10DCIS.com cells were described previously.Citation20 In brief, 5 x 105 cells were injected into the tail veins of nude mice. After 6 weeks, lungs were harvested, fixed, cut, and stained for Cullin-3 expression by immunohistochemical analysis.Citation19
Breast tissue arrays
Human breast tissue arrays were purchased from US Biomax Inc. This array (catalog BR1503) included 60 invasive ductal carcinomas, 7 intraductal carcinomas, 3 fibroadenomas, 2 cystosarcoma phyllodes, and 3 cases of normal breast tissue. Each specimen was represented by 2 cores from different tissue spots. Each array spot was 1 mm in diameter and 5 μm in thickness. Detailed information for this array can be viewed at http://www.biomax.us/tissue-arrays/Breast/BR1503. Sections were dewaxed and rehydrated and then underwent microwave antigen retrieval for 10 min. Immunohistochemistry for Cullin-3 was performed using the Dako staining system. Tissue cores with at least 40% of cells exhibiting Cullin-3 staining were considered positive.
Immunohistochemical analysis
Tissue sections from formalin-fixed, paraffin embedded MCF10DCIS.com xenografts were stained with hematoxylin and eosin (H&E) for morphological analysis, or incubated with anti-Cullin-3 antibody (BD Biosciences), followed by secondary antibody. The secondary antibodies were tagged with Avidin-biotinylated horseradish peroxidase (Vector Laboratories), colorized with 3′-3′-diaminobenzidine, and counterstained with hematoxylin. Visualization and documentation were accomplished with an OLYMPUS BX40 microscope supporting a Sony DXC-979MD 3CCCD video camera. Nuclei were counterstained with hematoxylin. Control sections were stained with corresponding normal IgG or secondary antibody only.
| Abbreviations: | ||
| HECT | = | homology to E6-associated protein carboxyl terminus |
| RING | = | Really Interesting New Gene |
| CRLs | = | Cullin-RING ubiquitin ligases |
| DCIS | = | ductal carcinoma in situ |
| IBC | = | invasive breast cancer |
| IHC | = | immunohistochemistry |
Acknowledgments
We thank Drs Fred Miller and Jing Xu for technical assistance and thoughtful discussion of this manuscript. We also acknowledge the DeRoy Testamentary Foundation Fellowship and the Ruth L. Kirschstein National Research Service Award T32-CA009531 for their support of Kelly K. Haagenson.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin 2011; 61:212 - 36; http://dx.doi.org/10.3322/caac.20121; PMID: 21685461
- Kuerer HM, Albarracin CT, Yang WT, Cardiff RD, Brewster AM, Symmans WF, et al. Ductal carcinoma in situ: state of the science and roadmap to advance the field. J Clin Oncol 2009; 27:279 - 88; http://dx.doi.org/10.1200/JCO.2008.18.3103; PMID: 19064970
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al. Cancer statistics, 2008. CA Cancer J Clin 2008; 58:71 - 96; http://dx.doi.org/10.3322/CA.2007.0010; PMID: 18287387
- Erbas B, Provenzano E, Armes J, Gertig D. The natural history of ductal carcinoma in situ of the breast: a review. Breast Cancer Res Treat 2006; 97:135 - 44; http://dx.doi.org/10.1007/s10549-005-9101-z; PMID: 16319971
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem 1998; 67:425 - 79; http://dx.doi.org/10.1146/annurev.biochem.67.1.425; PMID: 9759494
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 2005; 6:9 - 20; http://dx.doi.org/10.1038/nrm1547; PMID: 15688063
