Abstract
TRAIL is a member of the TNF superfamily that induces tumor-selective cell death by engaging the pro-apoptotic death receptors DR4 and DR5. The antitumor potential of the TRAIL pathway has been targeted by several therapeutic approaches including recombinant TRAIL and TRAIL-receptor agonist antibodies among others. Interest in sensitizing tumor cells to TRAIL-mediated apoptosis has driven investigations of TRAIL-receptor gene regulation, though regulation of the TRAIL gene has been less studied. Physiologically, TRAIL serves as a pro-apoptotic effector molecule in the immune surveillance of cancer that is conditionally expressed by immune cells upon stimulation via an interferon-response element that was identified in early studies of the TRAIL gene promoter. Here, we map the TRAIL gene promoter and review studies of TRAIL gene regulation that involve several modalities of gene regulation including transcription factors, epigenetics, single-nucleotide polymorphisms and functionally distinct isoforms.
Keywords: :
Introduction
TNF-related apoptosis-inducing ligand (TRAIL) was initially identified by its sequence homology with other tumor necrosis factor (TNF) family members.Citation1,Citation2 TRAIL has received considerable attention, primarily due to its tumor-selective apoptosis-inducing capability demonstrated in several human cancer cell lines.Citation3 TRAIL is expressed in a variety of human fetal and adult tissues including the spleen, thymus, prostate, small intestine and placenta.Citation1 Contrary to other TNF-family members, membrane-bound TRAIL is conditionally expressed in immune cells such as natural killer (NK) cells, B cells, monocytes and dendritic cells following cytokine stimulation.Citation4-Citation7 Intracellular stores of TRAIL have also been found in polymorphonuclear neutrophilsCitation8-Citation12 that are released after a variety of stimuli.Citation9,Citation13,Citation14
At physiological conditions, TRAIL is capable of binding to four distinct transmembrane receptors in humans: the proapoptotic death receptors DR4 and DR5 or the two decoy receptors DcR1 and DcR2. Both TRAIL and its receptors form homotrimers in a binary complex upon ligand binding.Citation15,Citation16 In mice, TRAIL-R is the only cognate death receptor.Citation17 TRAIL binds to two decoy receptors, DcR1Citation18-Citation20 that lacks an intracellular domain and DcR2Citation21-Citation23 that contains a truncated intracellular domain. The two decoy receptors compete for TRAIL binding with the death receptors at similar binding affinities.Citation19,Citation23 Additionally, TRAIL binds the soluble receptor osteoprotegerin that negatively regulates osteoclastogenesis and bone resorption, though the interaction is much weaker than that of the other TRAIL receptors.Citation24 The X-ray crystallographic structures of trimeric soluble TRAIL reveal a single zinc atom bound between the three monomers that is important for the integrity and activity of TRAIL ().Citation15,Citation16,Citation25-Citation27 Upon binding TRAIL, the two human proapoptotic death receptors DR4Citation18 and DR5Citation3,Citation21,Citation28-Citation30 initiate cell death signaling through colocalization of their intracellular death domains (DD) that occurs after ligand binding ().Citation31
Figure 1. Crystal structure of TRAIL:DR5 complex. Homotrimeric DR5 (gray, light pink,and yellow) bound to homotrimeric TRAIL (cyan, green and magenta). A single zinc atom (green) is found in the center of the complex. Figure was generated using PyMOL software with Protein Data Bank (PBD) accession number 1D4V.Citation26
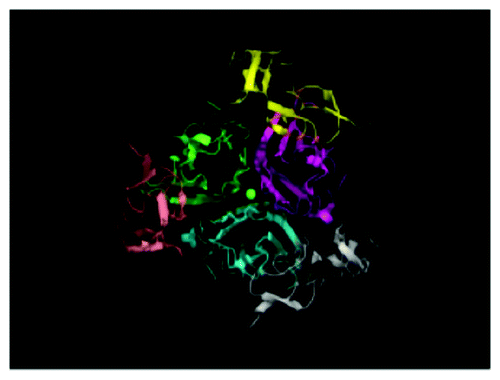
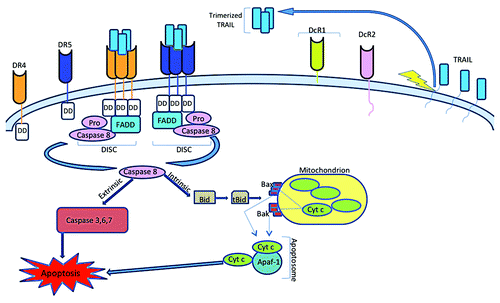
Figure 2. Apoptotic signaling induced by TRAIL. TRAIL initiates cell death by binding to the proapoptotic death receptors DR4 or DR5 that colocalizes their intracellular death domains. This clustering recruits the Fas-associated death domain (FADD) and pro-caspase-8 that results in its activation through autocatalytic cleavage. In type I cells, activate caspase-8 directly activates caspases-3, -6 and -7 to trigger the extrinsic cell death pathway. In type II cells, active caspase-8 cleaves Bid to a truncated form, tBid, which subsequently interacts with proapoptotic Bcl-2 family members Bax and Bak. This interaction leads to permeabilization of the mitochondrial membrane and release of cytochrome c. Cytosolic cytochrome c then combines with Apaf-1 and ATP to form the apoptosome that activates caspase-9 to trigger apoptosis through the caspase cascade.
