Abstract
PARP inhibitors, both as monotherapy and in combination with cytotoxic drugs, are currently undergoing clinical trials in several different cancer types. In this investigation, we compared the antiproliferative activity of two PARP/putative PARP inhibitors, i.e., olaparib and iniparib, in a panel of 14 breast cancer cell lines (seven tripe-negative and seven non-triple-negative). In almost all cell lines investigated, olaparib was a more potent inhibitor of cell growth than iniparib. Inhibition by both drugs was cell line-dependent and independent of the molecular subtype status of the cells, i.e., whether cells were triple-negative or non-triple negative. Although the primary target of PARP inhibitors is PARP1, no significant association was found between baseline levels of PARP1 activity and inhibition with either agent. Similarly, no significant correlation was evident between sensitivity and levels of CDK1, BRCA1 or miR-182. Combined addition of olaparib and either the CDK1 inhibitor, RO-3306 or a pan HER inhibitor (neratinib, afatinib) resulted in superior growth inhibition to that obtained with olaparib alone. We conclude that olaparib, in contrast to iniparib, is a strong inhibitor of breast cancer cell growth and may have efficacy in breast cancer irrespective of its molecular subtype, i.e., whether HER2-positive, estrogen receptor (ER)-positive or triple-negative. Olaparib, in combination with a selective CDK1 inhibitor or a pan HER inhibitor, is a potential new approach for treating breast cancer.
Introduction
PARP1 is a multifunctional enzyme, best known for its role in the repair of single strand DNA breaks.Citation1,Citation2 In recent years, several low molecular weight PARP inhibitors have been investigated as potential anti-cancer agents.Citation3 As anti-cancer drugs, PARP inhibitors can potentially be used in several different ways, such as monotherapy or as potentiators of the effects of specific cytotoxic drugs. To date, the efficacy of PARP inhibitors as monotherapy has been mostly confined to patients with BRCA1/2 mutation-associated breast and ovarian cancer.Citation4-Citation7 In these patients, the efficacy of the PARP inhibitor, olaparib appears to relate to a synthetic lethal interaction between defective BRCA1/2 function and inhibition of PARP1.Citation8,Citation9
Synthetic lethality occurs between two genes when loss of one gene is compatible with cell viability but loss of both results in cell death.Citation8,Citation9 The implication of synthetic lethality is that targeting of one such gene in a cancer (e.g., PARP1), where the other is non-functioning (e.g., in patients with BRCA1 or BRCA2 germline mutations), should in theory, be selectively lethal to tumor cells but not affect normal cells.Citation8,Citation9
Germline BRCA1/2 mutations however, are responsible for only a minority of all breast and ovarian cancers, constituting < 10% of these malignancies. Although phase I/II clinical trials have shown promising results with the PARP inhibitor olaparib, in patients with germline defects in these genes,Citation4-Citation7 it remains to be shown if PARP inhibitors have efficacy in patients with sporadic breast cancer.
A subgroup of patients with sporadic breast cancer possessing biological and clinical similarities with BRCA1/2-related malignancies is those with triple-negative (TN) disease, i.e., patients negative for estrogen receptors (ER), progesterone receptors and HER2. These similarities include a tendency for high tumor grade, expression of the basal cytokeratins 5 and 6, aberrant DNA repair and related gene expression signatures.Citation10-Citation12 These shared characteristics suggest that TN breast cancers have lower expression of BRCA1/2 or dysfunction in these or related DNA repair genes. If so, TN breast cancer may be susceptible to PARP inhibitors, as mentioned above for BRCA1/2 mutation related cancers. Although a phase II clinical trial showed that combinations of the putative PARP inhibitor, iniparib with cisplatin and gemcitabine was superior to cisplatin and gemcitabine alone in patients with advanced TNBC,Citation13 a follow-up phase III trial failed to meet the pre-specified criteria for significance with respect to progression free survival and overall survival.Citation14
Although iniparib has undergone evaluation in phase II/III clinical trials,Citation15 relatively little preclinical work appears to have been published on this agent prior to the initiation of these trials.Citation15-Citation17 We therefore decided to carry out a detailed preclinical study on the antiproliferative effects of iniparib in a panel of estrogen receptor-positive, HER2-positive and TN breast cancer cell lines and to compare response to this agent with that of olaparib. We also show that combined treatment with olaparib and either a CDK1 or a panHER inhibitor was superior to olaparib in a cell line-dependent manner.
Results
Comparative effects of olaparib and iniparib on cell growth in triple-negative and non-triple-negative breast cancer cell lines
and compare the relative growth inhibitory effects of 5 µM olaparib and iniparib on a panel of breast cancer cell lines. As can be seen, using both the MTT () and colony formation assays (), olaparib was a more effective growth inhibitor in almost all the cell lines investigated. Overall, the observed growth inhibition for both olaparib and iniparib was greater with the colony formation assay than with the MTT assay.