- Geyer R, Wee S, Anderson S, Yates J, Wolf DA. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol Cell 2003; 12:783 - 90; http://dx.doi.org/10.1016/S1097-2765(03)00341-1; PMID: 14527422
- Bosu DR, Kipreos ET. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div 2008; 3:7; http://dx.doi.org/10.1186/1747-1028-3-7; PMID: 18282298
- Furukawa M, He YJ, Borchers C, Xiong Y. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat Cell Biol 2003; 5:1001 - 7; http://dx.doi.org/10.1038/ncb1056; PMID: 14528312
- Pintard L, Willis JH, Willems A, Johnson JL, Srayko M, Kurz T, et al. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 2003; 425:311 - 6; http://dx.doi.org/10.1038/nature01959; PMID: 13679921
- Xu L, Wei Y, Reboul J, Vaglio P, Shin TH, Vidal M, et al. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 2003; 425:316 - 21; http://dx.doi.org/10.1038/nature01985; PMID: 13679922
- Chen LC, Manjeshwar S, Lu Y, Moore D, Ljung BM, Kuo WL, et al. The human homologue for the Caenorhabditis elegans cul-4 gene is amplified and overexpressed in primary breast cancers. Cancer Res 1998; 58:3677 - 83; PMID: 9721878
- Guardavaccaro D, Pagano M. Oncogenic aberrations of cullin-dependent ubiquitin ligases. Oncogene 2004; 23:2037 - 49; http://dx.doi.org/10.1038/sj.onc.1207413; PMID: 15021891
- Loignon M, Miao W, Hu L, Bier A, Bismar TA, Scrivens PJ, et al. Cul3 overexpression depletes Nrf2 in breast cancer and is associated with sensitivity to carcinogens, to oxidative stress, and to chemotherapy. Mol Cancer Ther 2009; 8:2432 - 40; http://dx.doi.org/10.1158/1535-7163.MCT-08-1186; PMID: 19638449
- Nioi P, Nguyen T. A mutation of Keap1 found in breast cancer impairs its ability to repress Nrf2 activity. Biochem Biophys Res Commun 2007; 362:816 - 21; http://dx.doi.org/10.1016/j.bbrc.2007.08.051; PMID: 17822677
- Wilkins A, Ping Q, Carpenter CL. RhoBTB2 is a substrate of the mammalian Cul3 ubiquitin ligase complex. Genes Dev 2004; 18:856 - 61; http://dx.doi.org/10.1101/gad.1177904; PMID: 15107402
- Miller FR, Soule HD, Tait L, Pauley RJ, Wolman SR, Dawson PJ, et al. Xenograft model of progressive human proliferative breast disease. J Natl Cancer Inst 1993; 85:1725 - 32; http://dx.doi.org/10.1093/jnci/85.21.1725; PMID: 8411256
- Miller FR, Santner SJ, Tait L, Dawson PJ. MCF10DCIS.com xenograft model of human comedo ductal carcinoma in situ. J Natl Cancer Inst 2000; 92:1185 - 6; http://dx.doi.org/10.1093/jnci/92.14.1185A; PMID: 10904098
- Tait LR, Pauley RJ, Santner SJ, Heppner GH, Heng HH, Rak JW, et al. Dynamic stromal-epithelial interactions during progression of MCF10DCIS.com xenografts. Int J Cancer 2007; 120:2127 - 34; http://dx.doi.org/10.1002/ijc.22572; PMID: 17266026
- Santner SJ, Dawson PJ, Tait L, Soule HD, Eliason J, Mohamed AN, et al. Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Res Treat 2001; 65:101 - 10; http://dx.doi.org/10.1023/A:1006461422273; PMID: 11261825
- Armitage P, Berry G, Matthews JNS. (2002). Statistical Methods in Medical Research (4th ed.). Oxford: Blackwell Science. See p. 506.
- R Development Core Team. (2011). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org/ <http://www.R-project.org/>.
- Shekhar MP, Tait L, Pauley RJ, Wu GS, Santner SJ, Nangia-Makker P, et al. Comedo-ductal carcinoma in situ: A paradoxical role for programmed cell death. Cancer Biol Ther 2008; 7:1774 - 82; http://dx.doi.org/10.4161/cbt.7.11.6781; PMID: 18787417