TRAIL Signaling
Apoptotic signaling
Upon binding to TRAIL, the colocalized DDs of DR4 or DR5 recruit Fas-associated death domain (FADD) and procaspase-8 to form the death inducing signaling complex (DISC). Capase-10 is also recruited to this DISC and is activated at the same rate as caspase-8.Citation32-Citation34 Mice lack a homolog of caspase-10, suggesting that capase-8 and caspase-10 may be functionally redundant. However, the ability of these two caspases to substitute for each other in TRAIL-mediated apoptosis in mammalian cells is unclear due to conflicting reports. The anti-apoptotic protein cellular FLICE-like inhibitory protein (c-FLIP) can also be recruited to DISC to inhibit binding activation of capase-8 and -10 by directly competing for binding to FADD.Citation35,Citation36
At the DISC, procaspase-8 is activated by autocatalytic cleavage to yield caspase-8, which can cleave the effector caspases-3, -6 and -7 to induce apoptosis by the intrinsic death pathway. Cells that undergo cell death via this pathway are known as type I cells. Activation of caspase-8 in type II cells, however, is not sufficient to trigger the caspase cascade. In type II cells, caspase-8 cleaves Bid to form tBid, which primarily interacts with Bax and Bak at the mitochondrial membrane to promote cytochrome c release.Citation37,Citation38In the cytosol, cytochrome c binds to apoptotic peptidase activating factor 1 (Apaf-1) and caspase-9 to form the apoptosome, which initiates the caspase cascade. Permeabilization of the mitochondrial membrane also releases Smac/DIABLO, which inhibits X-linked inhibitor of apoptosis protein (XIAP) to allow for complete activation of the effector caspases.Citation39-Citation42
Physiological Roles for TRAIL
TRAIL is expressed on the surface of immune effector cells such as natural killer cells, macrophages, dendritic cells and cytotoxic T cells in response to cytokines, particularly interferon-gamma that possesses a response element in the TRAIL gene promoter.Citation43 TRAIL-knockout mice or zebrafish do not display any gross developmental defects,Citation25,Citation44 though the mice are more susceptible to carcinogen-induced sarcomas and metastasis.Citation44 TRAIL-R knockout mice develop normally but have an enlarged thymus and decreased radiation-induced apoptosis in several tissues.Citation45 TRAIL-knockout mice also do not induce thymocyte apoptosis, which is deregulated in autoimmune diseases.Citation46 Clinical observations have also suggested a role for TRAIL in autoimmune diseases as patients with systemic lupus erythmatosus or multiple sclerosis have elevated serum levels of soluble TRAIL.Citation47,Citation48 TRAIL has also been implicated in cardiovascular problems such as atherosclerosisCitation49,Citation50and diabetes.Citation51
Regulation of the Human TRAIL Gene
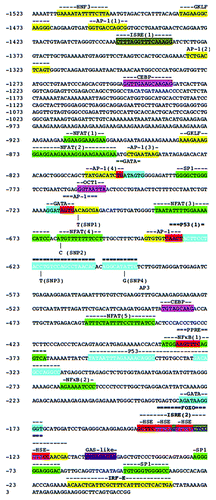
The human TRAIL gene is tightly regulated, potentially due to its considerable apoptotic potential and its involvement in some immune responses. The first report to clone the TRAIL gene promoter characterized a ~1.6kB promoter region, which is 97 bp upstream of the start of translation.Citation52 This report identified several putative transcription factor binding sites in the TRAIL gene promoter: NHF3, GKLF, AP-1, CEBP, NFAT, GATA and Interferon-γ-activated sequence (GAS), GSP1, GSP2 and GSP4. Sequential deletions of the human TRAIL gene promoter driving luciferase reporters in Caco-2 cells suggested that the regions between -1371 to -819 and -165 to -35 contain critical elements that are responsible for basal TRAIL gene transcription. Increased TRAIL gene transcription has been reported in responce to several cytokines and in some cases have been ascribed to transcription factors () with consensus binding sequences found in the TRAIL gene promoter ().
Figure 3. Molecules that alter human TRAIL gene transcription. Interferons (IFN) activate TRAIL gene transcription through ISRE and IRFE sequences in the promoter region. Mutant HRAS (G12V) silences TRAIL gene expression through hypermethylation of CpG islands in the TRAIL gene promoter. Green arrows indicate activating relationships and the red lines indicate inhibitory relationships.
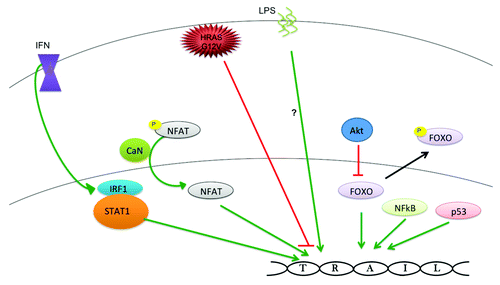
Figure 4. Sequence analysis of the human TRAIL gene promoter. Putative binding sites indicated by highlights above the appropriate sequences. Nucleotides contained in two or more putative binding sites are highlighted in red. Binding sites that have been empirically demonstrated to affect TRAIL promoter activity in experiment are bolded. Vertical lines below the sequence indicate SNPs. TRAIL sequence obtained from Accession number AF178756.
Interferons
Interferons are cytokines that are intimately involved in the immune response in a variety of response by causing immune cell recruitment and/or activation by several mechanisms. Luciferase reporter experiments found that interferon-γ (IFN-γ) was capable of upregulting TRAIL gene promoter activity by 2-fold in a region between -165 and -35,Citation52 which provided a molecular explanation for a previous report that found rapid induction of TRAIL following incubation with IFN-γ 5. STAT1 and IRF1 are thought to directly mediate the effects of IFN-γ on the TRAIL promoter. IFN-γ has also been shown to induce FasL- and TRAIL-dependent apoptosis in lung cancer cellsCitation53 and is responsible for IL18- and TLR3-induced TRAIL expression in NK cells.Citation54
There are reports that suggest type I interferons (IFN-α and -β) induce the TRAIL gene more strongly that IFN-γ.Citation43 IFN-α and -β potently induce TRAIL in CD4+ and CD8+ peripheral blood T cells following CD3-stimulation as well as Jurkat T cells.Citation55 IFN-α has also been shown to induce TRAIL in macrophagesCitation56 and in lymphoma cells in a JNK-dependent manner.Citation57 IFN-β induces TRAIL in colorectal cancer cell lines by a Stat-1-dependent mechanism.Citation58 Taking the evidence together, IFN- α, -β, -γ have been shown to induce TRAIL gene transcription, though the robustness of TRAIL induction among them seems to be context-dependent.
NFAT
The human TRAIL gene is also positively regulated by the transcription factor NFAT, which is activated by calcineurin-mediated dephosphorylation that occurs during T-cell activation. Wang et al. investigated the role of NFAT regulation of the TRAIL gene promoter due to the two putative binding sites they previously noted along with three other newly found potential NFAT binding sites.Citation52,Citation59 Among the five NFAT family members, NFATc1 was by far the most potent positive regulator of TRAIL gene transcription. Interestingly, NFAT-dependent TRAIL gene upregulation was not affected by deletion of their putative binding sites in TRAIL gene promoter luciferase reporters but instead was abrogated by deletion of the -165 to -35 region. Chromatin-immunoprecipitation and electrophoretic mobility shift assay (EMSA) experiments revealed that NFATc1 antagonizes SP-1 binding to the TRAIL gene promoter. This reports highlights a mechanism that immune cells such as cytotoxic T cells might utilize to upregulate TRAIL and underscores the need to directly test binding sites to accurately delineate transcriptional regulation. These observations also suggested that SP-1 might negatively regulate the TRAIL gene promoter.
SP-1
The study that described NFAT-mediated regulation of the TRAIL promoter also found that SP-1 represses TRAIL gene transcription, at least in human intestinal cells.Citation59 However, a recent study reported a positive regulation of TRAIL gene transcription by SP-1 in vascular smooth muscle cells that appears to involve SP-1 phosphorylated at Thr453.Citation60 Interestingly, phosphorylation of SP-1 at Thr453 and Thr739 by p38 mitogen-activated protein kinase appears to be essential for its positive regulation of the VEGF gene.Citation61 Future studies need to further examine if these differences are due to the difference in cell type or if this phosphorylation event modulates SP-1-mediated TRAIL gene regulation. The mechanism of HDACi-induced upregulation of TRAIL gene transcription appeared to involve SP-3.Citation62
NFκB
Inhibition of the prosurvival transcription factor NFκB in Jurkat T cells and primary T lymphocytes revealed that NFκB positively regulates TRAIL gene expression in a manner that depends on the NFκB binding site 1Citation63 (). NFκB also upregulates FasL and together, these proapoptotic ligands may be responsible for tumor-cell elimination in immune surveillance and/or attenuation of T-cell activation to prevent autoimmunity. NFκB is also responsible for constitutive TRAIL expression in human T-cell leukemia virus type I (HTLV-1)-induced leukemia, though other concomitant effects of NFκB seem to cause resistance to TRAIL-mediated apoptosis.Citation64 In accordance with a positive regulatory role for NFκB with respect to the TRAIL gene, the prostaglandin 15d-PGJ2 represses TRAIL gene transcription by inhibiting the binding of NFκB to the NFκB binding site 1 (). This study also identified the transcription factor HSF-1 as a negative regulator of the TRAIL gene that is involved in 15d-PGJ2-induced repression of TRAIL gene transcription. This negative regulation of HSF-1 is completely dependent on its DNA binding domain and is in contrast to its positive regulation of the FasL gene.Citation65 Cnb, the regulatory subunit of calcineurin, was recently shown to active NFκB-mediated TRAIL expression through direct binding to the integrin CD11b.Citation66
P53
Two potential p53 binding sites in the TRAIL gene promoter were identified following the observation that p53-inducing chemotherapies such as 5-fluorouracil and doxorubicin elevated TRAIL promoter activity.Citation67 One of these binding sites at the -630 position was shown to mediate p53-induced TRAIL promoter activity using luciferase reporter assays. Radiation has also been reported to induce TRAIL expression and it has been hypothesized that TRAIL mediates the bystander effects observed following radiation.Citation68,Citation69 The role of p53 in radiation-induced TRAIL expression should be explored since it is well accepted that ionizing radiation robustly induces p53.
