Abstract
MicroRNAs (miRNAs) are a class of post-transcriptional gene regulators with critical functions in normal cellular processes as well as disease processes. They are small molecules with 18~23 nucleotides in length. Since their early discovery in 1993, a large number of miRNAs have been characterized and analyzed to understand their pivotal role and their impact in a myriad of biological processes. Substantial research on miRNA highlights the involvement of these tiny RNAs in the etiopathogenesis of a variety of human diseases such as cancer, neuro-degenerative disorders, diabetes, cardiac hypertrophy and respiratory diseases. In this review, we update on recent advances of the emerging role of miRNAs in breast cancer and their clinical implications.
Keywords: :
Introduction
RNA was originally known to be a messenger, relaying information from DNA of genes to the cytoplasm for protein synthesis. However, the noncoding and the non-messenger part of the RNA molecule is what today’s scientists are fascinated. MicroRNAs (miRNAs) are a class of endogenous small RNAs that have revealed a new level of gene regulation in the cell. They also form vital components of the central dogma machinery and are found widely in the genome of high eukaryotic cells. It is believed that miRNA genes constitute about 1–2% of the known genes in eukaryotes.Citation1,Citation2 They have been shown to regulate key processes of gene activation and suppression. A single miRNA can coordinate multiple pathways involving the regulation of multiple genes. On the contrary, several different miRNAs can together control a single miRNA target. This multifarious role leads to the complexity of gene regulation, thus mediating complex network of eukaryotic cell function,Citation3 including hematopoietic cell differentiation, apoptosis, cell proliferation and organ development.Citation4
MicroRNAs as Master Gene Regulators
It is well known now that miRNAs regulate gene expression through a posttranscriptional mechanism, including mRNA degradation and translation repression. For example, miRNAs accelerate mRNA turnover by allowing the removal of the poly(A) tail from messages to which they are partially complementary.Citation5,Citation6 Deadenylation and the consequent loss of poly(A)-binding protein trigger 5′ decapping, thereby promoting exonucleolytic digestion from the 5′ end.Citation7,Citation8 A number of studies suggest that miRNA-mediated gene expression is also through translation repression. For example, studies have revealed a shift of targeted mRNAs to translationally active polysomes of lower mass, which indicates impaired initiation.Citation9 Other studies have suggested that miRNAs can only inhibit translation, initiated by a cap-dependent mechanism.Citation9,Citation10 Moreover, recent findings implicate that miRNAs cause an m7 G cap-dependent hindrance to the recruitment of 80S ribosomes to mRNA.Citation11-Citation13 Thus, the entire translational repression mechanism can be explained on the basis of the cap binding affinity of the miRNA-binding protein Ago, where the cap is concealed such that it cannot bind to the initiation factor eIF4E.Citation14
In fact, there are a number of proteins which help to mediate translational repression and mRNA degradation. In particular, some of these proteins are integral components of the RNA-induced silencing complex (RISC) that delivers those small RNAs to complementary sites within mRNA.Citation15,Citation16 There is overwhelming evidence suggesting that miRNAs exert their silencing function usually by interactions with the 3′-untranslated region (3′-UTR) of a target gene through imperfect base-pairing. This unique feature implies that a single miRNA can have multiple targets and cellular factors associated with RISC complex may influence miRNA targeting specificity. One consequence of the interactions is that miRNAs could regulate a large number of protein-coding genes. Since many of these miRNA targets are involved in various signaling pathways, their impact on gene expression can be significantly amplified. Because of this feature, miRNAs can function as master gene regulators, impacting a variety of cellular pathways important to normal cellular functions as well as cancer development and progression.
Breast Cancer Subtypes and MicroRNAs
Breast cancer is a complex disease characterized by heterogeneity of genetic alterations. According to American Cancer Society, excluding the cancers of skin, breast cancer is the most common cancer among women, accounting for nearly one in three cancers diagnosed in US women. Technological advancements in the last decade have helped the researches to understand this complex disease more thoroughly. Based on gene expression profile and the phenotype, the breast cancer can be divided into six major subtypes. The six major subtypes are luminal A, luminal B, tumor enriched with human epidermal growth factor receptor 2 (Her2), basal-like, normal-like and claudin-low subtype.Citation17-Citation20 Among them, luminal A is the most common subtype characterized by the expression of estrogen receptor (ER), progesterone receptor (PR), Bcl-2 and absence of Her2. It accounts for 50–60% of the total breast cancer cases.Citation18,Citation19 The luminal B subtype is characterized by the expression of ER, PR and absence of Her2. They can be differentiated from luminal A subtype on the basis of high Ki67 staining which indicates higher proliferation rate.Citation21 Both luminal A and luminal B subtypes are treated with adjuvant endocrine therapy such as tamoxifen and aromatase inhibitors, both of which target the ER signaling. These two tumor subtypes are associated with more favorable prognosis. Her2 positive subtype constitutes 15–20% of all the breast cancer. It is characterized by the high expression of the Her2 gene and high proliferation rate. Her2 positive tumors are treated using trastuzumab, an antibody against Her2. Although the survival rate has improved in the last decade the overall prognosis still remains poor. In contrast to other tumor subtypes, basal-like breast cancer expresses none of the three markers (ER, PR and Her2), accounting for 10–20% of all breast carcinomas, and is often associated with a poor prognosis. Although the treatment includes the conventional chemotherapeutic approach, promising strategies are being developed to treat basal-like breast cancer, such as poly-ADP ribose polymerase-1 inhibitors.Citation20,Citation22
Normal-like subtype is the rarest form of breast cancer subtype which accounts for only 5–10% of all breast carcinomas. Although poorly characterized this subtype has been shown to express ER, Her2 and PR and has a clinical outcome between basal like and luminal A subtype. It has a strong basal epithelial gene expression, are triple negative but lack expression for CK5 and EGFR.Citation20 Some researchers have also expressed their concern over the real existence of this breast cancer subtype as they could not confirm it by microarray approach. Their doubt is based on the possible contamination of cancerous cells with their surrounding normal tissue.Citation23 Claudin-low breast cancer subtype is triple negative in nature characterized by low expression of claudin-3, 4, 7, ocludin and E-cadherin with cancer stem like features. Claudin is a tight junction protein whereas E-cadherin is a calcium dependent cell-cell adhesion glycoprotein. This subtype has relatively low frequency in the range of 12–14% and a poor long-term prognosis.Citation24
Of considerable interest, a unique miRNA expression pattern can be associated with certain subtype. For example, miR-21, miR-210 and miR-221 are significantly overexpressed in triple-negative (ER, PR and Her2) breast cancer, whereas miR-10b, miR-145, miR-205, miR-122a are significantly underexpressed in these cancer types. Furthermore, miR-21, miR-210 and miR-221 correlate with worse patient disease-free and overall survival, and hence they play a significant role in triple-negative primary breast cancers.Citation22 In a separate study, comprehensive miRNA expression profiling was performed in normal basal and luminal breast epithelium cells isolated from plastic surgery. Out of 664 miRNAs analyzed, 116 are expressed in normal breast epithelium, among which eight miRNAs are significantly upregulated in normal basal cells, including let7c, miR-125b, miR-126, miR-127–3p, miR-143, miR-145, miR-146–5p and miR-199a-3p, whereas miR-200c and miR-429 are mostly luminal. Furthermore, miR-126, miR-127, miR-143, miR-145 and miR199 are significantly high in myoepithelioma as compared with luminal and basal-like subtypes. Levels of miR-200c and miR-429 are lower in malignant myoepithelioma than those in luminal and basal-like breast cancer.Citation25 Lowrey et al. used miRNA profiling in 29 early stage breast cancer specimens to determine the status of ER, PR and Her2 based on miRNA signature which could further help determine breast cancer subtype and prognosis. The artificial network analysis identified miR-342, miR-299, miR-217, miR-190, miR-135b, miR-218 as ER specific, miR-520 g, miR-377, miR-527–518a, miR-520f-520c as PR specific and miR-520d, miR-181c, miR-302c, miR-376b, miR-30e as Her2 specific. Further analysis revealed that miR-342 expression was highest in ER and Her2 positive luminal B tumors and lowest in triple-negative tumors whereas miR-520 g expression was downregulated in ER and PR-positive tumors. Importantly, this miRNA signature not only helps determine breast cancer phenotype but also indicate therapeutic targets and markers for prognosis.Citation26
Dysregulation of MicroRNAs in Human Breast Cancer
Several studies have been conducted to identify the miRNAs that are differentially expressed and regulate breast cancer initiation and progression in different breast cancer subtypes. miRNA analysis in 76 breast cancer and 10 normal breast samples led to the identification of 29 miRNAs whose expression is significantly dysregulated with the most significantly dysregulated miRNAs being miR-125b, miR-145, miR-21 and miR-155; 15 of them could correctly predict the nature of the sample analyzed (i.e., tumor or normal breast tissue).Citation27 In order to identify miRNAs capable of regulating transition from ductal carcinoma in situ to invasive ductal carcinoma, 94 biopsies were analyzed and of interest, a nine-miRNA signature was found to be specific for invasiveness and five miRNAs were associated with time to metastasis and overall survival. Specifically, let-7d, miR-210 and miR-221 were downregulated in the ductal carcinoma in situ and upregulated in the invasive transition.Citation28 In another study, 100 breast cancer patients and 89 healthy patients were recruited for miRNA genotyping and expression profiling using peripheral blood to detect and identify characteristic patterns. miR-499, miR-146a and miR-196a-2 in postmenopausal patients and miR-196a-2 in premenopausal patients may distinguish breast cancer patients from healthy individuals.Citation29 In an attempt to identify the specific miRNAs that are co-expressed in the serum and tissue of breast cancer (BC) patients, a genome-wide miRNA analysis by next-generation sequencing was conducted in 13 BC patients and 10 healthy controls. miRNAs found to be co-upregulated include miR-103, miR-23a, miR-29a, miR-222, miR-23b, miR-24 and miR-25. Among these, miR-222 is significantly increased in the serum of BC patients which may serve as a valuable biomarker for differentiating BC patients from normal individuals.Citation30
One of the major reasons for the mortality in breast cancer is metastasis.Citation31 Metastasis is a complex process during which a primary solid tumor invades the adjacent tissue and then spreads to the neighboring as well as the distant parts of the body.Citation32 Epithelial to mesenchymal transition (EMT) is thought to be an important component at the early stages of metastasis which is followed by extravasation, colonization of metastatic tissues and finally process of differentiation which is similar to mesenchymal to epithelial transition (MET).Citation33,Citation34 Several recent studies reported the involvement of miRNAs in the process of metastasis where some miRNAs are remarkably downregulated or eliminated, whereas other miRNAs are significantly accumulated in metastatic breast cancer specimen.Citation35 The miRNAs which promote the metastasis in breast cancer include miR-9,Citation36 miR-10b,Citation37,Citation38 miR-21,Citation39-Citation45 miR-29a,Citation46 miR-155Citation47 and miR-373/520 family.Citation48 For example, miR-9 contributes to the increased cell motility and invasion ability in breast cancer cells by targeting E-cadherin, activating the β-catenin pathway and increasing the vascular endothelial growth factor (VEGF) levels.Citation36 Tristetraprolin is a target for miR-29a, and it regulates EMT and metastasis in breast cancer.Citation46 CD44 is a cell surface molecule involved in multiple physiological cell functions including differentiation, proliferation, migration, angiogenesis and cell survival. Targeting of CD44 by miR-373/520 can enhance the migration and invasion ability in vitro and in vivo. Clinical metastasis samples also showed an inverse correlation between miR-373 and CD44 expression.Citation49 miR-10 family and miR-21 are very important miRNAs contributing to tumor metastasis, and we will discuss more details about them later.
