Abstract
Aberrant promoter region hypermethylation of upstream transcription factors may be responsible for silencing entire anti-neoplastic gene networks. In this study, we explored whether transcription factor coding gene, caudal-related homeobox 2 (CDX2), is silenced by promoter hypermethylation in lung cancer, and examined its potential tumor-suppressive functions. Semi-quantitative RT-PCR showed that four of six lung cancer cell lines exhibited no or weak CDX2 expression. Expression of CDX2 was correlated to CDX2 promoter region methylation status, as determined by methylation-specific PCR (MSP) and bisulfite sequencing. Restoration of CDX2 expression was induced by treatment with demethylating drug 5-aza-2'-deoxycytidine (5-AZA) in lung cancer cell lines. Methylation of CDX2 was common in human primary lung cancer (61 of 110 tumors, 55.45%), but no methylation was found in normal lung tissues. Re-expression of CDX2 suppressed lung cancer cell proliferation and blocked cells in G1 phase. β-catenin/TCF activity and downstream genes expression were inhibited by re-expression of CDX2, and increased by depletion of CDX2. In conclusion, CDX2 is frequently methylated in lung cancer, and expression of CDX2 is regulated by promoter region hypermethylation. CDX2 may serve as a tumor suppressor in lung cancer and inhibits lung cancer cell proliferation by suppressing Wnt signaling.
Introduction
Lung cancer is the most common cause of cancer-related death with 1.38 million deaths worldwide in 2008.Citation1 Tobacco smoking accounts for 80 and 50% of lung cancer deaths for males and females respectively.Citation2 The 5-year survival of lung cancer patients is only 15%, largely due to the fact that many lung cancer patients are diagnosed at advanced stage. Thus, to improve clinical outcomes, a better understanding of the molecular pathogenesis of lung cancer is required to identify new biomarkers and novel therapeutic targets.
Genetic and epigenetic factors play important roles in the initiation and progression of lung cancer. The genetic abnormalities in major components of several signaling pathways contribute to the pathogenesis of lung cancer.Citation3 Furthermore, in the last decade, aberrant transcriptional silencing of tumor suppressor genes by CpG island hypermethylation has also been recognized as key event in the onset and progression of cancers.Citation4,Citation5 In lung cancer, many genes involved in multiple signaling pathways have been found to be silenced by DNA methylation.Citation6-Citation8 For Wnt signaling pathway, mutations which target APC or β-catenin were rarely found in lung cancer.Citation9,Citation10 But disruption of the Wnt signaling pathway was found mainly by promoter region hypermethylation of Wnt signaling antagonizing components.Citation11,Citation12
Apart from epigenetic silencing of multiple genes by direct promoter methylation, aberrant methylation of upstream single regulatory transcription factors may be responsible for silencing entire anti-neoplastic gene networks.Citation13 Caudal-related homeobox 2 (CDX2) is an intestinal transcription factor that regulates expression of numerous intestine-specific genes associated with intestinal proliferation and differentiation.Citation14-Citation16 CDX2 is considered to be a tumor suppressor gene,Citation17-Citation19 which involved in Wnt/β-catenin signaling pathway in colon cancer.Citation20-Citation22 DNA methylation appears to cause silencing of CDX2 in squamous esophageal cancer and gastric cancer.Citation23,Citation24 Because the endoderm gives rise to the lining of the lung and the gastrointestinal tract, we hypothesized that CDX2 may be involved in lung carcinogenesis. In this study, we analyzed the epigenetic regulation and function of CDX2 in human lung cancer.