FOXO
Overexpression of Foxo3a in prostate cancer cells was found to induce TRAIL gene transcription by gene expression profiling.Citation70 In silico analysis revealed a binding site in the TRAIL gene promoter between -121 and -138 that was validated by luciferase reporter construct mutations and EMSAs to be completely responsible for Foxo3a-induced TRAIL gene promoter activity. This study also revealed that TRAIL expression is significantly decreased in metastatic prostate cancer. Foxo3a-dependent TRAIL upregulation has been recently described as the apoptotic mechanism responsible for memory B cell loss that results from chronic HIV infection.Citation71
Unexplored binding sites
Clearly there are several putative transcription factor-binding sites within the TRAIL gene promoter that have been unexplored. It is worth noting that only a few of the examined binding sites for a given transcription factor have turned out to be functionally important, as in the case of NFκB, AP-1 and SP-1, which have multiple putative binding sites in the TRAIL gene promoter. For example, Oct-1 is involved in regulating several housekeeping genes and has a putative binding site on the TRAIL promoter. Interestingly, HDACi have been found to induce Gadd45 expression through Oct-1.Citation72 This observation along with the report that HDACi induces TRAIL gene transcription suggests that the direct examination of Oct-1 binding to the TRAIL gene promoter may be worth further investigation. The role of the heat shock elements in the proximal region of the TRAIL gene promoter may also be worth investigation as heat shock has been linked to protection from TRAIL-mediated cell death.Citation73
The MAPK pathway
Cells transformed with HRASG12V were reported to have silenced expression of the TRAIL gene due to hypermethylation of CpG islands in the TRAIL gene promoter that lie ~2,000 bp upstream of the start of transcription.Citation74 Despite this silencing, TRAIL could still be induced by IFN-γ in these transformed cells and the silencing could be reversed with decitabine, a DNA methyltransferase inhibitor. Oncogenic mutations in KRAS during colon cancer progression are a common event in colon cancer.Citation75-Citation77 This finding may explain the silencing of TRAIL expression that has been noted in colon cancer, particularly during the progression from adenoma to carcinoma.Citation78 Interestingly, oncogenic RAS sensitizes transformed cells to TRAIL-mediated apoptosis, potentially through MEK-dependent upregulation of DR4 and DR5.Citation79 Thus, concomitant silencing of TRAIL expression may be required to prevent induction of TRAIL-mediated apoptosis during Ras transformation that commonly occurs in tumorigenesis.
Other inducers
A recent report found that pigment epithelium-derived factor (PEDF) induced the expression of TRAIL on the surface of macrophages.Citation80 Further analysis revealed that PEGF also induces peroxisome proliferator-activated receptor-gamma (PPARγ), which binds to the TRAIL promoter to upregulate transcription at a PPAR-response element (PPRE). This represents yet another mechanism utilized by immune cells to induce the expression of TRAIL. Other molecules have been reported to induce TRAIL gene transcription but the underlying direct transcriptional mechanism has not been elucidated. Lipopolysaccharide (LPS), which is a component of gram-negative bacteria, has been reported to induce TRAIL gene transcription at higher concentrations,Citation81 though an early report did not observe TRAIL-induction by LPS at lower doses.Citation5
Regulation of TRAIL activity by isoforms
The human TRAIL gene spans ~20 kb and contains five exons and four introns that contain typical splice acceptor-AG/GT-splice donor consensus sites at their boundaries.Citation43,Citation82 The first exon encodes for the 21 amino acid transmembrane domain and the 17 amino acid cytoplasmic domain. Exons 4 and 5 encode for the amino acids in the extracellular domain that are responsible for the interaction of TRAIL with its receptors. Exon 5 also encodes for the C-terminal amino acids along with containing the 3′-UTR and poly-A tail.
TRAIL is well known for its potent and cancer-selective apoptotic activity. However, it seems that this activity is unique to only one specific isoform of TRAIL among the nine variants that have been reported to date (). The first report of TRAIL isoforms identified three variants that involved variable inclusion of exons 2 and 3: the full length TRAILα, TRAILβ that lacks exon 3, and TRAILγ that lacks exons 2 and 3.Citation83 Computational analysis of exons 2 and 3 revealed that both exons were flanked by consensus splice donor and splice acceptor sequences that are involved in post-translational alternative splicing. Interestingly, TRAILα and TRAILβ where localized to the cytoplasm whereas TRAILγ was associated with the nuclear and cell surface membrane. Sequence analysis of TRAIL mRNA in granulosa tumor cells identified TRAILδ, which lacks exons 3 and 4.Citation84 The truncated TRAIL isoforms do not induce apoptosis like the full-length TRAIL (TRAILα) and it has been suggested that these variants are negative regulators of their full-length counterpart. No TRAIL variants other than the full length TRAIL have been reported in murine cells.
Figure 5. Genomic structure of human TRAIL variants. Exonic sequences of the full-length TRAIL are shown in green whereas novel sequences are shown in yellow.
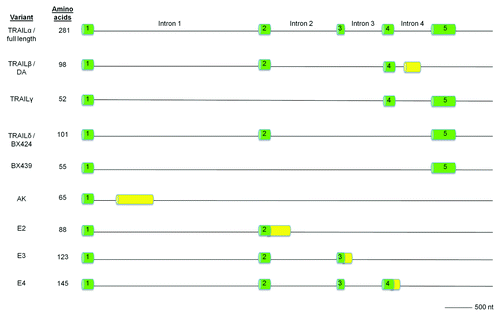
Recently, seven alternatively spliced TRAIL truncated variants were identified that were incapable of potentiating apoptosis: AK, E2, E3, E4, DA, BX424 and BX439.Citation85 All of these isoforms contain common N-terminal sequences and possess the transmembrane helix but vary in the C-terminal region. The DA isoform lacks exon 3 and encodes for the same protein as TRAILβ and BX424 lacks exons 3 and 4, yielding the same protein as TRAILδ. BX439 completely lacks exons 2–4 but possesses the same exons 1 and 5 as the full length TRAIL. AK and DA contain a unique exon not shared by any other reported isoforms. E2, E3 and E4 contain exons 1–2, 1–3 and 1–4, respectively, with an extended sequence at C-terminus. All of these variants, however, activated NFκB and the authors note the possibility that truncated TRAIL may have intracellular activity. Future studies should directly examine the ability of these various TRAIL isoforms to bind to TRAIL receptors through in vitro studies.