On the other hand, miRNAs as metastasis suppressors in breast cancer include miR-7,Citation50-Citation52 miR-17/20,Citation53,Citation54 miR-22,Citation55-Citation57 miR-30,Citation58,Citation59 miR-31,Citation60-Citation62 miR-126,Citation63-Citation68 miR-145,Citation69-Citation72 miR-146,Citation73,Citation74 miR-193b,Citation75 miR-205,Citation76,Citation77 miR-206,Citation78-Citation80 miR-335,Citation32,Citation81 miR-448,Citation82 miR-661Citation83,Citation84 and let-7.Citation85-Citation87 We select a few miRNA targets to illustrate the important role of these genes in metastasis. The metastasis suppressive function of miR-7 has been attributed to its ability to target epidermal growth factor receptor (EGFR) which regulates important cellular processes like proliferation and differentiation, and is frequently overexpressed in different cancers including breast cancer.Citation52 miR-7 can also inhibit p21-activated kinase 1 expression which is a signaling kinase involved in multiple cancers. Overexpression of miR-7 in highly invasive and tumorigenic breast cancer cell line, MDA-MB-231, results in suppression of invasion ability, motility, anchorage independence and tumorigenesis.Citation88 Although miR-17 may function as an oncogenic miRNA in other types of cancer, miR-17/20 has been reported to target cyclin D1, which is a cell cycle regulatory subunit promoting the G1-S phase transition. Increased cyclin D1 expression has been reported in ~50% of human breast cancers and has an inverse correlation with miR-17/20. Overexpression of miR-17/20 in breast cancer cell lines suppresses cell proliferation and arrests the cell cycle at the G1 phase.Citation54,Citation89 The metastasis suppressor action of this miRNA cluster has been reported in breast cancer where the cell conditioned medium from non-invasive breast cancer cell MCF-7 can inhibit invasion and migration ability of highly invasive MDA-MB-231 cell line. This effect is likely mediated through direct inhibition of IL-8 and inhibition of cytokeratin 8 through cyclin D1.Citation53 miR-22 was reported to inhibit tumor growth and metastasis in vivo using a mouse model of breast carcinoma. Upon overexpression, miR-22 can target the genes involved in senescence which includes CDK6, SIRT1 and Sp1, inducing senescence in addition to decreased cell motility and invasion in vitro.Citation55 A separate study showed that miR-22 can also target estrogen receptor α (ERα) and has inhibitory effect on ERα dependent breast cancer cell proliferation.Citation57 Work in our lab showed that miR-30 can directly target Ubc9, an E2-conjugating enzyme involved in sumoylation. Since Ubc9 has significant role in cell growth and cancer development, its inhibition by overexpressing miR-30 suppresses cell growth. A similar correlation is also seen in breast tumor specimens where Ubc9 expression is about 6-fold higher than in the matched normal breast tissue.Citation58 Moreover, enforced expression of miR-30 can inhibit self-renewal capacity of BT-ICs through Ubc9 and induces apoptosis by targeting ITGB3 expression. Particularly, BT-IC xenograft overexpressing miR-30 can reduce tumorigenesis and lung metastasis in non-obese diabetic/severe combined immunodeficient (NOD/SCID) mouse model.Citation59 One of the well-known tumor suppressor miRNAs is miR-145, which can target multiple gene like IRS-1, mucin-1, c-myc, JAM-A and fascin.Citation69-Citation72 More details about miR-145 will be discussed later. miR-146 is capable of targeting various important suppressors of tumorigenesis and metastasis. When ectopically expressed in metastatic breast cancer cell line MDA-MB-231, miR-146 negatively regulates NFκB via downregulating interleukin receptor associated kinase and TNF associated factor 6.Citation74 miR-146 can be upregulated by breast cancer metastasis suppressor 1 and can directly downregulate EGFR, thus significantly reducing the pulmonary metastasis in the MDA-MB-231 mouse model.Citation73
MicroRNAs and Breast Cancer Stem Cells
One of the major obstacles toward treatment of breast cancer is the fraction of the cells that are able to initiate new tumor growth and are often resistant to standard chemotherapeutic drugs.Citation90 These are the breast cancer stem cells (BCSCs) which are often characterized based on their ability to initiate tumors in immunocompromised or syngeneic mice, self-renewal capacity as measured by tumor formation in secondary mice and also the ability to differentiate into the non-self-renewing cells, which constitute the tumor bulk.Citation91 BCSCs are commonly identified using the characteristic CD44+/CD24-/low marker profile or Aldefluor assays.Citation92,Citation93 It has been proposed that miRNAs contribute to the maintenance of tumor-initiating or stem cell properties and characteristic miRNA expression signature can predict the various features of breast cancer including metastasis.Citation94 miRNA expression profiling studies in the breast cancer stem cells indicates differential expression pattern as compared with normal mammary stem and/or progenitor cells. The study performed by Yu et al. used chemotherapy in mice undergoing passaging of breast cancer SKBR3 cells to selectively enrich for self-renewing BCSCs. The tumors raised are enriched with a high percentage of CD44+/CD24-/low cells with high ability to form mammospheres in vitro and a high rate of forming tumors capable of serial transplantation as xenografts. Importantly, the miRNAs that were significantly downregulated in the BCSC-enriched cell population includes let-7 along with miR-16, miR-107, miR-128 and miR-20b, as compared with the parental and differentiated cells. Furthermore, they provide evidence that let-7 is also reduced in BCSC from clinical cancer specimens and reduced let-7 expression is required to maintain mammospheres. Increased let-7 level inhibits tumor formation in NOD/SCID mice and the tumors are less likely to metastasize.Citation95
Shimono et al. reported that 37 miRNAs are differentially expressed in CD44+/CD24-/low BCSCs as compared with non-tumorigenic cancer cells. In particular, three clusters, miR-200c-141, miR-200b-200a-429 and miR-183–96–182, are significantly downregulated. Importantly, miR-200c inhibits the clonogenicity of breast cancer cells and suppresses the cancer cell growth and induces differentiation. Moreover, miR-200c strongly suppresses the normal mammary outgrowth in vivo and tumor formation driven by human BCSCs. Their work indicates that both normal and BCSCs share common molecular mechanisms regulating stem cell functions.Citation92 Furthermore, Chang et al. showed that p53 directly binds to the miR-200c promoter and transactivates it. Loss of p53 leads to a decreased level of miR-200c and an increase in the expression of EMT and stemness markers, leading to the development of a high tumor grade.Citation96
Han et al. cultured and isolated ALDH1+ and CD44+/CD24-/low cells from breast cancer MCF-7 parental cells to evaluate the role of miR-21 in regulating the BCSC-like cell biological features, especially EMT. They found that HIF-1α and miR-21 are upregulated in the BCSC-like cells and reduction in miR-21 expression by antagomir leads to reversal of EMT, downexpression of HIF-1α, as well as suppression of invasion and migration. This indicates that miR-21 regulates EMT transition in BCSC as well as overexpression of HIF-1α.Citation97
Transforming growth factor and its family has a significant regulatory contribution in normal as well as CSCs. Wang et al. demonstrated that upon exposure to TGF-β there is an increase in the population of BCSCs and this effect is mediated via miR-181. The target of miR-181, ataxia telangiectasia mutated (ATM), is reduced in mammospheres upon TGF-β treatment. When miR-181 is upregulated or ATM is depleted in breast cancer cells, there is an induction in sphere formation. These results led to the conclusion that TGF-β pathway regulates the BCSCs property by interfering with the tumor suppressor ATM through miR-181 which itself is upregulated by TGF-β at the post-transcriptional level.Citation98
Of interest, Hwang et al. isolated a distinct subpopulation of breast cancer stem cells which were PROCR+ /ESA+ instead of usual CD44+/CD24-/low cells and performed miRNA expression profiling which showed miR-495 as the most highly upregulated miRNA in these cells. This miRNA is also upregulated in CD44+/CD24-/low population, highlighting its significance in maintaining BCSC properties. E-cadherin is a direct target of miR-495 expression and upon overexpression, it promotes cell invasion and inhibits REDD1 expression to increase cell proliferation through a posttranscriptional mechanism.Citation99
Key MicroRNAs That Play an Important Role in Breast Cancer
Although a plethora of miRNAs have been reported to be implicated in breast cancer initiation, progression and/or metastasis, we will discuss three groups of miRNAs which can act as oncogene or tumor suppressor or play a role in breast cancer therapy resistance.