Results
CDX2 expression is regulated by promoter region hypermethylation in human lung cancer
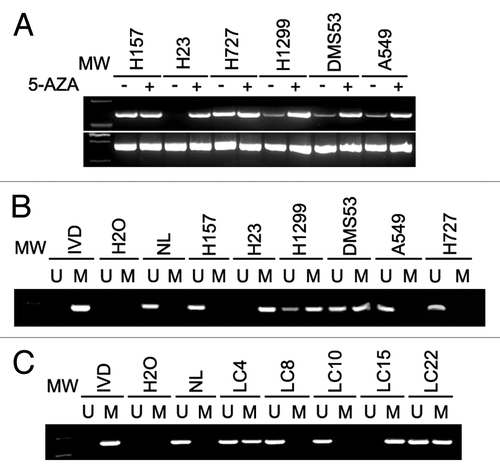
CDX2 expression was examined by RT-PCR in six established lung cancer cell lines. CDX2 was not expressed in one cell line (H23), weakly expressed in three cell lines (H1299, DMS53 and A549), and strongly expressed in two cell lines (H157 and H727) (). To explore if CDX2 expression was regulated by DNA methylation, methylation-specific PCR (MSP) was performed. As expected, complete methylation was found in H23 cells, and partial methylation was found in H1299 and DMS53 cells (). However, no methylation was observed in H157 and H727 cells. These results indicate that loss or reduction of CDX2 expression is correlated with promoter region hypermethylation in human lung cancer cell lines. To further validate the regulation of CDX2 expression by DNA methylation, 5-aza-2-deoxycytidine (5-AZA), a DNA methylation transferase inhibitor, was applied to treat lung cancer cell lines. As shown in , re-expression of CDX2 was found after 5-AZA treatment of H23 cells. Increased expression was observed in H1299 and DMS53 cells. However, no expression changes were found before and after 5-AZA treatment in H157 and H727 cell lines. The results described above suggest that CDX2 expression is regulated by promoter region hypermethylation in human lung cancer.
Figure 1. CDX2 expression and MSP results in lung cancer cells and primary lung cancer. (A) Expression of CDX2 was examined by semi-quantitative RT-PCR in lung cancer cell lines in the presence or absence of 5-AZA treatment. GAPDH was used as an internal control. -, Untreated; +, 5-AZA treated; MW, molecular weight marker. (B) MSP results of CDX2 in lung cancer cell lines. IVD, in vitro methylated DNA; NL, normal blood lymphocyte DNA; M, methylated bands; U, unmethylated bands. (C) Representative MSP results of CDX2 in human primary lung cancer tissues. LC, lung cancer.
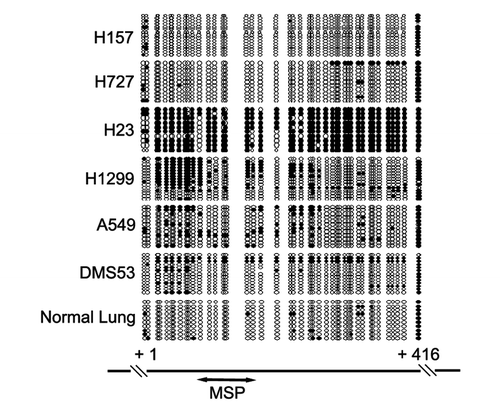
To verify MSP results representing the methylation status of CDX2 promoter region, bisulfite sequencing was performed in lung cancer cell lines. Consistent with the MSP results, dense methylation in the promoter region was found in H23 cells. Scattered or allele methylation was found in H1299, A549 and DMS53 cells, while scarce CpG methylation was revealed in H157 and H727 cells ().
CDX2 is frequently methylated in human primary lung cancer
To explore CDX2 methylation status in human primary lung cancer, 110 cases of lung cancer and 5 cases of normal lung tissue were analyzed by MSP. CDX2 methylation was detected in 55.45% of lung cancer, but no methylation was found in normal lung tissues. Representative examples of gel analysis of MSP were shown in . Above results eliminate the possibility of lung tissue-specific CDX2 methylation and indicate that CDX2 methylation is a potential lung cancer marker. The association of CDX2 methylation and clinical factors was analyzed in this study, including age, gender, tumor size, smoking history, tissue phenotype, stage and differentiation. However, no association was found among CDX2 methylation and above clinical factors (P > 0.05).