The abundance of these isoforms varied significantly across the tested cancer cell lines with THP-1 human leukemia cell line and BT-325 human glioma cell line expressing most of these variants. A notable exception among the cell line panel was the human colon cancer cell line HCT116 that only expressed the full-length isoform.
SNPs in the TRAIL gene
Due to the altered expression of TRAIL in various disease settings, there have been multiple efforts to identify single nucleotide polymorphisms (SNPs) in various patient populations. SNP analysis of peripheral blood samples found that having a T instead of a C at position -723 was significantly associated with sporadic breast cancer and decreased TRAIL mRNA levels (SNP1)Citation86 (–). Luciferase reporter assays in cell lines indicated that this mutation from C to T indeed repressed TRAIL transcriptional activity. In silico analysis found that this mutation is predicted to create an SP-1 binding site. The authors propose that SP3 is negatively regulating this SP1 site, though future studies will need to validate the mutation-induced putative binding site and directly evaluate this possibility.
Another SNP analysis of the TRAIL gene promoter revealed four SNPs that were highly polymorphic in healthy individuals.Citation87 However, these SNPs were not significantly associated with changes in TRAIL mRNA levels or with multiple sclerosis as a diagnostic or prognostic marker. The authors of the study note that SNP(1) is in an AP1 binding site, though they do not comment on other binding sites predicted at the other SNP sites. Interestingly, both SNP3 and SNP4 each lie in p53 response element half-sites that was previously shown to be important for TRAIL gene upregulation in response to chemotherapy.Citation67 While these SNPs were not associated with basal TRAIL levels in this population, it would be interesting to determine if these SNPs affect p53-induced TRAIL gene expression and response in chemotherapy-treated cancer patients.
A SNP at the -716 position of the TRAIL gene promoter was identified but not associated with prostate cancer. Other TRAIL gene promoter SNP associations have been identified in other disease settings but have not been evaluated for its functional effects on TRAIL gene transcription such as -1525/-1595 in fatty liver disease.Citation88 SNPs have been identified in coding regions such as position 1595 in exon 5 being linked with multiple sclerosis.Citation89 A small cohort study of healthy volunteers found five SNPs in TRAIL exons: three in the 3′-UTR at 1525, 1588 and 1595 whereas the other two were in in exon 1 and two at positions 192 and 912.Citation90 These two polymorphisms in the coding region do not alter the encoded amino acid sequence. Another SNP study in bronchial asthma patients found five SNPs in the TRAIL gene with one SNP being in a coding region at 825 and five SNPs being in the 3′UTR at 1053, 1202, 1438, 1501 and 1508.Citation91 Future studies should further investigate the functional effects of identified SNPs on TRAIL gene transcription and therapeutic response to chemotherapies.
TRAIL expression in disease and physiology
Altered expression levels of TRAIL been noted in several diseases relative to healthy controls. For instance, TRAIL mRNA levels are elevated in patients with multiple sclerosis.Citation92 Patients with systemic lupus erythematous or multiple sclerosis have elevated serum levels of soluble TRAIL.Citation47,Citation48 These clinical observations support a critical role for TRAIL in preventing autoimmune disorders, which is in line with the observation that TRAIL-knockout mice cannot induce thymocyte apoptosis.Citation46 Breast cancer patients with brain metastases have downregulated TRAIL mRNA levels.Citation93 TRAIL gene expression silencing has also been noted in metastatic prostate cancerCitation70 and colon cancer.Citation78 A role for TRAIL has been implicated in particular types of cellular differentiation such as colonic epithelial cells through a reciprocal expression relationship with PKCε.Citation94 TRAIL-receptor signaling has also been implicated in the DNA damage response to radiation as well as the late effects of radiation.Citation45,Citation95
Conclusion
The role of TRAIL as an effector molecule in the immune system and its apoptotic potential is reflected in the regulation of the TRAIL gene. Regulation of the TRAIL gene appears tightly controlled by transcriptional mechanisms that respond to interferon stimulation or are involved in immune cell activation as well as transcription factors that are tumor suppressors such as p53 and Foxo3a. Assimilating our current knowledge of TRAIL gene regulation, it is clear that the regulation is multimodal and can be highly context-dependent. The emerging evidence on TRAIL isoforms and TRAIL gene SNPs add other layers of complexity to how cells can tune the TRAIL gene to alter its effects. Mapping transcription factor binding sites and SNPs in the TRAIL gene promoter revealed several overlapping sites that should be examined in future studies. A detailed understanding of TRAIL gene upregulation will be better defined how TRAIL is utilized by the immune system and how it is altered in diseases. Such studies also have the potential to yield drug targets that can harness the antitumor potential of TRAIL.
References
- Wiley SR Sr., Schooley K, Smolak PJ, Din WS, Huang CP, Nicholl JK, et al. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity 1995; 3:673 - 82; http://dx.doi.org/10.1016/1074-7613(95)90057-8; PMID: 8777713
- Pitti RM, Marsters SA, Ruppert S, Donahue CJ, Moore A, Ashkenazi A. Induction of apoptosis by Apo-2 ligand, a new member of the tumor necrosis factor cytokine family. J Biol Chem 1996; 271:12687 - 90; http://dx.doi.org/10.1074/jbc.271.22.12687; PMID: 8663110
- Walczak H, Degli-Esposti MA, Johnson RS, Smolak PJ, Waugh JY, Boiani N, et al. TRAIL-R2: a novel apoptosis-mediating receptor for TRAIL. EMBO J 1997; 16:5386 - 97; http://dx.doi.org/10.1093/emboj/16.17.5386; PMID: 9311998