Oncogenic miRNAs
Oncogenic miRNAs are frequently upregulated in cancer and they act by repressing the expression of tumor suppressor gene/s mainly involved in apoptosis, cell proliferation, cell migration and invasion and metastasis.Citation100 Following are the few important miRNAs and miRNA families which have been extensively studied and established as oncomirs in breast cancer.
miR-10 family
The miR-10 family constitutes of miR-10a and miR-10b and found to be retained within the Hox cluster of developmental regulators. Since they regulate Hox transcripts, it is likely that this miRNA family may play pivotal roles during the development process.Citation101 miR-10 family has been reported to be dysregulated in various cancers including glioblastoma,Citation102 colon cancer,Citation103 acute myeloid leukemia,Citation104 melanoma,Citation105 pancreatic cancerCitation106 and hepatocellular carcinoma.Citation107 In case of breast cancer, miR-10 family is reported to be involved both in the development and metastasis through miR-10a and miR-10b, respectively.Citation105
miR-10b is highly overexpressed in metastatic breast cancer cells and induces cell migration and invasion in murine xenograft model of breast cancer via targeting HOXD10 gene.Citation37 A recent report indicated that E-cadherin is a potential target for miR-10b and expression level of miR-10b has a positive correlation with tumor size, pathological grading, clinical staging, lymph node metastasis, Her2-positivity and tumor proliferation, but is negatively correlated with ER+, PR+ and E-cadherin levels. miR-10b is induced by transcription factor twist which is able to enhance invasion ability and metastasis in breast cancer cells and animal model by targeting HOXD10 in addition to E-cadherin and Tiam1.Citation37,Citation38,Citation108 miR-10b suppression of HOXD10 results in elevated level of pro-metastatic gene, RHOC, which is a member of Rho GTPase family, known to regulate actin dynamics and hence cell shape and motility.Citation109 miR-10b inhibits Tiam1 (T lymphoma invasion and metastasis)-mediated Rac activation and controls cell-cell adhesion and EMT through E-cadherin and suppresses the ability of breast cancer cells to invade and migrate.Citation110 Hence, suppression of E-cadherin by miR-10b may modulate breast cancer metastasis. Thus, miR-10b could be a biomarker of advanced progression and breast cancer metastasis.Citation108 However, caution should be taken in this regard, because a study conducted in patients with primary breast cancer reveals no significant association between miR-10b levels and metastasis or prognosis.Citation111
miR-21
miR-21 is one of the important miRNAs associated with cell migration and invasion of breast cancer cells and hence contributes to tumor progression and metastasis.Citation112,Citation113 Aberrant expression of miR-21 was first reported by Chan et al. showing markedly elevated levels in human glioblastoma tumor tissues and established glioblastoma cell lines compared with non-neoplastic fetal and adult brain tissues.Citation114 During the same year, Iorio et al. reported that miR-21, along with other miRNAs including miR-125b, miR-145 and miR-155 are also aberrantly expressed in human breast cancer, as shown by microarray and then confirmed by northern blot analyses.Citation115 In consistent with these reports, our work with real-time RT-PCR array analysis showed that miR-21 is highly overexpressed in breast tumors compared with the matched normal breast tissues.Citation116 Through two-dimensional differentiation in-gel electrophoresis of tumors treated with anti-miR-21, we identified the tumor suppressor tropomyosin 1 (TPM1) as a potential target of miR-21.Citation43 Subsequently, we identified two additional direct miR-21 targets, programmed cell death 4 (PDCD4) and maspin, both of which have been implicated in invasion and metastasis.Citation44 The role of maspin (mammary serine protease inhibitor) in breast cancer as invasion and metastasis suppressor is well established because it can suppress invasion ability, angiogenesis and induce apoptosis.Citation117-Citation119 Expression of miR-21 is negatively correlated with expression of PTEN in breast cancer and also correlates with advanced stage and metastasis and poor survival.Citation120,Citation121 It has been observed in breast cancer that transforming growth factor-β (TGF-β) can upregulate the expression of miR-21. Elevated miR-21 expression is associated with aggressive disease status which includes high tumor grade, negative hormone receptor status and ductal carcinoma. Samples collected from 344 patients diagnosed with primary breast cancer indicate that despite no value for prognosis, a high level of miR-21 is associated with poor disease-free survival in early stage patients.Citation122
miR-17~92 cluster
miR-17~92 is a polycistron and is located in a region of DNA that is amplified in human B-cell lymphomas.Citation123 It comprises six mature miRNAs including miR-18b, miR-19b, miR-20a, miR-92, miR-93 and miR-106.Citation124 This miRNA cluster was found to be amplified in lymphomas.Citation125 It was also reported that miR-17~92 cluster is markedly overexpressed in lung cancers, especially with small-cell lung cancer.Citation126 As discussed earlier, this miRNA cluster may play a metastasis suppression role.Citation53 However, miR-17–5p was shown to be highly expressed in invasive MDA-MB-231 breast cancer cells but not in non-invasive MCF-7 breast cancer cells. Ectopic expression of this miRNA in MCF-7 cells can lead to more invasive and migratory phenotypes by targeting HBP1/β-catenin pathway. Similarly, downregulation of miR-17–5p suppresses the migration and invasion of MDA-MB-231 cells in vitro.Citation127
Tumor Suppressor MicroRNAs
let-7 family
Reinhart et al. identified let-7 in Caenorhabditis elegans as an essential factor for cell type determination during embryogenesis.Citation128 The let-7 family comprises of 10 members identified in humans, including let-7a, let-7b, let-7c, let-7d, let-7e, let-7f, let-7g, let-7i, miR-98 and miR-202. Normal physiological functions of let-7 include muscle formation, cell adhesion and regulation of gene expression and development. let-7 biogenesis is regulated by Lin28 which have been shown to act as a post-transcriptional repressor.Citation128,Citation129 let-7 expression is downregulated in numerous types of cancer, including lung cancer,Citation130 gastric cancer,Citation131 colon cancer,Citation132 Burkitt's lymphoma.Citation133 In breast cancer it is lost at an early stage of disease progression.Citation134 Yu et al. investigated the role of let-7 in breast cancer stem cells and used breast tumor initiating cells (BT-IC) and non-BT-IC from primary breast cancer for their self-renewal, differentiation and tumorigenicity. They found that let-7 family miRNAs were markedly reduced in BT-IC and increased with differentiation. Upon overexpression, let-7 reduces proliferation, mammosphere formation of BT-IC in vitro and tumor formation and metastasis in NOD/SCID mice. Targets for let-7 include H-RAS and HMGA2, suggesting that let-7 regulates BT-IC stem cell-like properties by silencing multiple targets.Citation85
miR-200 family
miR-200 family consists of five members which include miR-200a, miR-200b, miR-200c, miR-141 and miR-429. miR-200 family are the suppressors of EMT, which is in part mediated through the regulation of E-cadherin transcriptional repressors ZEB1 and ZEB2 (also known as SIP1).Citation135-Citation137 miR-200 family was found to be lost in invasive breast cancer cell lines with mesenchymal phenotypes and also in regions of metaplastic breast cancer specimens lacking E-cadherin. Ectopic expression of the miR-200 family significantly increases E-cadherin expression and alters cell morphology to an epithelial phenotype.Citation136,Citation137 In this regard, miR-200c regulates breast cancer cell migration, stress □ber formation, migration, invasion and elongation and thus, metastasis by targeting FHOD1 and PPM1F which are direct regulators of the actin cytoskeleton.Citation138 Downregulation of miR-200c is also associated with drug resistance in human breast cancer.Citation139
However, the role of miR-200 family in breast cancer metastasis still remains controversial. Although accumulating evidence suggests that miR-200 family regulates the E-cadherin expression and EMT transition, this does not necessarily predict the metastatic potential. miR-200 family has been shown to regulate PLCG1, BMI1, TGF-β2, FAP-1, ZEB and Suz12, hence acting as tumors suppressor.Citation137,Citation140-Citation148 On the other hand, this miRNA family regulates transcription factor ZEB (Zinc-finger enhancer binding) which is activator of EMT and overexpression of this miRNA in mouse breast cancer cell lines unexpectedly leads to macroscopic metastasis.Citation149 Korpal et al. also strengthen this notion in clinical and experimental models of breast cancer metastasis. Overexpression of miR-200 family in weakly metastatic 4TO7 cells promotes metastatic colonization in part through inhibition of Sec23a mediated secretion of IGFBP4 and Tinag1, both of which act as metastasis suppressors.Citation150 Evidently, further investigation is needed to define its role in metastasis. However, cellular content may also play a role in this aspect.