Lung cancer cell proliferation is inhibited by re-expression of CDX2
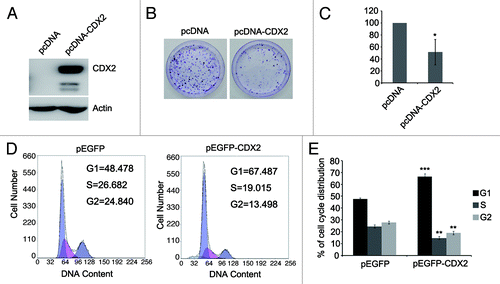
To investigate the effect of CDX2 on lung cancer cell proliferation, colony formation assay was employed in H23 cells. Re-expression of CDX2 was confirmed by western blotting after transfection of CDX2 expression vector (). The colony formation efficiency was reduced in CDX2-expressing cells compared with empty vector control (down to 51%; ). These data demonstrate that CDX2 inhibits lung cancer cell proliferation.
Figure 3. Effects of CDX2 on lung cancer cell proliferation and cell cycle distribution. (A) Confirmation of re-expression of CDX2 in H23 cells by western blotting. Actin was used as internal reference. (B) Typical colony formation assay for CDX2 and control vector are presented. H23 cells were transfected with pcDNA-CDX2 or control vector. After selection with G418 for 2–3 weeks, the surviving colonies were stained with 0.5% crystal violet and counted. (C) Quantitative analysis of colony formation assays. The numbers of G418 resistant colonies in control vector transfected cells were set to 100%. The results are given as the mean ± SD from triplicate experiments. (*P < 0.05). (D) Representative results of cell cycle distribution for CDX2 and control vector. H23 cells were transfected with the pEGFP-CDX2 or control vector. Transfected cells were sorted by GFP expression. Cell cycle distributions were measured by propidium iodide (PI) staining followed by flow cytometry after transfection for 48 h. (E) Quantitative analysis of cell cycle distribution. The percentages of G1, S and G2 cells transfected with control vector served as controls. The results are given as the mean ± SD from triplicate experiments. (**P < 0.01; ***P < 0.001).
G1 arrest is induced by re-expression of CDX2 in H23 cells
The effect of CDX2 on cell cycle was evaluated by flow cytometry. The ratio of G1 phase was increased but S and G2 phase were reduced after re-expression of CDX2 in H23 cells (G1: 47.7 ± 1.01 vs 66.4 ± 2.61, P < 0.001; S: 27.83 ± 1.67 vs 18.93 ± 1.45, P < 0.01; G2: 24.46 ± 1.40 vs 14.68 ± 1.42, P <0.01) (). These results indicate that CDX2 induces G1 phase arrest in lung cancer.
CDX2 inhibits Wnt/β-catenin signaling in lung cancer cells
CDX2 was reported to suppress colon cancer proliferation by inhibiting Wnt/β-catenin signaling pathway.Citation20-Citation22 In this study, we analyzed the effect of CDX2 on β-catenin/TCF-4 by determining the activity of luciferase reporters. As shown in , the activity of β-catenin/TCF-4 was much lower in CDX2-expressing H23 cells than in empty vector control. Further studies revealed that the expression of c-Myc, Cyclin D1 and Survivin, downstream targets of β-catenin/TCF-4, was reduced after re-expression of CDX2 in H23 cells ().
Figure 4. CDX2 antagonizes Wnt/β-catenin signaling. (A) CDX2 inhibits TCF-4 reporter activity. CDX2 expression vector and TCF-4 reporter pTOPFlash (OT) or control inactive reporter pFOPFlash (OF) were transfected into H23 cells. After 48 h of transfection, the relative luciferase activity (a ratio of firefly luciferase to Renilla luciferase) was measured by GLOMAX luminometer. The activity of OF in control vector-transfected cells was defined as 1. The results are given as the mean ± SD from triplicate experiments. (***P < 0.001). (B) Re-expression of CDX2 represses c-Myc, Cyclin D1 and Survivin, as measured by western blotting in H23 cells. Actin was used as internal reference. (C) Knockdown of CDX2 increases the activity of TCF-4 reporter. TCF-4 reporter pTOPFlash (OT) and control siRNA (CTL) or two individual siRNAs targeting CDX2 were transfected into H157 cells. After 48 h of transfection, the relative luciferase activity was measured. The results are given as the mean ± SD from triplicate experiments. (**P < 0.01). (D) Knockdown of CDX2 increases the expression of Cyclin D1 in H157 cells. Actin was used as internal reference.