- Fanger NA, Maliszewski CR, Schooley K, Griffith TS. Human dendritic cells mediate cellular apoptosis via tumor necrosis factor-related apoptosis-inducing ligand (TRAIL). J Exp Med 1999; 190:1155 - 64; http://dx.doi.org/10.1084/jem.190.8.1155; PMID: 10523613
- Griffith TS, Wiley SR, Kubin MZ, Sedger LM, Maliszewski CR, Fanger NA. Monocyte-mediated tumoricidal activity via the tumor necrosis factor-related cytokine, TRAIL. J Exp Med 1999; 189:1343 - 54; http://dx.doi.org/10.1084/jem.189.8.1343; PMID: 10209050
- Kemp TJ, Moore JM, Griffith TS. Human B cells express functional TRAIL/Apo-2 ligand after CpG-containing oligodeoxynucleotide stimulation. J Immunol 2004; 173:892 - 9; PMID: 15240676
- Zamai L, Ahmad M, Bennett IM, Azzoni L, Alnemri ES, Perussia B. Natural killer (NK) cell-mediated cytotoxicity: differential use of TRAIL and Fas ligand by immature and mature primary human NK cells. J Exp Med 1998; 188:2375 - 80; http://dx.doi.org/10.1084/jem.188.12.2375; PMID: 9858524
- Cassatella MA, Huber V, Calzetti F, Margotto D, Tamassia N, Peri G, et al. Interferon-activated neutrophils store a TNF-related apoptosis-inducing ligand (TRAIL/Apo-2 ligand) intracellular pool that is readily mobilizable following exposure to proinflammatory mediators. J Leukoc Biol 2006; 79:123 - 32; http://dx.doi.org/10.1189/jlb.0805431; PMID: 16244105
- Kemp TJ, Ludwig AT, Earel JK, Moore JM, Vanoosten RL, Moses B, et al. Neutrophil stimulation with Mycobacterium bovis bacillus Calmette-Guerin (BCG) results in the release of functional soluble TRAIL/Apo-2L. Blood 2005; 106:3474 - 82; http://dx.doi.org/10.1182/blood-2005-03-1327; PMID: 16037389
- Koga Y, Matsuzaki A, Suminoe A, Hattori H, Hara T. Neutrophil-derived TNF-related apoptosis-inducing ligand (TRAIL): a novel mechanism of antitumor effect by neutrophils. Cancer Res 2004; 64:1037 - 43; http://dx.doi.org/10.1158/0008-5472.CAN-03-1808; PMID: 14871835
- Ludwig AT, Moore JM, Luo Y, Chen X, Saltsgaver NA, O’Donnell MA, et al. Tumor necrosis factor-related apoptosis-inducing ligand: a novel mechanism for Bacillus Calmette-Guérin-induced antitumor activity. Cancer Res 2004; 64:3386 - 90; http://dx.doi.org/10.1158/0008-5472.CAN-04-0374; PMID: 15150089
- Tecchio C, Huber V, Scapini P, Calzetti F, Margotto D, Todeschini G, et al. IFNalpha-stimulated neutrophils and monocytes release a soluble form of TNF-related apoptosis-inducing ligand (TRAIL/Apo-2 ligand) displaying apoptotic activity on leukemic cells. Blood 2004; 103:3837 - 44; http://dx.doi.org/10.1182/blood-2003-08-2806; PMID: 14726404
- Simons MP, Leidal KG, Nauseef WM, Griffith TS. TNF-related apoptosis-inducing ligand (TRAIL) is expressed throughout myeloid development, resulting in a broad distribution among neutrophil granules. J Leukoc Biol 2008; 83:621 - 9; http://dx.doi.org/10.1189/jlb.0707452; PMID: 18063697
- Simons MP, Moore JM, Kemp TJ, Griffith TS. Identification of the mycobacterial subcomponents involved in the release of tumor necrosis factor-related apoptosis-inducing ligand from human neutrophils. Infect Immun 2007; 75:1265 - 71; http://dx.doi.org/10.1128/IAI.00938-06; PMID: 17194806
- Cha S-S, Kim M-S, Choi YH, Sung B-J, Shin NK, Shin H-C, et al. 2.8 A resolution crystal structure of human TRAIL, a cytokine with selective antitumor activity. Immunity 1999; 11:253 - 61; http://dx.doi.org/10.1016/S1074-7613(00)80100-4; PMID: 10485660
- Hymowitz SG, Christinger HW, Fuh G, Ultsch M, O’Connell M, Kelley RF, et al. Triggering cell death: the crystal structure of Apo2L/TRAIL in a complex with death receptor 5. Mol Cell 1999; 4:563 - 71; http://dx.doi.org/10.1016/S1097-2765(00)80207-5; PMID: 10549288
- Wu GS, Burns TF, Zhan Y, Alnemri ES, El-Deiry WS. Molecular cloning and functional analysis of the mouse homologue of the KILLER/DR5 tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) death receptor. Cancer Res 1999; 59:2770 - 5; PMID: 10383128
- Pan G, Ni J, Wei Y-F, Yu G, Gentz R, Dixit VM. An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science 1997; 277:815 - 8; http://dx.doi.org/10.1126/science.277.5327.815; PMID: 9242610
- Degli-Esposti MA, Smolak PJ, Walczak H, Waugh J, Huang C-P, DuBose RF, et al. Cloning and characterization of TRAIL-R3, a novel member of the emerging TRAIL receptor family. J Exp Med 1997; 186:1165 - 70; http://dx.doi.org/10.1084/jem.186.7.1165; PMID: 9314565
- Mongkolsapaya J, Cowper AE, Xu X-N, Morris G, McMichael AJ, Bell JI, et al. Lymphocyte inhibitor of TRAIL (TNF-related apoptosis-inducing ligand): a new receptor protecting lymphocytes from the death ligand TRAIL. J Immunol 1998; 160:3 - 6; PMID: 9551946
- Pan G, Ni J, Yu G, Wei Y-F, Dixit VM. TRUNDD, a new member of the TRAIL receptor family that antagonizes TRAIL signalling. FEBS Lett 1998; 424:41 - 5; http://dx.doi.org/10.1016/S0014-5793(98)00135-5; PMID: 9537512
- Marsters SA, Sheridan JP, Pitti RM, Huang A, Skubatch M, Baldwin D, et al. A novel receptor for Apo2L/TRAIL contains a truncated death domain. Curr Biol 1997; 7:1003 - 6; http://dx.doi.org/10.1016/S0960-9822(06)00422-2; PMID: 9382840
- Degli-Esposti MA, Dougall WC, Smolak PJ, Waugh JY, Smith CA, Goodwin RG. The novel receptor TRAIL-R4 induces NF-kappaB and protects against TRAIL-mediated apoptosis, yet retains an incomplete death domain. Immunity 1997; 7:813 - 20; http://dx.doi.org/10.1016/S1074-7613(00)80399-4; PMID: 9430226
- Emery JG, McDonnell P, Burke MB, Deen KC, Lyn S, Silverman C, et al. Osteoprotegerin is a receptor for the cytotoxic ligand TRAIL. J Biol Chem 1998; 273:14363 - 7; http://dx.doi.org/10.1074/jbc.273.23.14363; PMID: 9603945
- Eimon PM, Kratz E, Varfolomeev E, Hymowitz SG, Stern H, Zha J, et al. Delineation of the cell-extrinsic apoptosis pathway in the zebrafish. Cell Death Differ 2006; 13:1619 - 30; http://dx.doi.org/10.1038/sj.cdd.4402015; PMID: 16888647