miR-205
Like miR-200 family, miR-205 is also significantly downregulated in cells that had undergone EMT in response to TGF-β and in the triple-negative primary breast cancers.Citation137,Citation151 In situ hybridization (ISH) analysis of formalin-fixed and paraffin-embedded specimens representing normal and tumor tissue from more than 100 patient cases revealed that expression of miR-205 is restricted to basal epithelium of normal mammary ducts and lobules as compared with reduced or lost expression in matching tumor specimens.Citation152 We also reported that miR-205 is significantly underexpressed in breast tumor compared with the matched normal breast tissue.Citation76 Ectopic expression of miR-205 in breast cancer cells inhibits invasion, proliferation and anchorage independent growth. This effect is in part through direct targeting of Her3 and VEGF-A.Citation76 A separate study by a different group reported that miR-205 can also target Her3 and interferes with the PI3K/Akt survival pathway in addition to improving responsiveness to tyrosine kinase inhibitor drugs like gefitinib and lapatinib.Citation77 p53-induced miR-205 can act as a tumor suppressor in triple negative breast cancer and its re-expression can strongly reduce cell proliferation, cell cycle progression and clonogenic potential in vitro and inhibit tumor growth in vivo. The experimentally validated targets for miR-205 include E2F1 and LAMC1 which are regulators of cell cycle progression and cell adhesion, proliferation and migration, respectively.Citation153
miR-145
Iorio et al. reported that miR-145 is significantly downregulated in breast cancer specimen compared with normal breast tissue, as demonstrated by microarray and northern blot analyses. Like miR-205, miR-145 is significantly underexpressed in breast tumor compared with the matched normal breast tissue. Due to early manifestation of altered miR-145 expression in breast cancer, this miRNA may have a clinical diagnostic potential as a novel biomarker for early cancer detection.Citation152 Spizzo et al. demonstrated the tumor suppression function of miR-145 in breast cancer cell lines wherein miR-145 has pro-apoptotic potential which is dependent on TP53 activation. Furthermore, TP53 activation can, in turn, stimulate miR-145 expression promoting a positive regulatory death loop. In addition, miR-145 can directly target estrogen receptor-α (ER-α) protein expression through direct interaction and promotes apoptosis in both ER-α positive and wild type TP53-expressing breast cancer cells.Citation154 Our lab showed that miR-145 can directly target and silence a well-known oncogene c-Myc, which regulates cell growth and proliferation leading to inhibition of tumor cell growth both in vitro and in vivo. Also, expression of miR-145 is induced transcriptionally by p53 through interaction of p53 response element in the promoter region of miR-145.Citation70 miR-145 also plays a role in suppression of metastasis in breast cancer. For example, while miR-145 has no significant effect on cell growth in metastatic breast cancer cell lines it can inhibit invasion ability of these cells, partly due to the silencing of the metastasis gene mucin 1 (MUC1). This silencing of MUC1 further causes a reduction of β-catenin as well as the cadherin 11, which has oncogenic potential.Citation71 In addition, miR-145 has been shown to target cell-cell adhesion protein, JAM-A and fascin in breast cancer cell lines; suppression of these targets can drastically decrease breast cancer cell motility.Citation72 Kim et al. evaluated the therapeutic potency of miR-145 against breast cancer and found that adenoviral construct of miR-145 (Ad-miR-145) can suppress cell growth and motility in both the in vitro and in vivo systems. Furthermore, a treatment of Ad-miR-145 combined with 5-FU showed a signi□cant anti-tumor effect, as compared with 5-FU alone.Citation155 A study conducted in 106 cases of normal and breast cancer tissues revealed that miR-145 was significantly downregulated in breast tumors. miR-145 can target and regulate N-RAS and VEGF-A at the post-transcriptional level, leading to inhibition of tumor angiogenesis, tumor growth and invasion as N-RAS and VEGF-A are required for these phenotypes.Citation156
MicroRNAs Involved in Resistance to Breast Cancer Therapy
One of the major drawback and obstacle toward breast cancer therapy is chemoresistance. This resistance is largely attributed to genetic and epigenetic modifications. The key genes that are involved in breast cancer drug resistance include multidrug resistance 1 (MDR1) which codes for P-glycoprotein, multidrug resistance-associated protein (MRPs), breast cancer resistant protein (BCRP), glutathione S-transferase π (GSTπ) and DNA repair genes such as OCitation6-methylguanine DNA methyltransferase (MGMT).Citation157,Citation158 The first three genes belong to ATP-binding cassette (ABC)-superfamily multidrug efflux pumps. Their normal physiological function is to pump out the toxic metabolites and xenobiotics to protect the cells.Citation157 The human GSTπ is a phase II detoxification enzyme and has been reported to contribute to chemoresistance in many cancers by mediating the efflux of chemotherapeutic drugs especially cisplatin from the cells.Citation159 MGMT is a DNA repair enzyme and often found to be methylated in brain tumors. It confers chemoresistance by targeting alkylating agents as demonstrated in several cell lines and xenograft models.Citation160,Citation161 Accumulating evidence suggests that chemoresistance and miRNAs are closely related as these miRNAs can target and modulate the key genes involved in breast cancer therapy resistance. For example, MDR1 gene has been shown to be directly regulated by miR-451 and miR-298 in doxorubicin resistant and sensitive MCF-7 and MDA-MB-231 cells. Upon overexpression, these miRNAs can downregulate P-glycoprotein, thereby increasing nuclear localization and cytotoxicity of doxorubicin in resistant breast cancer cells.Citation162,Citation163 Similarly MRP-1 has been reported to be overexpressed in VP-16 resistant MCF-7 cell line as compared with parent cell line. Also, miR-326 expression was shown to be downregulated in MCF-7/VP cells as determined by miRNA microarray, but upon overexpression of miR-326, these cells become sensitive to VP-16 and doxorubicin.Citation164 Other miRNAs regulating the MRP-1 gene includes miR-7 and miR-345; MRP-2 is shown to be regulated by miR-489.Citation162,Citation165 Other important miRNAs that are involved in breast cancer therapy resistance are summarized in along with their drugs and potential targets.
Table 1. miRNAs involved in breast cancer treatment resistance and chemosensitivity along with their target molecules
Another major hurdle toward breast cancer chemotherapy is the involvement of breast cancer stem cells (BCSCs). It is believed that cancer stem cells also contribute to breast cancer development, progression and chemoresistance. These BCSCs are often characterized by the expression of CD44 but absence or low expression of CD24.Citation166 In particular, they can also be distinguished from non-BCSCs in tumor by increased expression of ABC transporter proteins which in turn are responsible for protecting cells from chemotherapy through efflux pumping mechanisms.Citation167 Hence, BCSCs, although they comprise a small proportion of breast cancer population, could be a major contributor for chemoresistance. In this regard, miRNAs play a very significant role as they not only can regulate the formation of cancer stem cells but also they directly impact expression of resistance genes.
Perspective
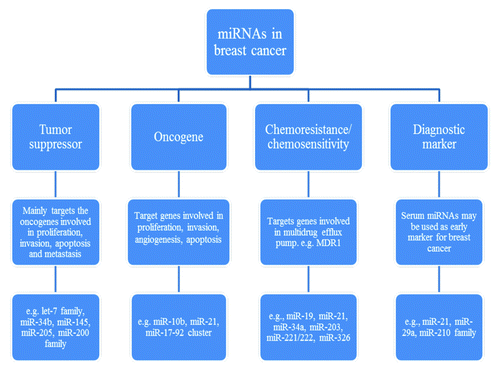
Breast cancer is the best studied cancer in terms of pathobiology, subtypes and treatment, and thus, the role of miRNAs in breast cancer is well characterized. Here, we have discussed three groups of miRNAs, however, due to multiple targeting feature, miRNAs can often regulate several pathways. More importantly, miRNAs exert their silencing function through a complex process which may depend on cellular content, revealing tissue or cell type-specific phenotypes. A better understanding of the miRNA-guided network will open new windows for diagnostics () and therapy of breast cancer. In this regard, miRNAs could be potential biomarkers for predicting a response to systemic therapy and prognosis in clinical settings. For instance, targeting specific miRNAs of the drug-resistant network is promising in overcoming drug resistance in breast cancer. In particular, combinations of different anticancer agents with miRNA reagents such as anti-miR-21 may provide more effective therapeutic approaches. Therefore, the clinical applications of miRNA studies may rely on successful identification of miRNA signatures as cancer biomarkers and development of miRNA-based targeted therapeutics.
| Abbreviations: | ||
| ataxia telangiectasia mutated | = | ATM |
| breast cancer | = | breast cancer resistant protein, BCRP |
| breast cancer | = | BC |
| breast cancer stem cells | = | BCSCs |
| breast tumor initiating cells | = | BT-IC |
| epidermal growth factor receptor | = | EGFR |
| epithelial to mesenchymal transition | = | EMT estrogen receptor, ER |
| human epidermal growth factor receptor 2 | = | Her2 |
| mesenchymal to epithelial transition | = | MET |