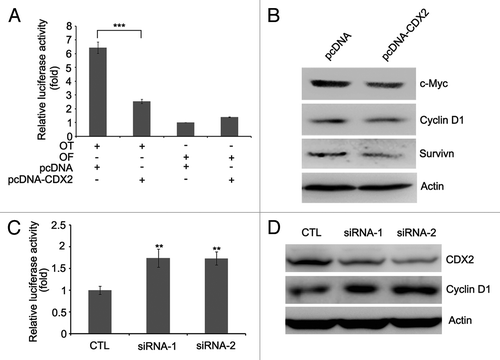
To further investigate the effect of CDX2 on Wnt/β-catenin signaling pathway, RNA interference (RNAi) was employed to knockdown endogenous CDX2 in H157 cells. As shown in , significant decrease of CDX2 expression was achieved. The activity of β-catenin/TCF-4 was increased in CDX2-depleted cells compared with siRNA control (). Furthermore, the expression of Cyclin D1 was reduced when depletion of CDX2 in H157 cells (). These results suggest that CDX2 is a Wnt signaling inhibitor in lung cancer.
Discussion
CDX2 was found frequently methylated in squamous esophageal cancer and gastric cancer, and the expression of CDX2 was regulated by promoter region methylation.Citation23,Citation24 Though CDX2 methylation was also screened by MethyLight in primary lung adenocarcinomas, the regulation and function of CDX2 remained unclear in human lung cancer.Citation27,Citation28 In this study, we found that promoter region methylation and CDX2 expression were well correlated, and CDX2 expression was reactivated by 5-AZA treatment in silenced cells, suggesting that methylation constitutes a primary mechanism in regulating CDX2 expression in lung cancer cell lines. However, A549 cell line is an exceptional case. MSP showed unmethylated CDX2 but RT-PCR demonstrated weak expression. This phenomenon can be explained by promoter region scattered methylation, as revealed by bisulfite sequencing ().
CDX2 methylation was not limited to lung cancer cell lines. Similarly, methylation of CDX2 was frequently found in human primary lung cancer, indicating that CDX2 may serve as a lung cancer detection marker. In addition, both methylated and unmethylated alleles were found in primary lung cancer. This finding likely represents the presence of normal, non-neoplastic lung epithelium in the human tissue samples. Thus, normal tissue surrounding the tumor sites accounts for the unmethylated alleles among the cancer cases.
Previous studies indicated that CDX2 is a potential tumor suppressor gene in colon and gastric cancer.Citation17-Citation19,Citation29 To explore the effect of CDX2 on lung cancer proliferation, colony formation assay was employed in H23 cells. The colony formation efficiency was reduced after re-expression of CDX2. Furthermore, consistent with the finding in colon cancer cells,Citation19,Citation30 we observed that re-expression of CDX2 caused G1 arrest in H23 cells, which may contribute to the proliferation inhibition of lung cancer cells induced by CDX2. To understand the mechanism, the effect of CDX2 on Wnt/β-catenin signaling pathway was analyzed. Our study demonstrates that re-expression of CDX2 inhibited the activity of β-catenin/TCF-4 significantly in H23 cells, and knockdown of CDX2 increased the activity of β-catenin/TCF-4 in H157 cells. In addition, re-expression of CDX2 suppressed the expression of c-Myc, Cyclin D1 and Survivin in H23 cells and knockdown of CDX2 increased Cyclin D1 expression in H157 cells. Taken together, these results suggest that CDX2 may serve as a tumor suppressor by inhibiting Wnt signaling in lung cancer.
In conclusion, CDX2 is frequently methylated in human lung cancer and may serve as lung cancer detection marker. Promoter region methylation of CDX2 silences its expression in lung cancer. Methylation of CDX2 may lead to active Wnt signaling, which then promotes lung carcinogenesis.