- Mongkolsapaya J, Grimes JM, Chen N, Xu X-N, Stuart DI, Jones EY, et al. Structure of the TRAIL-DR5 complex reveals mechanisms conferring specificity in apoptotic initiation. Nat Struct Biol 1999; 6:1048 - 53; http://dx.doi.org/10.1038/14935; PMID: 10542098
- Bodmer J-L, Meier P, Tschopp J, Schneider P. Cysteine 230 is essential for the structure and activity of the cytotoxic ligand TRAIL. J Biol Chem 2000; 275:20632 - 7; http://dx.doi.org/10.1074/jbc.M909721199; PMID: 10748154
- Pan G, O’Rourke K, Chinnaiyan AM, Gentz R, Ebner R, Ni J, et al. The receptor for the cytotoxic ligand TRAIL. Science 1997; 276:111 - 3; http://dx.doi.org/10.1126/science.276.5309.111; PMID: 9082980
- MacFarlane M, Ahmad M, Srinivasula SM, Fernandes-Alnemri T, Cohen GM, Alnemri ES. Identification and molecular cloning of two novel receptors for the cytotoxic ligand TRAIL. J Biol Chem 1997; 272:25417 - 20; http://dx.doi.org/10.1074/jbc.272.41.25417; PMID: 9325248
- Wu GS, Burns TF, McDonald ER 3rd, Jiang W, Meng R, Krantz ID, et al. KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat Genet 1997; 17:141 - 3; http://dx.doi.org/10.1038/ng1097-141; PMID: 9326928
- Tartaglia LA, Ayres TM, Wong GHW, Goeddel DV. A novel domain within the 55 kd TNF receptor signals cell death. Cell 1993; 74:845 - 53; http://dx.doi.org/10.1016/0092-8674(93)90464-2; PMID: 8397073
- Kischkel FC, Lawrence DA, Tinel A, LeBlanc H, Virmani A, Schow P, et al. Death receptor recruitment of endogenous caspase-10 and apoptosis initiation in the absence of caspase-8. J Biol Chem 2001; 276:46639 - 46; http://dx.doi.org/10.1074/jbc.M105102200; PMID: 11583996
- Wang J, Chun HJ, Wong W, Spencer DM, Lenardo MJ. Caspase-10 is an initiator caspase in death receptor signaling. Proc Natl Acad Sci U S A 2001; 98:13884 - 8; http://dx.doi.org/10.1073/pnas.241358198; PMID: 11717445
- Sprick MR, Rieser E, Stahl H, Grosse-Wilde A, Weigand MA, Walczak H. Caspase-10 is recruited to and activated at the native TRAIL and CD95 death-inducing signalling complexes in a FADD-dependent manner but can not functionally substitute caspase-8. EMBO J 2002; 21:4520 - 30; http://dx.doi.org/10.1093/emboj/cdf441; PMID: 12198154
- Micheau O, Thome M, Schneider P, Holler N, Tschopp J, Nicholson DW, et al. The long form of FLIP is an activator of caspase-8 at the Fas death-inducing signaling complex. J Biol Chem 2002; 277:45162 - 71; http://dx.doi.org/10.1074/jbc.M206882200; PMID: 12215447
- Thome M, Tschopp J. Regulation of lymphocyte proliferation and death by FLIP. Nat Rev Immunol 2001; 1:50 - 8; http://dx.doi.org/10.1038/35095508; PMID: 11905814
- Li H, Zhu H, Xu CJ, Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 1998; 94:491 - 501; http://dx.doi.org/10.1016/S0092-8674(00)81590-1; PMID: 9727492
- Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 1998; 94:481 - 90; http://dx.doi.org/10.1016/S0092-8674(00)81589-5; PMID: 9727491
- Maas C, Verbrugge I, de Vries E, Savich G, van de Kooij LW, Tait SWG, et al. Smac/DIABLO release from mitochondria and XIAP inhibition are essential to limit clonogenicity of Type I tumor cells after TRAIL receptor stimulation. Cell Death Differ 2010; 17:1613 - 23; http://dx.doi.org/10.1038/cdd.2010.39; PMID: 20395960
- Jost PJ, Grabow S, Gray D, McKenzie MD, Nachbur U, Huang DCS, et al. XIAP discriminates between type I and type II FAS-induced apoptosis. Nature 2009; 460:1035 - 9; http://dx.doi.org/10.1038/nature08229; PMID: 19626005
- Albeck JG, Burke JM, Aldridge BB, Zhang M, Lauffenburger DA, Sorger PK. Quantitative analysis of pathways controlling extrinsic apoptosis in single cells. Mol Cell 2008; 30:11 - 25; http://dx.doi.org/10.1016/j.molcel.2008.02.012; PMID: 18406323
- Sun X-M, Bratton SB, Butterworth M, MacFarlane M, Cohen GM. Bcl-2 and Bcl-xL inhibit CD95-mediated apoptosis by preventing mitochondrial release of Smac/DIABLO and subsequent inactivation of X-linked inhibitor-of-apoptosis protein. J Biol Chem 2002; 277:11345 - 51; http://dx.doi.org/10.1074/jbc.M109893200; PMID: 11801595
- Gong B, Almasan A. Genomic organization and transcriptional regulation of human Apo2/TRAIL gene. Biochem Biophys Res Commun 2000; 278:747 - 52; http://dx.doi.org/10.1006/bbrc.2000.3872; PMID: 11095979
- Cretney E, Takeda K, Yagita H, Glaccum M, Peschon JJ, Smyth MJ. Increased susceptibility to tumor initiation and metastasis in TNF-related apoptosis-inducing ligand-deficient mice. J Immunol 2002; 168:1356 - 61; PMID: 11801676
- Finnberg N, Gruber JJ, Fei P, Rudolph D, Bric A, Kim S-H, et al. DR5 knockout mice are compromised in radiation-induced apoptosis. Mol Cell Biol 2005; 25:2000 - 13; http://dx.doi.org/10.1128/MCB.25.5.2000-2013.2005; PMID: 15713653
- Lamhamedi-Cherradi S-E, Zheng S-J, Maguschak KA, Peschon J, Chen YH. Defective thymocyte apoptosis and accelerated autoimmune diseases in TRAIL-/- mice. Nat Immunol 2003; 4:255 - 60; http://dx.doi.org/10.1038/ni894; PMID: 12577054
- Wandinger K-P, Lünemann JD, Wengert O, Bellmann-Strobl J, Aktas O, Weber A, et al. TNF-related apoptosis inducing ligand (TRAIL) as a potential response marker for interferon-beta treatment in multiple sclerosis. Lancet 2003; 361:2036 - 43; http://dx.doi.org/10.1016/S0140-6736(03)13641-0; PMID: 12814715
- Lub-de Hooge MN, de Vries EGE, de Jong S, Bijl M. Soluble TRAIL concentrations are raised in patients with systemic lupus erythematosus. Ann Rheum Dis 2005; 64:854 - 8; http://dx.doi.org/10.1136/ard.2004.029058; PMID: 15564310
- Schoppet M, Sattler AM, Schaefer JR, Hofbauer LC. Osteoprotegerin (OPG) and tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) levels in atherosclerosis. Atherosclerosis 2006; 184:446 - 7; http://dx.doi.org/10.1016/j.atherosclerosis.2005.10.028; PMID: 16325821
- Michowitz Y, Goldstein E, Roth A, Afek A, Abashidze A, Ben Gal Y, et al. The involvement of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) in atherosclerosis. J Am Coll Cardiol 2005; 45:1018 - 24; http://dx.doi.org/10.1016/j.jacc.2004.12.065; PMID: 15808757