| microRNA | = | miRNA |
| multidrug resistance 1 | = | MDR1 |
| multidrug resistance-associated protein | = | MRP |
| non-obese diabetic/severe combined immunodeficient | = | NOD/SCID |
| programmed cell death 4 | = | PDCD4 |
| progesterone receptor | = | PR |
| RNA-induced silencing complex | = | RISC |
| transforming growth factor-beta | = | TGF-β |
| tropomyosin 1 | = | TPM1 |
| vascular endothelial growth factor | = | VEGF |
| Zinc-finger enhancer binding | = | ZEB |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed
References
- Chalfie M, Horvitz HR, Sulston JE. Mutations that lead to reiterations in the cell lineages of C. elegans. Cell 1981; 24:59 - 69; http://dx.doi.org/10.1016/0092-8674(81)90501-8; PMID: 7237544
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75:843 - 54; http://dx.doi.org/10.1016/0092-8674(93)90529-Y; PMID: 8252621
- Kim VN, Nam JW. Genomics of microRNA. Trends Genet 2006; 22:165 - 73; http://dx.doi.org/10.1016/j.tig.2006.01.003; PMID: 16446010
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281 - 97; http://dx.doi.org/10.1016/S0092-8674(04)00045-5; PMID: 14744438
- Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A 2006; 103:4034 - 9; http://dx.doi.org/10.1073/pnas.0510928103; PMID: 16495412
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 2006; 312:75 - 9; http://dx.doi.org/10.1126/science.1122689; PMID: 16484454
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev 2006; 20:1885 - 98; http://dx.doi.org/10.1101/gad.1424106; PMID: 16815998
- Eulalio A, Rehwinkel J, Stricker M, Huntzinger E, Yang SF, Doerks T, et al. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev 2007; 21:2558 - 70; http://dx.doi.org/10.1101/gad.443107; PMID: 17901217
- Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science 2005; 309:1573 - 6; http://dx.doi.org/10.1126/science.1115079; PMID: 16081698
- Humphreys DT, Westman BJ, Martin DI, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci U S A 2005; 102:16961 - 6; http://dx.doi.org/10.1073/pnas.0506482102; PMID: 16287976
- Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science 2007; 317:1764 - 7; http://dx.doi.org/10.1126/science.1146067; PMID: 17656684
- Thermann R, Hentze MW. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature 2007; 447:875 - 8; http://dx.doi.org/10.1038/nature05878; PMID: 17507927
- Wakiyama M, Takimoto K, Ohara O, Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev 2007; 21:1857 - 62; http://dx.doi.org/10.1101/gad.1566707; PMID: 17671087
- Kiriakidou M, Tan GS, Lamprinaki S, De Planell-Saguer M, Nelson PT, Mourelatos Z. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell 2007; 129:1141 - 51; http://dx.doi.org/10.1016/j.cell.2007.05.016; PMID: 17524464
- Höck J, Weinmann L, Ender C, Rüdel S, Kremmer E, Raabe M, et al. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep 2007; 8:1052 - 60; http://dx.doi.org/10.1038/sj.embor.7401088; PMID: 17932509
- Zhang L, Ding L, Cheung TH, Dong MQ, Chen J, Sewell AK, et al. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol Cell 2007; 28:598 - 613; http://dx.doi.org/10.1016/j.molcel.2007.09.014; PMID: 18042455
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A 2003; 100:8418 - 23; http://dx.doi.org/10.1073/pnas.0932692100; PMID: 12829800
- Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 2001; 98:10869 - 74; http://dx.doi.org/10.1073/pnas.191367098; PMID: 11553815
- Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature 2000; 406:747 - 52; http://dx.doi.org/10.1038/35021093; PMID: 10963602
- Eroles P, Bosch A, Pérez-Fidalgo JA, Lluch A. Molecular biology in breast cancer: intrinsic subtypes and signaling pathways. Cancer Treat Rev 2012; 38:698 - 707; http://dx.doi.org/10.1016/j.ctrv.2011.11.005; PMID: 22178455
- Cheang MC, Chia SK, Voduc D, Gao D, Leung S, Snider J, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst 2009; 101:736 - 50; http://dx.doi.org/10.1093/jnci/djp082; PMID: 19436038
- Radojicic J, Zaravinos A, Vrekoussis T, Kafousi M, Spandidos DA, Stathopoulos EN. MicroRNA expression analysis in triple-negative (ER, PR and Her2/neu) breast cancer. Cell Cycle 2011; 10:507 - 17; http://dx.doi.org/10.4161/cc.10.3.14754; PMID: 21270527
- Weigelt B, Mackay A, A’hern R, Natrajan R, Tan DS, Dowsett M, et al. Breast cancer molecular profiling with single sample predictors: a retrospective analysis. Lancet Oncol 2010; 11:339 - 49; http://dx.doi.org/10.1016/S1470-2045(10)70008-5; PMID: 20181526
- Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res 2010; 12:R68; http://dx.doi.org/10.1186/bcr2635; PMID: 20813035
- Bockmeyer CL, Christgen M, Müller M, Fischer S, Ahrens P, Länger F, et al. MicroRNA profiles of healthy basal and luminal mammary epithelial cells are distinct and reflected in different breast cancer subtypes. Breast Cancer Res Treat 2011; 130:735 - 45; http://dx.doi.org/10.1007/s10549-010-1303-3; PMID: 21409395
- Lowery AJ, Miller N, Devaney A, McNeill RE, Davoren PA, Lemetre C, et al. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res 2009; 11:R27; http://dx.doi.org/10.1186/bcr2257; PMID: 19432961
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005; 65:7065 - 70; http://dx.doi.org/10.1158/0008-5472.CAN-05-1783; PMID: 16103053
- Volinia S, Galasso M, Sana ME, Wise TF, Palatini J, Huebner K, et al. Breast cancer signatures for invasiveness and prognosis defined by deep sequencing of microRNA. Proc Natl Acad Sci U S A 2012; 109:3024 - 9; http://dx.doi.org/10.1073/pnas.1200010109; PMID: 22315424
- Alshatwi AA, Shafi G, Hasan TN, Syed NA, Al-Hazzani AA, Alsaif MA, et al. Differential expression profile and genetic variants of microRNAs sequences in breast cancer patients. PLoS One 2012; 7:e30049; http://dx.doi.org/10.1371/journal.pone.0030049; PMID: 22363415
- Wu Q, Wang C, Lu Z, Guo L, Ge Q. Analysis of serum genome-wide microRNAs for breast cancer detection. Clin Chim Acta 2012; 413:1058 - 65; http://dx.doi.org/10.1016/j.cca.2012.02.016; PMID: 22387599
- Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin 2010; 60:277 - 300; http://dx.doi.org/10.3322/caac.20073; PMID: 20610543
- Tavazoie SF, Alarcón C, Oskarsson T, Padua D, Wang Q, Bos PD, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 2008; 451:147 - 52; http://dx.doi.org/10.1038/nature06487; PMID: 18185580
- Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell 2008; 14:818 - 29; http://dx.doi.org/10.1016/j.devcel.2008.05.009; PMID: 18539112
- Baum B, Settleman J, Quinlan MP. Transitions between epithelial and mesenchymal states in development and disease. Semin Cell Dev Biol 2008; 19:294 - 308; http://dx.doi.org/10.1016/j.semcdb.2008.02.001; PMID: 18343170
- Wang L, Wang J. MicroRNA-mediated breast cancer metastasis: from primary site to distant organs. Oncogene 2012; 31:2499 - 511; http://dx.doi.org/10.1038/onc.2011.444; PMID: 21963843
- Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol 2010; 12:247 - 56; PMID: 20173740
- Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007; 449:682 - 8; http://dx.doi.org/10.1038/nature06174; PMID: 17898713
- Moriarty CH, Pursell B, Mercurio AM. miR-10b targets Tiam1: implications for Rac activation and carcinoma migration. J Biol Chem 2010; 285:20541 - 6; http://dx.doi.org/10.1074/jbc.M110.121012; PMID: 20444703
- Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene 2007; 26:2799 - 803; http://dx.doi.org/10.1038/sj.onc.1210083; PMID: 17072344
- Song B, Wang C, Liu J, Wang X, Lv L, Wei L, et al. MicroRNA-21 regulates breast cancer invasion partly by targeting tissue inhibitor of metalloproteinase 3 expression. J Exp Clin Cancer Res 2010; 29:29; http://dx.doi.org/10.1186/1756-9966-29-29; PMID: 20346171
- Connolly EC, Van Doorslaer K, Rogler LE, Rogler CE. Overexpression of miR-21 promotes an in vitro metastatic phenotype by targeting the tumor suppressor RHOB. Mol Cancer Res 2010; 8:691 - 700; http://dx.doi.org/10.1158/1541-7786.MCR-09-0465; PMID: 20460403
- Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene 2008; 27:4373 - 9; http://dx.doi.org/10.1038/onc.2008.72; PMID: 18372920
- Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol Chem 2007; 282:14328 - 36; http://dx.doi.org/10.1074/jbc.M611393200; PMID: 17363372
- Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res 2008; 18:350 - 9; http://dx.doi.org/10.1038/cr.2008.24; PMID: 18270520
- Qi L, Bart J, Tan LP, Platteel I, Sluis Tv, Huitema S, et al. Expression of miR-21 and its targets (PTEN, PDCD4, TM1) in flat epithelial atypia of the breast in relation to ductal carcinoma in situ and invasive carcinoma. BMC Cancer 2009; 9:163; http://dx.doi.org/10.1186/1471-2407-9-163; PMID: 19473551
- Gebeshuber CA, Zatloukal K, Martinez J. miR-29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Rep 2009; 10:400 - 5; http://dx.doi.org/10.1038/embor.2009.9; PMID: 19247375
- Kong W, Yang H, He L, Zhao JJ, Coppola D, Dalton WS, et al. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol 2008; 28:6773 - 84; http://dx.doi.org/10.1128/MCB.00941-08; PMID: 18794355
- Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol 2008; 10:202 - 10; http://dx.doi.org/10.1038/ncb1681; PMID: 18193036
- Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol 2008; 10:202 - 10; http://dx.doi.org/10.1038/ncb1681; PMID: 18193036
- Foekens JA, Sieuwerts AM, Smid M, Look MP, de Weerd V, Boersma AW, et al. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci U S A 2008; 105:13021 - 6; http://dx.doi.org/10.1073/pnas.0803304105; PMID: 18755890