Materials and Methods
Human tissue samples
One hundred and ten cases of primary lung cancer were collected as fresh frozen tissue from China (General Hospital of PLA). All malignancies were examined and tumor staging was classified according to the TNM classification of American Joint Committee on Cancer (AJCC). The lung cancer cases included 46 adenocarcinoma, 42 squamous cell cancer, 12 large cell cancer, 3 carcinoid, 3 small cell cancer, 2 BAC and 2 miscellaneous cases. Of these, 22 were stage IA, 24 were stage IB, 4 were stage IIA, 30 were stage IIB, 18 were stage IIIA, 7 were stage IIIB and 5 were stage IV. Five cases of normal lung tissue were obtained at autopsy from patients without associated lung carcinoma as normal comparisons. All samples were collected under the guidelines approved by Chinese PLA General Hospital’s institutional review board and with informed consent from patients.
Lung cancer cell lines
Six lung cancer cell lines were included in this study. H157, H23, H727, H1299 and A549 are non-small cell lung cancer cell lines. DMS53 is a small cell lung cancer cell line. All cell lines were established from primary lung cancer. Each of the cell lines was maintained in 90% RPMI 1640 supplemented with 10% fetal bovine serum. Cells were passaged 1:3 once full confluence was reached.
5-aza-2’-deoxycytidine (5-AZA) treatment
Lung cancer cell lines were split to low density (30% confluence) 12 h before treatment. Cells were treated with 5-AZA (Sigma) at a concentration of 2 μM. Growth medium, conditioned with 5-AZA, was exchanged every 24 h for a total treatment interval of 96 h. At the end of the treatment course, DNA and RNA were harvested and isolated as described below.
RNA isolation and RT-PCR
Total RNA was isolated by Trizol (Invitrogen). First strand cDNA was synthesized by using SuperScript First-Strand Synthesis System for RT-PCR kit (Invitrogen) according to manufacturer’s instructions. PCR was conducted using the Taq polymerase (Takara). Primers to CDX2 were as follows: 5’-GGCTGGAGCTGGAGAAGGAG-3’ (F) and 5’-CAGACACTGAGGCTTGCAGG-3’ (R). GAPDH was as an internal control using the following primer set: 5’-GACCACAGTCCATGCCATCAC-3’ (F) and 5’-GTCCACCACCCTGTTGCTGTA-3’ (R). Amplified products were analyzed on 1.5% agarose gels.
Methylation-specific PCR (MSP) and bisulfite sequencing
Bisulfite modification of DNA, MSP and bisulfite sequencing were performed as previously described.Citation25,Citation26 The bisulfite-treated DNA was amplified with methylation-specific primer set to CDX2: 5’-TTTTCGTGTTTTTCGGTAGTTTTTAGC-3’ (MF) and 5’-ACTCACGTACATAATAACGAAAATCCG-3’ (MR); or unmethylation-specific primer set: 5’-TTTTTTGTGTTTTTTGGTAGTTTTTAGT-3’ (UF) and 5’-TAACTCACATACATAATAACAAAAATCCA-3’ (UR). MSP products were analyzed using 3% agarose gels. For Bisulfite sequencing, bisulfite-treated DNA was subjected to PCR using primers flanking the targeted MSP regions described above. Sequencing primer set was as follows: 5’-GATYGTTTYGGAGGTAGAAGAG-3’ (F) and 5’-CRCRTAACCATTCCAATCCTCC-3’ (R). PCR products were gel purified and cloned into pCR2.1 vector (Invitrogen) for sequencing.
Colony formation assay
H23 cells were seeded in 6-well culture plates 24 h prior to transfection. pcDNA-CDX2 or empty vector was transfected into H23 cells using FuGENE 6 (Roche) according to manufacturer’s instructions. Twenty-four hours after transfection, cells were reseeded at 3,000 cells/60mm-dish in triplicate. After selected for 2–3 weeks with G418 (200 μg/mL), surviving colonies (≥ 50 cells per colony) were counted after staining with 0.2% crystal violet.
Cell cycle analysis
pEGFP-CDX2 or empty vector was transfected into H23 cells using FuGENE 6. Forty-eight hours after transfection, cells were harvested, washed with phosphate-buffered saline (PBS) and fixed with ice cold 70% ethanol at -20 °C overnight. Samples were then washed with PBS and stained with propidium iodide (Sigma) containing RNase A (Sigma) for 30 min at 37 °C. GFP-positive cells were isolated and cell cycle distribution in different phases was determined using flow cytometry (Becton Dickinson).