- Corallini F, Celeghini C, Rizzardi C, Pandolfi A, Di Silvestre S, Vaccarezza M, et al. Insulin down-regulates TRAIL expression in vascular smooth muscle cells both in vivo and in vitro. J Cell Physiol 2007; 212:89 - 95; http://dx.doi.org/10.1002/jcp.21006; PMID: 17352408
- Wang Q, Ji Y, Wang X, Evers BM. Isolation and molecular characterization of the 5′-upstream region of the human TRAIL gene. Biochem Biophys Res Commun 2000; 276:466 - 71; http://dx.doi.org/10.1006/bbrc.2000.3512; PMID: 11027498
- Kim K-B, Choi Y-H, Kim I-K, Chung C-W, Kim BJ, Park Y-M, et al. Potentiation of Fas- and TRAIL-mediated apoptosis by IFN-gamma in A549 lung epithelial cells: enhancement of caspase-8 expression through IFN-response element. Cytokine 2002; 20:283 - 8; http://dx.doi.org/10.1006/cyto.2003.2008; PMID: 12633570
- Tu Z, Hamalainen-Laanaya HK, Crispe IN, Orloff MS. Synergy between TLR3 and IL-18 promotes IFN-γ dependent TRAIL expression in human liver NK cells. Cell Immunol 2011; 271:286 - 91; http://dx.doi.org/10.1016/j.cellimm.2011.07.006; PMID: 21802664
- Kayagaki N, Yamaguchi N, Nakayama M, Eto H, Okumura K, Yagita H. Type I interferons (IFNs) regulate tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) expression on human T cells: A novel mechanism for the antitumor effects of type I IFNs. J Exp Med 1999; 189:1451 - 60; http://dx.doi.org/10.1084/jem.189.9.1451; PMID: 10224285
- Solis M, Goubau D, Romieu-Mourez R, Genin P, Civas A, Hiscott J. Distinct functions of IRF-3 and IRF-7 in IFN-alpha gene regulation and control of anti-tumor activity in primary macrophages. Biochem Pharmacol 2006; 72:1469 - 76; http://dx.doi.org/10.1016/j.bcp.2006.06.002; PMID: 16846591
- Yanase N, Hata K, Shimo K, Hayashida M, Evers BM, Mizuguchi J. Requirement of c-Jun NH2-terminal kinase activation in interferon-alpha-induced apoptosis through upregulation of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) in Daudi B lymphoma cells. Exp Cell Res 2005; 310:10 - 21; http://dx.doi.org/10.1016/j.yexcr.2005.06.021; PMID: 16099454
- Choi EA, Lei H, Maron DJ, Wilson JM, Barsoum J, Fraker DL, et al. Stat1-dependent induction of tumor necrosis factor-related apoptosis-inducing ligand and the cell-surface death signaling pathway by interferon beta in human cancer cells. Cancer Res 2003; 63:5299 - 307; PMID: 14500361
- Wang Q, Zhou Y, Weiss HL, Chow C-W, Evers BM. NFATc1 regulation of TRAIL expression in human intestinal cells. PLoS One 2011; 6:e19882; http://dx.doi.org/10.1371/journal.pone.0019882; PMID: 21603612
- Chan J, Prado-Lourenco L, Khachigian LM, Bennett MR, Di Bartolo BA, Kavurma MM. TRAIL promotes VSMC proliferation and neointima formation in a FGF-2-, Sp1 phosphorylation-, and NFkappaB-dependent manner. Circ Res 2010; 106:1061 - 71; http://dx.doi.org/10.1161/CIRCRESAHA.109.206029; PMID: 20150555
- Lin H-H, Lai S-C, Chau L-Y. Heme oxygenase-1/carbon monoxide induces vascular endothelial growth factor expression via p38 kinase-dependent activation of Sp1. J Biol Chem 2011; 286:3829 - 38; http://dx.doi.org/10.1074/jbc.M110.168831; PMID: 21115498
- Nebbioso A, Clarke N, Voltz E, Germain E, Ambrosino C, Bontempo P, et al. Tumor-selective action of HDAC inhibitors involves TRAIL induction in acute myeloid leukemia cells. Nat Med 2005; 11:77 - 84; http://dx.doi.org/10.1038/nm1161; PMID: 15619633
- Baetu TM, Kwon H, Sharma S, Grandvaux N, Hiscott J. Disruption of NF-kappaB signaling reveals a novel role for NF-kappaB in the regulation of TNF-related apoptosis-inducing ligand expression. J Immunol 2001; 167:3164 - 73; PMID: 11544302
- Matsuda T, Almasan A, Tomita M, Uchihara JN, Masuda M, Ohshiro K, et al. Resistance to Apo2 ligand (Apo2L)/tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-mediated apoptosis and constitutive expression of Apo2L/TRAIL in human T-cell leukemia virus type 1-infected T-cell lines. J Virol 2005; 79:1367 - 78; http://dx.doi.org/10.1128/JVI.79.3.1367-1378.2005; PMID: 15650163
- Cippitelli M, Fionda C, Di Bona D, Lupo A, Piccoli M, Frati L, et al. The cyclopentenone-type prostaglandin 15-deoxy-delta 12,14-prostaglandin J2 inhibits CD95 ligand gene expression in T lymphocytes: interference with promoter activation via peroxisome proliferator-activated receptor-gamma-independent mechanisms. J Immunol 2003; 170:4578 - 92; PMID: 12707336
- Su Z, Xin S, Xu L, Cheng J, Guo J, Li L, et al. The calcineurin B subunit induces TNF-related apoptosis-inducing ligand (TRAIL) expression via CD11b-NF-κB pathway in RAW264.7 macrophages. Biochem Biophys Res Commun 2012; 417:777 - 83; http://dx.doi.org/10.1016/j.bbrc.2011.12.034; PMID: 22197822
- Kuribayashi K, Krigsfeld G, Wang W, Xu J, Mayes PA, Dicker DT, et al. TNFSF10 (TRAIL), a p53 target gene that mediates p53-dependent cell death. Cancer Biol Ther 2008; 7:2034 - 8; http://dx.doi.org/10.4161/cbt.7.12.7460; PMID: 19106633
- Unnithan J, Macklis RM. TRAIL induction by radiation in lymphoma patients. Cancer Invest 2004; 22:522 - 5; http://dx.doi.org/10.1081/CNV-200026397; PMID: 15565809
- Shareef MM, Cui N, Burikhanov R, Gupta S, Satishkumar S, Shajahan S, et al. Role of tumor necrosis factor-α and TRAIL in high-dose radiation-induced bystander signaling in lung adenocarcinoma. Cancer Res 2007; 67:11811 - 20; http://dx.doi.org/10.1158/0008-5472.CAN-07-0722; PMID: 18089811
- Modur V, Nagarajan R, Evers BM, Milbrandt J. FOXO proteins regulate tumor necrosis factor-related apoptosis inducing ligand expression. Implications for PTEN mutation in prostate cancer. J Biol Chem 2002; 277:47928 - 37; http://dx.doi.org/10.1074/jbc.M207509200; PMID: 12351634
- van Grevenynghe J, Cubas RA, Noto A, DaFonseca S, He Z, Peretz Y, et al. Loss of memory B cells during chronic HIV infection is driven by Foxo3a- and TRAIL-mediated apoptosis. J Clin Invest 2011; 121:3877 - 88; http://dx.doi.org/10.1172/JCI59211; PMID: 21926463
- Hirose T, Sowa Y, Takahashi S, Saito S, Yasuda C, Shindo N, et al. p53-independent induction of Gadd45 by histone deacetylase inhibitor: coordinate regulation by transcription factors Oct-1 and NF-Y. Oncogene 2003; 22:7762 - 73; http://dx.doi.org/10.1038/sj.onc.1207091; PMID: 14586402