- Reddy SD, Ohshiro K, Rayala SK, Kumar R. MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Res 2008; 68:8195 - 200; http://dx.doi.org/10.1158/0008-5472.CAN-08-2103; PMID: 18922890
- Webster RJ, Giles KM, Price KJ, Zhang PM, Mattick JS, Leedman PJ. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem 2009; 284:5731 - 41; http://dx.doi.org/10.1074/jbc.M804280200; PMID: 19073608
- Yu Z, Willmarth NE, Zhou J, Katiyar S, Wang M, Liu Y, et al. microRNA 17/20 inhibits cellular invasion and tumor metastasis in breast cancer by heterotypic signaling. Proc Natl Acad Sci U S A 2010; 107:8231 - 6; http://dx.doi.org/10.1073/pnas.1002080107; PMID: 20406904
- Yu Z, Wang C, Wang M, Li Z, Casimiro MC, Liu M, et al. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J Cell Biol 2008; 182:509 - 17; http://dx.doi.org/10.1083/jcb.200801079; PMID: 18695042
- Xu D, Takeshita F, Hino Y, Fukunaga S, Kudo Y, Tamaki A, et al. miR-22 represses cancer progression by inducing cellular senescence. J Cell Biol 2011; 193:409 - 24; http://dx.doi.org/10.1083/jcb.201010100; PMID: 21502362
- Patel JB, Appaiah HN, Burnett RM, Bhat-Nakshatri P, Wang G, Mehta R, et al. Control of EVI-1 oncogene expression in metastatic breast cancer cells through microRNA miR-22. Oncogene 2011; 30:1290 - 301; http://dx.doi.org/10.1038/onc.2010.510; PMID: 21057539
- Pandey DP, Picard D. miR-22 inhibits estrogen signaling by directly targeting the estrogen receptor alpha mRNA. Mol Cell Biol 2009; 29:3783 - 90; http://dx.doi.org/10.1128/MCB.01875-08; PMID: 19414598
- Wu F, Zhu S, Ding Y, Beck WT, Mo YY. MicroRNA-mediated regulation of Ubc9 expression in cancer cells. Clin Cancer Res 2009; 15:1550 - 7; http://dx.doi.org/10.1158/1078-0432.CCR-08-0820; PMID: 19223510
- Yu F, Deng H, Yao H, Liu Q, Su F, Song E. Mir-30 reduction maintains self-renewal and inhibits apoptosis in breast tumor-initiating cells. Oncogene 2010; 29:4194 - 204; http://dx.doi.org/10.1038/onc.2010.167; PMID: 20498642
- Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szász AM, Wang ZC, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell 2009; 137:1032 - 46; http://dx.doi.org/10.1016/j.cell.2009.03.047; PMID: 19524507
- Valastyan S, Chang A, Benaich N, Reinhardt F, Weinberg RA. Concurrent suppression of integrin alpha5, radixin, and RhoA phenocopies the effects of miR-31 on metastasis. Cancer Res 2010; 70:5147 - 54; http://dx.doi.org/10.1158/0008-5472.CAN-10-0410; PMID: 20530680
- Valastyan S, Benaich N, Chang A, Reinhardt F, Weinberg RA. Concomitant suppression of three target genes can explain the impact of a microRNA on metastasis. Genes Dev 2009; 23:2592 - 7; http://dx.doi.org/10.1101/gad.1832709; PMID: 19875476
- Tavazoie SF, Alarcón C, Oskarsson T, Padua D, Wang Q, Bos PD, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 2008; 451:147 - 52; http://dx.doi.org/10.1038/nature06487; PMID: 18185580
- Sempere LF, Preis M, Yezefski T, Ouyang H, Suriawinata AA, Silahtaroglu A, et al. Fluorescence-based codetection with protein markers reveals distinct cellular compartments for altered MicroRNA expression in solid tumors. Clin Cancer Res 2010; 16:4246 - 55; http://dx.doi.org/10.1158/1078-0432.CCR-10-1152; PMID: 20682703
- Lim PK, Bliss SA, Patel SA, Taborga M, Dave MA, Gregory LA, et al. Gap junction-mediated import of microRNA from bone marrow stromal cells can elicit cell cycle quiescence in breast cancer cells. Cancer Res 2011; 71:1550 - 60; http://dx.doi.org/10.1158/0008-5472.CAN-10-2372; PMID: 21343399
- Li X, Shen Y, Ichikawa H, Antes T, Goldberg GS. Regulation of miRNA expression by Src and contact normalization: effects on nonanchored cell growth and migration. Oncogene 2009; 28:4272 - 83; http://dx.doi.org/10.1038/onc.2009.278; PMID: 19767772
- Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci U S A 2008; 105:1516 - 21; http://dx.doi.org/10.1073/pnas.0707493105; PMID: 18227515
- Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell 2008; 15:272 - 84; http://dx.doi.org/10.1016/j.devcel.2008.07.008; PMID: 18694566
- Spizzo R, Nicoloso MS, Lupini L, Lu Y, Fogarty J, Rossi S, et al. miR-145 participates with TP53 in a death-promoting regulatory loop and targets estrogen receptor-alpha in human breast cancer cells. Cell Death Differ 2010; 17:246 - 54; http://dx.doi.org/10.1038/cdd.2009.117; PMID: 19730444
- Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, et al. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci U S A 2009; 106:3207 - 12; http://dx.doi.org/10.1073/pnas.0808042106; PMID: 19202062
- Sachdeva M, Mo YY. MicroRNA-145 suppresses cell invasion and metastasis by directly targeting mucin 1. Cancer Res 2010; 70:378 - 87; http://dx.doi.org/10.1158/0008-5472.CAN-09-2021; PMID: 19996288
- Götte M, Mohr C, Koo CY, Stock C, Vaske AK, Viola M, et al. miR-145-dependent targeting of junctional adhesion molecule A and modulation of fascin expression are associated with reduced breast cancer cell motility and invasiveness. Oncogene 2010; 29:6569 - 80; http://dx.doi.org/10.1038/onc.2010.386; PMID: 20818426
- Hurst DR, Edmonds MD, Scott GK, Benz CC, Vaidya KS, Welch DR. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res 2009; 69:1279 - 83; http://dx.doi.org/10.1158/0008-5472.CAN-08-3559; PMID: 19190326
- Bhaumik D, Scott GK, Schokrpur S, Patil CK, Campisi J, Benz CC. Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells. Oncogene 2008; 27:5643 - 7; http://dx.doi.org/10.1038/onc.2008.171; PMID: 18504431
- Li XF, Yan PJ, Shao ZM. Downregulation of miR-193b contributes to enhance urokinase-type plasminogen activator (uPA) expression and tumor progression and invasion in human breast cancer. Oncogene 2009; 28:3937 - 48; http://dx.doi.org/10.1038/onc.2009.245; PMID: 19701247
- Wu H, Zhu S, Mo YY. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res 2009; 19:439 - 48; http://dx.doi.org/10.1038/cr.2009.18; PMID: 19238171
- Iorio MV, Casalini P, Piovan C, Di Leva G, Merlo A, Triulzi T, et al. microRNA-205 regulates HER3 in human breast cancer. Cancer Res 2009; 69:2195 - 200; http://dx.doi.org/10.1158/0008-5472.CAN-08-2920; PMID: 19276373
- Leivonen SK, Mäkelä R, Ostling P, Kohonen P, Haapa-Paananen S, Kleivi K, et al. Protein lysate microarray analysis to identify microRNAs regulating estrogen receptor signaling in breast cancer cell lines. Oncogene 2009; 28:3926 - 36; http://dx.doi.org/10.1038/onc.2009.241; PMID: 19684618
- Di Leva G, Gasparini P, Piovan C, Ngankeu A, Garofalo M, Taccioli C, et al. MicroRNA cluster 221-222 and estrogen receptor alpha interactions in breast cancer. J Natl Cancer Inst 2010; 102:706 - 21; http://dx.doi.org/10.1093/jnci/djq102; PMID: 20388878
- Song G, Zhang Y, Wang L. MicroRNA-206 targets notch3, activates apoptosis, and inhibits tumor cell migration and focus formation. J Biol Chem 2009; 284:31921 - 7; http://dx.doi.org/10.1074/jbc.M109.046862; PMID: 19723635
- Edmonds MD, Hurst DR, Vaidya KS, Stafford LJ, Chen D, Welch DR. Breast cancer metastasis suppressor 1 coordinately regulates metastasis-associated microRNA expression. Int J Cancer 2009; 125:1778 - 85; http://dx.doi.org/10.1002/ijc.24616; PMID: 19585508
- Li QQ, Chen ZQ, Cao XX, Xu JD, Xu JW, Chen YY, et al. Involvement of NF-κB/miR-448 regulatory feedback loop in chemotherapy-induced epithelial-mesenchymal transition of breast cancer cells. Cell Death Differ 2011; 18:16 - 25; http://dx.doi.org/10.1038/cdd.2010.103; PMID: 20798686
- Vetter G, Saumet A, Moes M, Vallar L, Le Béchec A, Laurini C, et al. miR-661 expression in SNAI1-induced epithelial to mesenchymal transition contributes to breast cancer cell invasion by targeting Nectin-1 and StarD10 messengers. Oncogene 2010; 29:4436 - 48; http://dx.doi.org/10.1038/onc.2010.181; PMID: 20543867
- Reddy SD, Pakala SB, Ohshiro K, Rayala SK, Kumar R. MicroRNA-661, a c/EBPalpha target, inhibits metastatic tumor antigen 1 and regulates its functions. Cancer Res 2009; 69:5639 - 42; http://dx.doi.org/10.1158/0008-5472.CAN-09-0898; PMID: 19584269
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 2007; 131:1109 - 23; http://dx.doi.org/10.1016/j.cell.2007.10.054; PMID: 18083101
- Dangi-Garimella S, Yun J, Eves EM, Newman M, Erkeland SJ, Hammond SM, et al. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J 2009; 28:347 - 58; http://dx.doi.org/10.1038/emboj.2008.294; PMID: 19153603
- Cabodi S, Taverna D. Interfering with inflammation: a new strategy to block breast cancer self-renewal and progression?. Breast Cancer Res 2010; 12:305; http://dx.doi.org/10.1186/bcr2563; PMID: 20459595
- Reddy SD, Ohshiro K, Rayala SK, Kumar R. MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Res 2008; 68:8195 - 200; http://dx.doi.org/10.1158/0008-5472.CAN-08-2103; PMID: 18922890
- Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: Cyclin D1: normal and abnormal functions. Endocrinology 2004; 145:5439 - 47; http://dx.doi.org/10.1210/en.2004-0959; PMID: 15331580
- Smalley M, Piggott L, Clarkson R. Breast cancer stem cells: Obstacles to therapy. Cancer Lett 2012; http://dx.doi.org/10.1016/j.canlet.2012.04.023; PMID: 22554712
- McDermott SP, Wicha MS. Targeting breast cancer stem cells. Mol Oncol 2010; 4:404 - 19; http://dx.doi.org/10.1016/j.molonc.2010.06.005; PMID: 20599450
- Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 2009; 138:592 - 603; http://dx.doi.org/10.1016/j.cell.2009.07.011; PMID: 19665978
- Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007; 1:555 - 67; http://dx.doi.org/10.1016/j.stem.2007.08.014; PMID: 18371393
- Wang L, Zhang D, Zhang C, Zhang S, Wang Z, Qu C, et al. A microRNA expression signature characterizing the properties of tumor-initiating cells for breast cancer. Oncol Lett 2012; 3:119 - 24; PMID: 22740866
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 2007; 131:1109 - 23; http://dx.doi.org/10.1016/j.cell.2007.10.054; PMID: 18083101
- Chang CJ, Chao CH, Xia W, Yang JY, Xiong Y, Li CW, et al. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol 2011; 13:317 - 23; http://dx.doi.org/10.1038/ncb2173; PMID: 21336307
- Han M, Wang Y, Liu M, Bi X, Bao J, Zeng N, et al. MiR-21 regulates epithelial-mesenchymal transition phenotype and hypoxia-inducible factor-1α expression in third-sphere forming breast cancer stem cell-like cells. Cancer Sci 2012; 103:1058 - 64; http://dx.doi.org/10.1111/j.1349-7006.2012.02281.x; PMID: 22435731
- Wang Y, Yu Y, Tsuyada A, Ren X, Wu X, Stubblefield K, et al. Transforming growth factor-β regulates the sphere-initiating stem cell-like feature in breast cancer through miRNA-181 and ATM. Oncogene 2011; 30:1470 - 80; http://dx.doi.org/10.1038/onc.2010.531; PMID: 21102523
- Hwang-Verslues WW, Chang PH, Wei PC, Yang CY, Huang CK, Kuo WH, et al. miR-495 is upregulated by E12/E47 in breast cancer stem cells, and promotes oncogenesis and hypoxia resistance via downregulation of E-cadherin and REDD1. Oncogene 2011; 30:2463 - 74; http://dx.doi.org/10.1038/onc.2010.618; PMID: 21258409
- Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol 2007; 302:1 - 12; http://dx.doi.org/10.1016/j.ydbio.2006.08.028; PMID: 16989803
- Lund AH. miR-10 in development and cancer. Cell Death Differ 2010; 17:209 - 14; http://dx.doi.org/10.1038/cdd.2009.58; PMID: 19461655
- Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, et al. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res 2007; 67:2456 - 68; http://dx.doi.org/10.1158/0008-5472.CAN-06-2698; PMID: 17363563
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006; 103:2257 - 61; http://dx.doi.org/10.1073/pnas.0510565103; PMID: 16461460
- Jongen-Lavrencic M, Sun SM, Dijkstra MK, Valk PJ, Löwenberg B. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood 2008; 111:5078 - 85; http://dx.doi.org/10.1182/blood-2008-01-133355; PMID: 18337557
- Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci U S A 2006; 103:9136 - 41; http://dx.doi.org/10.1073/pnas.0508889103; PMID: 16754881
- Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA 2007; 297:1901 - 8; http://dx.doi.org/10.1001/jama.297.17.1901; PMID: 17473300
- Varnholt H, Drebber U, Schulze F, Wedemeyer I, Schirmacher P, Dienes HP, et al. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology 2008; 47:1223 - 32; http://dx.doi.org/10.1002/hep.22158; PMID: 18307259
- Liu Y, Zhao J, Zhang PY, Zhang Y, Sun SY, Yu SY, et al. MicroRNA-10b targets E-cadherin and modulates breast cancer metastasis. Med Sci Monit 2012; 18:BR299 - 308; PMID: 22847191
- Ridley AJ. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol 2006; 16:522 - 9; http://dx.doi.org/10.1016/j.tcb.2006.08.006; PMID: 16949823
- Minard ME, Ellis LM, Gallick GE. Tiam1 regulates cell adhesion, migration and apoptosis in colon tumor cells. Clin Exp Metastasis 2006; 23:301 - 13; http://dx.doi.org/10.1007/s10585-006-9040-z; PMID: 17086355
- Gee HE, Camps C, Buffa FM, Colella S, Sheldon H, Gleadle JM, et al. MicroRNA-10b and breast cancer metastasis. Nature 2008; 455:E8 - 9, author reply E9; http://dx.doi.org/10.1038/nature07362; PMID: 18948893
- Han M, Liu M, Wang Y, Mo Z, Bi X, Liu Z, et al. Re-expression of miR-21 contributes to migration and invasion by inducing epithelial-mesenchymal transition consistent with cancer stem cell characteristics in MCF-7 cells. Mol Cell Biochem 2012; 363:427 - 36; http://dx.doi.org/10.1007/s11010-011-1195-5; PMID: 22187223
- Han M, Liu M, Wang Y, Chen X, Xu J, Sun Y, et al. Antagonism of miR-21 reverses epithelial-mesenchymal transition and cancer stem cell phenotype through AKT/ERK1/2 inactivation by targeting PTEN. PLoS One 2012; 7:e39520; http://dx.doi.org/10.1371/journal.pone.0039520; PMID: 22761812
- Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 2005; 65:6029 - 33; http://dx.doi.org/10.1158/0008-5472.CAN-05-0137; PMID: 16024602
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005; 65:7065 - 70; http://dx.doi.org/10.1158/0008-5472.CAN-05-1783; PMID: 16103053
- Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene 2007; 26:2799 - 803; http://dx.doi.org/10.1038/sj.onc.1210083; PMID: 17072344
- Brew K, Dinakarpandian D, Nagase H. Tissue inhibitors of metalloproteinases: evolution, structure and function. Biochim Biophys Acta 2000; 1477:267 - 83; http://dx.doi.org/10.1016/S0167-4838(99)00279-4; PMID: 10708863
- Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol 2012; 13:283 - 96; PMID: 22473468
- Bailey CM, Khalkhali-Ellis Z, Seftor EA, Hendrix MJ. Biological functions of maspin. J Cell Physiol 2006; 209:617 - 24; http://dx.doi.org/10.1002/jcp.20782; PMID: 17001697
- Huang GL, Zhang XH, Guo GL, Huang KT, Yang KY, Shen X, et al. Clinical significance of miR-21 expression in breast cancer: SYBR-Green I-based real-time RT-PCR study of invasive ductal carcinoma. Oncol Rep 2009; 21:673 - 9; PMID: 19212625
- Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 2008; 14:2348 - 60; http://dx.doi.org/10.1261/rna.1034808; PMID: 18812439
- Qian B, Katsaros D, Lu L, Preti M, Durando A, Arisio R, et al. High miR-21 expression in breast cancer associated with poor disease-free survival in early stage disease and high TGF-beta1. Breast Cancer Res Treat 2009; 117:131 - 40; http://dx.doi.org/10.1007/s10549-008-0219-7; PMID: 18932017
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature 2005; 435:828 - 33; http://dx.doi.org/10.1038/nature03552; PMID: 15944707
- Fassina A, Marino F, Siri M, Zambello R, Ventura L, Fassan M, et al. The miR-17-92 microRNA cluster: a novel diagnostic tool in large B-cell malignancies. Lab Invest 2012; 92:1574 - 82; http://dx.doi.org/10.1038/labinvest.2012.129; PMID: 22964854
- Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res 2004; 64:3087 - 95; http://dx.doi.org/10.1158/0008-5472.CAN-03-3773; PMID: 15126345
- Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res 2005; 65:9628 - 32; http://dx.doi.org/10.1158/0008-5472.CAN-05-2352; PMID: 16266980
- Li H, Bian C, Liao L, Li J, Zhao RC. miR-17-5p promotes human breast cancer cell migration and invasion through suppression of HBP1. Breast Cancer Res Treat 2011; 126:565 - 75; http://dx.doi.org/10.1007/s10549-010-0954-4; PMID: 20505989
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000; 403:901 - 6; http://dx.doi.org/10.1038/35002607; PMID: 10706289
- Wang X, Cao L, Wang Y, Wang X, Liu N, You Y. Regulation of let-7 and its target oncogenes (Review). [Review] Oncol Lett 2012; 3:955 - 60; PMID: 22783372
- Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 2004; 64:3753 - 6; http://dx.doi.org/10.1158/0008-5472.CAN-04-0637; PMID: 15172979
- Zhang HH, Wang XJ, Li GX, Yang E, Yang NM. Detection of let-7a microRNA by real-time PCR in gastric carcinoma. World J Gastroenterol 2007; 13:2883 - 8; PMID: 17569129
- Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull 2006; 29:903 - 6; http://dx.doi.org/10.1248/bpb.29.903; PMID: 16651716
- Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, et al. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res 2007; 67:9762 - 70; http://dx.doi.org/10.1158/0008-5472.CAN-07-2462; PMID: 17942906
- Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, et al. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res 2007; 67:11612 - 20; http://dx.doi.org/10.1158/0008-5472.CAN-07-5019; PMID: 18089790
- Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 2008; 22:894 - 907; http://dx.doi.org/10.1101/gad.1640608; PMID: 18381893
- Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem 2008; 283:14910 - 4; http://dx.doi.org/10.1074/jbc.C800074200; PMID: 18411277
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 2008; 10:593 - 601; http://dx.doi.org/10.1038/ncb1722; PMID: 18376396
- Jurmeister S, Baumann M, Balwierz A, Keklikoglou I, Ward A, Uhlmann S, et al. MicroRNA-200c Represses Migration and Invasion of Breast Cancer Cells by Targeting Actin-Regulatory Proteins FHOD1 and PPM1F. Mol Cell Biol 2011; 32:633 - 51; http://dx.doi.org/10.1128/MCB.06212-11; PMID: 22144583
- Chen J, Tian W, Cai H, He H, Deng Y. Down-regulation of microRNA-200c is associated with drug resistance in human breast cancer. Med Oncol 2011; 29:2527 - 34; http://dx.doi.org/10.1007/s12032-011-0117-4; PMID: 22101791
- Iliopoulos D, Lindahl-Allen M, Polytarchou C, Hirsch HA, Tsichlis PN, Struhl K. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol Cell 2010; 39:761 - 72; http://dx.doi.org/10.1016/j.molcel.2010.08.013; PMID: 20832727
- Uhlmann S, Zhang JD, Schwäger A, Mannsperger H, Riazalhosseini Y, Burmester S, et al. miR-200bc/429 cluster targets PLCgamma1 and differentially regulates proliferation and EGF-driven invasion than miR-200a/141 in breast cancer. Oncogene 2010; 29:4297 - 306; http://dx.doi.org/10.1038/onc.2010.201; PMID: 20514023