Luciferase reporter assays
H23 cells were seeded at 2 × 104 cells/well in 96-well culture plates 24 h prior to transfection. A luciferase reporter construct, pTOPFlash (OT), containing 4 × TCF-binding sites, or pFOPFlash (OF), containing mutant TCF/LEF binding sites was co-transfected with either pcDNA-CDX2 or empty vector, together with an internal control Renilla luciferase reporter pRL-CMV vector using FuGENE 6. Forty-eight hours after transfection, cells were harvested and analyzed using a dual-luciferase assay kit (Promega).
Western blotting
Cell protein lysates were prepared using 1× PBS supplemented with 1% Nonidet P-40, 1× protease inhibitors cocktail (Roche) and 50 μg/ml phenylmethylsulfonyl fluoride. Proteins were resolved by 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride membrane. Membranes were incubated with the indicated primary antibodies and anti-mouse or anti-rabbit secondary antibodies conjugated to horseradish peroxidase (HRP). After addition of HRP substrate, the chemiluminescence signal was detected using a Luminescent Image Analyzer LAS-4000 (Fujifilm). Antibodies used were as follows: rabbit anti-CDX2 (Zhongshan Goldenbridge, ZA-0520, 1:500), rabbit anti-Cylin D1 (Santa Cruz, sc-753, 1:500), rabbit anti-c-Myc (Bioworld, BS2462, 1:500) and rabbit anti-Survivin (Bioworld, BS1771, 1:300).
RNA interference
Fifty to sixty percent confluent H157 cells were transfected with 50 nM of siRNAs using Lipofectamine 2000 (Invitrogen) following the manufacturer's direction. The target sequences for CDX2 were the following: siRNA-1, 5’-GAG AGG GAC UCA AGG GAA A dGdG-3’ and siRNA-2, 5’-GAA GAA GUU GCA GCA GCA ATT-3’.
Statistical analysis
All experiments were repeated more than three times. Statistical analysis was carried out by Student’s t-test. P values of < 0.05 were considered to be statistically significant.
| Abbreviations: | ||
| CDX2 | = | caudal-related homeobox 2 |
| CpG | = | cytosine-guanine dinucleotide |
| RT-PCR | = | reverse transcription-polymerase chain reaction |
| cDNA | = | complementary DNA |
| mRNA | = | messenger RNA |
| MSP | = | methylation-specific PCR |
| GAPDH | = | glyceraldehyde-3-phosphate dehydrogenase |
| 5-AZA | = | 5-aza-2’-deoxycytidine |
| NL | = | normal blood lymphocyte DNA |
| IVD | = | in vitro methylated DNA |
| OT | = | pTOPFlash |
| OF | = | pFOPFlash |
| TCF-4 | = | T-cell factor-4 |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are very grateful to Prof. Youyong Lu for providing us the pEGFP-CDX2 and pcDNA-CDX2 vectors. This work was supported by grants from the National Basic Research Program (973 program no. 2012CB934002, 2010CB912802, 2009CB521801), National HighTechnology R&D Program of China (863 program no. SS2012AA020821, SS2012AA02A203, SS2012AA02A209), National Key Scientific instrument Special Programme of China (grant no. 2011YQ03013405) and National Science Foundation of China (grant no. 81121004, 81071953, 81161120432)
Notes
† These authors contributed equally to this work.