- Ozören N, El-Deiry W. Heat shock protects HCT116 and H460 cells from TRAIL-induced apoptosis. Exp Cell Res 2002; 281:175 - 81; http://dx.doi.org/10.1006/excr.2002.5660; PMID: 12460647
- Lund P, Kotova I, Kedinger V, Khanwalkar H, Voltz E, Hahn WC, et al. Transformation-dependent silencing of tumor-selective apoptosis-inducing TRAIL by DNA hypermethylation is antagonized by decitabine. Mol Cancer Ther 2011; 10:1611 - 23; http://dx.doi.org/10.1158/1535-7163.MCT-11-0140; PMID: 21697397
- Forrester K, Almoguera C, Han K, Grizzle WE, Perucho M. Detection of high incidence of K-ras oncogenes during human colon tumorigenesis. Nature 1987; 327:298 - 303; http://dx.doi.org/10.1038/327298a0; PMID: 2438556
- Bos JL, Fearon ER, Hamilton SR, Verlaan-de Vries M, van Boom JH, van der Eb AJ, et al. Prevalence of ras gene mutations in human colorectal cancers. Nature 1987; 327:293 - 7; http://dx.doi.org/10.1038/327293a0; PMID: 3587348
- Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, et al. Genetic alterations during colorectal-tumor development. N Engl J Med 1988; 319:525 - 32; http://dx.doi.org/10.1056/NEJM198809013190901; PMID: 2841597
- Koornstra JJ, Kleibeuker JH, van Geelen CMM, Rijcken FEM, Hollema H, de Vries EGE, et al. Expression of TRAIL (TNF-related apoptosis-inducing ligand) and its receptors in normal colonic mucosa, adenomas, and carcinomas. J Pathol 2003; 200:327 - 35; http://dx.doi.org/10.1002/path.1364; PMID: 12845629
- Nesterov A, Nikrad M, Johnson T, Kraft AS. Oncogenic Ras sensitizes normal human cells to tumor necrosis factor-alpha-related apoptosis-inducing ligand-induced apoptosis. Cancer Res 2004; 64:3922 - 7; http://dx.doi.org/10.1158/0008-5472.CAN-03-2219; PMID: 15173003
- Ho T-C, Chen S-L, Shih S-C, Chang S-J, Yang S-L, Hsieh J-W, et al. Pigment epithelium-derived factor (PEDF) promotes tumor cell death by inducing macrophage membrane tumor necrosis factor-related apoptosis-inducing ligand (TRAIL). J Biol Chem 2011; 286:35943 - 54; http://dx.doi.org/10.1074/jbc.M111.266064; PMID: 21846721
- Halaas O, Vik R, Ashkenazi A, Espevik T. Lipopolysaccharide induces expression of APO2 ligand/TRAIL in human monocytes and macrophages. Scand J Immunol 2000; 51:244 - 50; http://dx.doi.org/10.1046/j.1365-3083.2000.00671.x; PMID: 10736093
- Shapiro MB, Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res 1987; 15:7155 - 74; http://dx.doi.org/10.1093/nar/15.17.7155; PMID: 3658675
- Krieg A, Krieg T, Wenzel M, Schmitt M, Ramp U, Fang B, et al. TRAIL-beta and TRAIL-gamma: two novel splice variants of the human TNF-related apoptosis-inducing ligand (TRAIL) without apoptotic potential. Br J Cancer 2003; 88:918 - 27; http://dx.doi.org/10.1038/sj.bjc.6600772; PMID: 12644830
- Woods DC, Alvarez C, Johnson AL. Cisplatin-mediated sensitivity to TRAIL-induced cell death in human granulosa tumor cells. Gynecol Oncol 2008; 108:632 - 40; http://dx.doi.org/10.1016/j.ygyno.2007.11.034; PMID: 18191995
- Wang P, Lu Y, Li C, Li N, Yu P, Ma D. Novel transcript variants of TRAIL show different activities in activation of NF-κB and apoptosis. Life Sci 2011; 89:839 - 46; http://dx.doi.org/10.1016/j.lfs.2011.09.003; PMID: 21952139
- Pal R, Gochhait S, Chattopadhyay S, Gupta P, Prakash N, Agarwal G, et al. Functional implication of TRAIL -716 C/T promoter polymorphism on its in vitro and in vivo expression and the susceptibility to sporadic breast tumor. Breast Cancer Res Treat 2011; 126:333 - 43; http://dx.doi.org/10.1007/s10549-010-0900-5; PMID: 20443055
- Weber A, Wandinger K-P, Mueller W, Aktas O, Wengert O, Grundström E, et al. Identification and functional characterization of a highly polymorphic region in the human TRAIL promoter in multiple sclerosis. J Neuroimmunol 2004; 149:195 - 201; http://dx.doi.org/10.1016/j.jneuroim.2003.12.014; PMID: 15020080
- Yan X, Xu L, Qi J, Liang X, Ma C, Guo C, et al. sTRAIL levels and TRAIL gene polymorphisms in Chinese patients with fatty liver disease. Immunogenetics 2009; 61:551 - 6; http://dx.doi.org/10.1007/s00251-009-0389-4; PMID: 19629467
- Kikuchi S, Miyagishi R, Fukazawa T, Yabe I, Miyazaki Y, Sasaki H. TNF-related apoptosis inducing ligand (TRAIL) gene polymorphism in Japanese patients with multiple sclerosis. J Neuroimmunol 2005; 167:170 - 4; http://dx.doi.org/10.1016/j.jneuroim.2005.06.021; PMID: 16040132
- Gray HL, Sorensen EL, Hunt JS, Ober C. Three polymorphisms in the 3′ UTR of the TRAIL (TNF-related apoptosis-inducing ligand) gene. Genes Immun 2001; 2:469 - 70; http://dx.doi.org/10.1038/sj.gene.6363806; PMID: 11781716
- Unoki M, Furuta S, Onouchi Y, Watanabe O, Doi S, Fujiwara H, et al. Association studies of 33 single nucleotide polymorphisms (SNPs) in 29 candidate genes for bronchial asthma: positive association a T924C polymorphism in the thromboxane A2 receptor gene. Hum Genet 2000; 106:440 - 6; http://dx.doi.org/10.1007/s004390000267; PMID: 10830912
- Huang MP, Gomes MA, Hillert J, Hillert J, Huang W-X. Apoptosis mediators fasL and TRAIL are upregulated in peripheral blood mononuclear cells in MS. Neurology 2000; 55:928 - 34; http://dx.doi.org/10.1212/WNL.55.7.928; PMID: 11061246
- Bos PD, Zhang XHF, Nadal C, Shu W, Gomis RR, Nguyen DX, et al. Genes that mediate breast cancer metastasis to the brain. Nature 2009; 459:1005 - 9; http://dx.doi.org/10.1038/nature08021; PMID: 19421193
- Gobbi G, Di Marcantonio D, Micheloni C, Carubbi C, Galli D, Vaccarezza M, et al. TRAIL up-regulation must be accompanied by a reciprocal PKCε down-regulation during differentiation of colonic epithelial cell: implications for colorectal cancer cell differentiation. J Cell Physiol 2012; 227:630 - 8; http://dx.doi.org/10.1002/jcp.22765; PMID: 21465464
- Finnberg N, Klein-Szanto AJP, El-Deiry WS. TRAIL-R deficiency in mice promotes susceptibility to chronic inflammation and tumorigenesis. J Clin Invest 2008; 118:111 - 23; http://dx.doi.org/10.1172/JCI29900; PMID: 18079962