- Schickel R, Park SM, Murmann AE, Peter ME. miR-200c regulates induction of apoptosis through CD95 by targeting FAP-1. Mol Cell 2010; 38:908 - 15; http://dx.doi.org/10.1016/j.molcel.2010.05.018; PMID: 20620960
- Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 2008; 22:894 - 907; http://dx.doi.org/10.1101/gad.1640608; PMID: 18381893
- Li S, Wang Q, Wang Y, Chen X, Wang Z. PLC-gamma1 and Rac1 coregulate EGF-induced cytoskeleton remodeling and cell migration. Mol Endocrinol 2009; 23:901 - 13; http://dx.doi.org/10.1210/me.2008-0368; PMID: 19264842
- Iliopoulos D, Polytarchou C, Hatziapostolou M, Kottakis F, Maroulakou IG, Struhl K, et al. MicroRNAs differentially regulated by Akt isoforms control EMT and stem cell renewal in cancer cells. Sci Signal 2009; 2:ra62; http://dx.doi.org/10.1126/scisignal.2000356; PMID: 19825827
- Gibbons DL, Lin W, Creighton CJ, Rizvi ZH, Gregory PA, Goodall GJ, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev 2009; 23:2140 - 51; http://dx.doi.org/10.1101/gad.1820209; PMID: 19759262
- Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep 2008; 9:582 - 9; http://dx.doi.org/10.1038/embor.2008.74; PMID: 18483486
- Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res 2008; 68:7846 - 54; http://dx.doi.org/10.1158/0008-5472.CAN-08-1942; PMID: 18829540
- Dykxhoorn DM, Wu Y, Xie H, Yu F, Lal A, Petrocca F, et al. miR-200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS One 2009; 4:e7181; http://dx.doi.org/10.1371/journal.pone.0007181; PMID: 19787069
- Korpal M, Ell BJ, Buffa FM, Ibrahim T, Blanco MA, Celià-Terrassa T, et al. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat Med 2011; 17:1101 - 8; http://dx.doi.org/10.1038/nm.2401; PMID: 21822286
- Jelena R, Apostolos Z, Thomas V, Maria K, Demetrios AS, Efstathios NS. MicroRNA expression analysis in triple-negative (ER, PR and Her2/neu) breast cancer. Cell Cycle 2011; 10:10 - 2; PMID: 21191182
- Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, et al. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res 2007; 67:11612 - 20; http://dx.doi.org/10.1158/0008-5472.CAN-07-5019; PMID: 18089790
- Piovan C, Palmieri D, Di Leva G, Braccioli L, Casalini P, Nuovo G, et al. Oncosuppressive role of p53-induced miR-205 in triple negative breast cancer. Mol Oncol 2012; 6:458 - 72; http://dx.doi.org/10.1016/j.molonc.2012.03.003; PMID: 22578566
- Spizzo R, Nicoloso MS, Lupini L, Lu Y, Fogarty J, Rossi S, et al. miR-145 participates with TP53 in a death-promoting regulatory loop and targets estrogen receptor-alpha in human breast cancer cells. Cell Death Differ 2010; 17:246 - 54; http://dx.doi.org/10.1038/cdd.2009.117; PMID: 19730444
- Kim SJ, Oh JS, Shin JY, Lee KD, Sung KW, Nam SJ, et al. Development of microRNA-145 for therapeutic application in breast cancer. J Control Release 2011; 155:427 - 34; http://dx.doi.org/10.1016/j.jconrel.2011.06.026; PMID: 21723890
- Zou C, Xu Q, Mao F, Li D, Bian C, Liu LZ, et al. MiR-145 inhibits tumor angiogenesis and growth by N-RAS and VEGF. Cell Cycle 2012; 11:2137 - 45; http://dx.doi.org/10.4161/cc.20598; PMID: 22592534
- Sharom FJ. ABC multidrug transporters: structure, function and role in chemoresistance. Pharmacogenomics 2008; 9:105 - 27; http://dx.doi.org/10.2217/14622416.9.1.105; PMID: 18154452
- Baker EK, El-Osta A. The rise of DNA methylation and the importance of chromatin on multidrug resistance in cancer. Exp Cell Res 2003; 290:177 - 94; http://dx.doi.org/10.1016/S0014-4827(03)00342-2; PMID: 14567978
- Idelman G, Jacobson EM, Tuttle TR, Ben-Jonathan N. Lactogens and estrogens in breast cancer chemoresistance. Expert Rev Endocrinol Metab 2011; 6:411 - 22; http://dx.doi.org/10.1586/eem.11.19; PMID: 21731573
- Wilson T, Longley D, Johnston P.. Chemoresistance in solid tumours. Ann Onc 2006; 17:Suppl 10:x315 - 24
- Pegg AE, Dolan ME, Moschel RC. Structure, function, and inhibition of O6-alkylguanine-DNA alkyltransferase. Prog Nucleic Acid Res Mol Biol 1995; 51:167 - 223; http://dx.doi.org/10.1016/S0079-6603(08)60879-X; PMID: 7659775
- Kovalchuk O, Filkowski J, Meservy J, Ilnytskyy Y, Tryndyak VP, Chekhun VF, et al. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther 2008; 7:2152 - 9; http://dx.doi.org/10.1158/1535-7163.MCT-08-0021; PMID: 18645025
- Bao L, Hazari S, Mehra S, Kaushal D, Moroz K, Dash S. Increased expression of P-glycoprotein and doxorubicin chemoresistance of metastatic breast cancer is regulated by miR-298. Am J Pathol 2012; 180:2490 - 503; http://dx.doi.org/10.1016/j.ajpath.2012.02.024; PMID: 22521303
- Liang Z, Wu H, Xia J, Li Y, Zhang Y, Huang K, et al. Involvement of miR-326 in chemotherapy resistance of breast cancer through modulating expression of multidrug resistance-associated protein 1. Biochem Pharmacol 2010; 79:817 - 24; http://dx.doi.org/10.1016/j.bcp.2009.10.017; PMID: 19883630
- Pogribny IP, Filkowski JN, Tryndyak VP, Golubov A, Shpyleva SI, Kovalchuk O. Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int J Cancer 2010; 127:1785 - 94; http://dx.doi.org/10.1002/ijc.25191; PMID: 20099276
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 2003; 100:3983 - 8; http://dx.doi.org/10.1073/pnas.0530291100; PMID: 12629218
- Hirschmann-Jax C, Foster AE, Wulf GG, Nuchtern JG, Jax TW, Gobel U, et al. A distinct “side population” of cells with high drug efflux capacity in human tumor cells. Proc Natl Acad Sci U S A 2004; 101:14228 - 33; http://dx.doi.org/10.1073/pnas.0400067101; PMID: 15381773
- Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem 2008; 283:29897 - 903; http://dx.doi.org/10.1074/jbc.M804612200; PMID: 18708351
- Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, et al. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem 2008; 283:31079 - 86; http://dx.doi.org/10.1074/jbc.M806041200; PMID: 18790736
- Rao X, Di Leva G, Li M, Fang F, Devlin C, Hartman-Frey C, et al. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene 2011; 30:1082 - 97; http://dx.doi.org/10.1038/onc.2010.487; PMID: 21057537
- Pan YZ, Morris ME, Yu AM. MicroRNA-328 negatively regulates the expression of breast cancer resistance protein (BCRP/ABCG2) in human cancer cells. Mol Pharmacol 2009; 75:1374 - 9; http://dx.doi.org/10.1124/mol.108.054163; PMID: 19270061
- Liang Z, Wu H, Xia J, Li Y, Zhang Y, Huang K, et al. Involvement of miR-326 in chemotherapy resistance of breast cancer through modulating expression of multidrug resistance-associated protein 1. Biochem Pharmacol 2010; 79:817 - 24; http://dx.doi.org/10.1016/j.bcp.2009.10.017; PMID: 19883630
- Masri S, Liu Z, Phung S, Wang E, Yuan YC, Chen S. The role of microRNA-128a in regulating TGFbeta signaling in letrozole-resistant breast cancer cells. Breast Cancer Res Treat 2010; 124:89 - 99; http://dx.doi.org/10.1007/s10549-009-0716-3; PMID: 20054641
- Pogribny IP, Filkowski JN, Tryndyak VP, Golubov A, Shpyleva SI, Kovalchuk O. Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int J Cancer 2010; 127:1785 - 94; http://dx.doi.org/10.1002/ijc.25191; PMID: 20099276
- Kong W, He L, Coppola M, Guo J, Esposito NN, Coppola D, et al. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J Biol Chem 2010; 285:17869 - 79; http://dx.doi.org/10.1074/jbc.M110.101055; PMID: 20371610
- Zhou M, Liu Z, Zhao Y, Ding Y, Liu H, Xi Y, et al. MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J Biol Chem 2010; 285:21496 - 507; http://dx.doi.org/10.1074/jbc.M109.083337; PMID: 20460378
- Cittelly DM, Das PM, Spoelstra NS, Edgerton SM, Richer JK, Thor AD, et al. Downregulation of miR-342 is associated with tamoxifen resistant breast tumors. Mol Cancer 2010; 9:317; http://dx.doi.org/10.1186/1476-4598-9-317; PMID: 21172025
- Kastl L, Brown I, Schofield AC. miRNA-34a is associated with docetaxel resistance in human breast cancer cells. Breast Cancer Res Treat 2012; 131:445 - 54; http://dx.doi.org/10.1007/s10549-011-1424-3; PMID: 21399894
- Gong C, Yao Y, Wang Y, Liu B, Wu W, Chen J, et al. Up-regulation of miR-21 Mediates Resistance to Trastuzumab Therapy for Breast Cancer. J Biol Chem 2011; 287:286 - 98; PMID: 22069308
- Wang ZX, Lu BB, Wang H, Cheng ZX, Yin YM. MicroRNA-21 modulates chemosensitivity of breast cancer cells to doxorubicin by targeting PTEN. Arch Med Res 2011; 42:281 - 90; http://dx.doi.org/10.1016/j.arcmed.2011.06.008; PMID: 21820606
- Liang Z, Li Y, Huang K, Wagar N, Shim H. Regulation of miR-19 to breast cancer chemoresistance through targeting PTEN. Pharm Res 2011; 28:3091 - 100; http://dx.doi.org/10.1007/s11095-011-0570-y; PMID: 21853360
- Zhu Y, Yu F, Jiao Y, Feng J, Tang W, Yao H, et al. Reduced miR-128 in breast tumor-initiating cells induces chemotherapeutic resistance via Bmi-1 and ABCC5. Clin Cancer Res 2011; 17:7105 - 15; http://dx.doi.org/10.1158/1078-0432.CCR-11-0071; PMID: 21953503
- Chen J, Tian W, Cai H, He H, Deng Y. Down-regulation of microRNA-200c is associated with drug resistance in human breast cancer. Med Oncol 2012; 29:2527 - 34; http://dx.doi.org/10.1007/s12032-011-0117-4; PMID: 22101791
- Ru P, Steele R, Hsueh EC, Ray RB. Anti-miR-203 Upregulates SOCS3 Expression in Breast Cancer Cells and Enhances Cisplatin Chemosensitivity. Genes Cancer 2011; 2:720 - 7; http://dx.doi.org/10.1177/1947601911425832; PMID: 22207897
- Bao L, Hazari S, Mehra S, Kaushal D, Moroz K, Dash S. Increased expression of P-glycoprotein and doxorubicin chemoresistance of metastatic breast cancer is regulated by miR-298. Am J Pathol 2012; 180:2490 - 503; http://dx.doi.org/10.1016/j.ajpath.2012.02.024; PMID: 22521303