References
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010; 127:2893 - 917; http://dx.doi.org/10.1002/ijc.25516; PMID: 21351269
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61:69 - 90; http://dx.doi.org/10.3322/caac.20107; PMID: 21296855
- Brambilla E, Gazdar A. Pathogenesis of lung cancer signalling pathways: roadmap for therapies. Eur Respir J 2009; 33:1485 - 97; http://dx.doi.org/10.1183/09031936.00014009; PMID: 19483050
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002; 3:415 - 28; PMID: 12042769
- Esteller M. Epigenetics in cancer. N Engl J Med 2008; 358:1148 - 59; http://dx.doi.org/10.1056/NEJMra072067; PMID: 18337604
- Shames DS, Girard L, Gao B, Sato M, Lewis CM, Shivapurkar N, et al. A genome-wide screen for promoter methylation in lung cancer identifies novel methylation markers for multiple malignancies. PLoS Med 2006; 3:e486; http://dx.doi.org/10.1371/journal.pmed.0030486; PMID: 17194187
- Zöchbauer-Müller S, Fong KM, Virmani AK, Geradts J, Gazdar AF, Minna JD. Aberrant promoter methylation of multiple genes in non-small cell lung cancers. Cancer Res 2001; 61:249 - 55; PMID: 11196170
- Brock MV, Hooker CM, Ota-Machida E, Han Y, Guo M, Ames S, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med 2008; 358:1118 - 28; http://dx.doi.org/10.1056/NEJMoa0706550; PMID: 18337602
- Königshoff M, Eickelberg O. WNT signaling in lung disease: a failure or a regeneration signal?. Am J Respir Cell Mol Biol 2010; 42:21 - 31; http://dx.doi.org/10.1165/rcmb.2008-0485TR; PMID: 19329555
- Van Scoyk M, Randall J, Sergew A, Williams LM, Tennis M, Winn RA. Wnt signaling pathway and lung disease. Transl Res 2008; 151:175 - 80; http://dx.doi.org/10.1016/j.trsl.2007.12.011; PMID: 18355764
- Fukui T, Kondo M, Ito G, Maeda O, Sato N, Yoshioka H, et al. Transcriptional silencing of secreted frizzled related protein 1 (SFRP 1) by promoter hypermethylation in non-small-cell lung cancer. Oncogene 2005; 24:6323 - 7; http://dx.doi.org/10.1038/sj.onc.1208777; PMID: 16007200
- Sato K, Tomizawa Y, Iijima H, Saito R, Ishizuka T, Nakajima T, et al. Epigenetic inactivation of the RUNX3 gene in lung cancer. Oncol Rep 2006; 15:129 - 35; PMID: 16328045
- Akiyama Y, Watkins N, Suzuki H, Jair KW, van Engeland M, Esteller M, et al. GATA-4 and GATA-5 transcription factor genes and potential downstream antitumor target genes are epigenetically silenced in colorectal and gastric cancer. Mol Cell Biol 2003; 23:8429 - 39; http://dx.doi.org/10.1128/MCB.23.23.8429-8439.2003; PMID: 14612389
- Boudreau F, Rings EH, van Wering HM, Kim RK, Swain GP, Krasinski SD, et al. Hepatocyte nuclear factor-1 alpha, GATA-4, and caudal related homeodomain protein Cdx2 interact functionally to modulate intestinal gene transcription. Implication for the developmental regulation of the sucrase-isomaltase gene. J Biol Chem 2002; 277:31909 - 17; http://dx.doi.org/10.1074/jbc.M204622200; PMID: 12060663
- Sakaguchi T, Gu X, Golden HM, Suh E, Rhoads DB, Reinecker HC. Cloning of the human claudin-2 5′-flanking region revealed a TATA-less promoter with conserved binding sites in mouse and human for caudal-related homeodomain proteins and hepatocyte nuclear factor-1alpha. J Biol Chem 2002; 277:21361 - 70; http://dx.doi.org/10.1074/jbc.M110261200; PMID: 11934881
- Suh E, Wang Z, Swain GP, Tenniswood M, Traber PG. Clusterin gene transcription is activated by caudal-related homeobox genes in intestinal epithelium. Am J Physiol Gastrointest Liver Physiol 2001; 280:G149 - 56; PMID: 11123208
- Bonhomme C, Duluc I, Martin E, Chawengsaksophak K, Chenard MP, Kedinger M, et al. The Cdx2 homeobox gene has a tumour suppressor function in the distal colon in addition to a homeotic role during gut development. Gut 2003; 52:1465 - 71; http://dx.doi.org/10.1136/gut.52.10.1465; PMID: 12970140
- Gross I, Duluc I, Benameur T, Calon A, Martin E, Brabletz T, et al. The intestine-specific homeobox gene Cdx2 decreases mobility and antagonizes dissemination of colon cancer cells. Oncogene 2008; 27:107 - 15; http://dx.doi.org/10.1038/sj.onc.1210601; PMID: 17599044
- Aoki K, Tamai Y, Horiike S, Oshima M, Taketo MM. Colonic polyposis caused by mTOR-mediated chromosomal instability in Apc+/Delta716 Cdx2+/- compound mutant mice. Nat Genet 2003; 35:323 - 30; http://dx.doi.org/10.1038/ng1265; PMID: 14625550
- Guo RJ, Huang E, Ezaki T, Patel N, Sinclair K, Wu J, et al. Cdx1 inhibits human colon cancer cell proliferation by reducing beta-catenin/T-cell factor transcriptional activity. J Biol Chem 2004; 279:36865 - 75; http://dx.doi.org/10.1074/jbc.M405213200; PMID: 15215241
- Ezaki T, Guo RJ, Li H, Reynolds AB, Lynch JP. The homeodomain transcription factors Cdx1 and Cdx2 induce E-cadherin adhesion activity by reducing beta- and p120-catenin tyrosine phosphorylation. Am J Physiol Gastrointest Liver Physiol 2007; 293:G54 - 65; http://dx.doi.org/10.1152/ajpgi.00533.2006; PMID: 17463179
- Guo RJ, Funakoshi S, Lee HH, Kong J, Lynch JP. The intestine-specific transcription factor Cdx2 inhibits beta-catenin/TCF transcriptional activity by disrupting the beta-catenin-TCF protein complex. Carcinogenesis 2010; 31:159 - 66; http://dx.doi.org/10.1093/carcin/bgp213; PMID: 19734199
- Guo M, House MG, Suzuki H, Ye Y, Brock MV, Lu F, et al. Epigenetic silencing of CDX2 is a feature of squamous esophageal cancer. Int J Cancer 2007; 121:1219 - 26; http://dx.doi.org/10.1002/ijc.22828; PMID: 17534889
- Yuasa Y, Nagasaki H, Akiyama Y, Sakai H, Nakajima T, Ohkura Y, et al. Relationship between CDX2 gene methylation and dietary factors in gastric cancer patients. Carcinogenesis 2005; 26:193 - 200; http://dx.doi.org/10.1093/carcin/bgh304; PMID: 15498792
- Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A 1996; 93:9821 - 6; http://dx.doi.org/10.1073/pnas.93.18.9821; PMID: 8790415
- Jia Y, Yang Y, Liu S, Herman JG, Lu F, Guo M. SOX17 antagonizes WNT/β-catenin signaling pathway in hepatocellular carcinoma. Epigenetics 2010; 5:743 - 9; http://dx.doi.org/10.4161/epi.5.8.13104; PMID: 20716954
- Tsou JA, Galler JS, Siegmund KD, Laird PW, Turla S, Cozen W, et al. Identification of a panel of sensitive and specific DNA methylation markers for lung adenocarcinoma. Mol Cancer 2007; 6:70; http://dx.doi.org/10.1186/1476-4598-6-70; PMID: 17967182
- Selamat SA, Galler JS, Joshi AD, Fyfe MN, Campan M, Siegmund KD, et al. DNA methylation changes in atypical adenomatous hyperplasia, adenocarcinoma in situ, and lung adenocarcinoma. PLoS One 2011; 6:e21443; http://dx.doi.org/10.1371/journal.pone.0021443; PMID: 21731750
- Park Y, Srivastava A, Kim GH, Mino-Kenudson M, Deshpande V, Zukerberg LR, et al. CDX2 expression in the intestinal-type gastric epithelial neoplasia: frequency and significance. Mod Pathol 2010; 23:54 - 61; http://dx.doi.org/10.1038/modpathol.2009.135; PMID: 19820687
- Aoki K, Kakizaki F, Sakashita H, Manabe T, Aoki M, Taketo MM. Suppression of colonic polyposis by homeoprotein CDX2 through its nontranscriptional function that stabilizes p27Kip1. Cancer Res 2011; 71:593 - 602; http://dx.doi.org/10.1158/0008-5472.CAN-10-2842; PMID: 21224344