Table 1. Comparative antiproliferative effects of olaparib and iniparib at 5 µM concentration, using the MTT assay
Table 2. Comparative antiproliferative effects of olaparib and iniparib at 5 µM concentration, using the colony formation assay
and compare the IC50 values for olaparib and iniparib in the same panel of cell lines. Consistent with above findings, IC50 values for olaparib were considerably lower than those found for iniparib. Thus, with the MTT assay, IC50 values for olaparib across the cell lines, varied from 4.2 to 19.8 µM (), whereas the corresponding IC50 values for iniparib were all > 10 µM. Using the colony formation assays, IC50 values were lower than those found with the MTT assay, ranging from 0.6 to 3.2 µM with olaparib and from 5.7 to > 20 µM with iniparib (). No significant correlation however, was found between the IC50 values obtained with the MTT and colony formation assays.
Table 3. Comparative IC50 concentration for olaparib and iniparib, using the MTT assay
Table 4. Comparative IC50 concentration for olaparib and iniparib using the colony formation assay
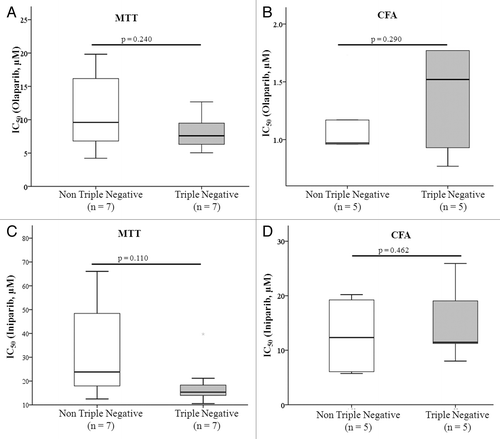
Since TN breast cancers have been shown to exhibit similar molecular and histological characteristics to BRCA1/2 associated cancers (see above), we compared response to both olaparib and iniparib in TN and non-TN cell lines. However, using both the MTT and colony formation assay, no significant difference in response was found between the 2 groups of cell lines, i.e., TN and non-TN cell lines exhibited similar responses to olaparib and iniparib ().
Figure 1. Boxplots illustrating the relationship between triple negative status and response to olaparib and iniparib as measured by determining IC50 values by (A and C) MTT assay and (B and D) colony formation assay in a panel of breast cancer cell lines. Boxes represent the 25th and 75th percentile with the median indicated. The bars indicate the 10th and 90th percentile. Data was analyzed using the Mann Whitney U test. CFA, colony formation assay.
Attempts to identify biomarkers predictive of iniparib and olaparib sensitivity
Since response to both olaparib and iniparib was variable from cell line to cell line and independent of the TN status of the cells, we attempted to identify potential markers of sensitivity. As preliminary data had previously suggested that CDK1Citation18, BRCA1 protein and miR-182 levelsCitation19 were associated with response to PARP inhibitors, we investigated these as potential predictors of response to olaparib and iniparib. However, BRCA1, CDK1 and miR-182 levels were unrelated to olaparib (Figs. S2 and S3) or iniparib sensitivity (Figs. S4 and S5). Similar results were obtained when BRCA1, CDK1 and miR levels were expressed as continuous variables.
Since PARP1 is the primary target for PARP inhibitors, we also investigated if a relationship existed between PARP1 activity and sensitivity to iniparib and olaparib. Although PARP1 activity varied widely in the cell lines investigated, no relationship was found between basal activity levels, and response to either olaparib (Figs. S2A and S3A) or iniparib (Figs. S4A and S5A).
Effect of combined treatment with olaparib and a CDK1 inhibitor on cell growth
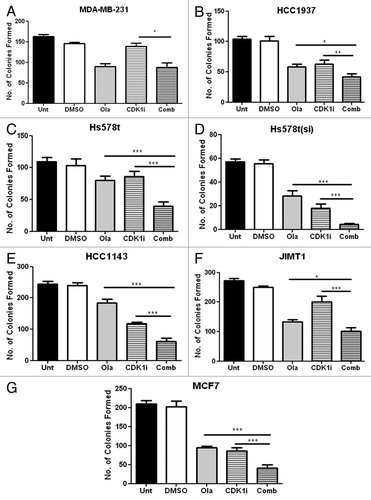
As olaparib was the more potent cell growth inhibitor, we decided to investigate its effects in combination with novel drugs. Since reduced CDK1 activity was previously shown to impair both BRCA1 function and homologous recombination repair,Citation18 we investigated combined inhibition with olaparib and the selective CDK1 inhibitor, RO-3306 (Merck). As shown in , combined addition of both olaparib and RO-3306 significantly enhanced growth inhibition compared with either olaparib or RO-3306 alone in 6/7 cell lines investigated.
Figure 2. Barcharts illustrating the effect of olaparib (1 µM) alone or in combination with the CDK1 inhibitor, RO-3306 (Merck) (1 µM) on clonogenic survival of a panel of breast cancer cell lines. *p < 0.05, **p < 0.01 and *** p < 0.005, Student’s paired t-test. Unt, Untreated; Ola, Olaparib; CKDi, CDK inhibitor.
Effect of combined treatment with olaparib and panHER inhibitors on cell growth
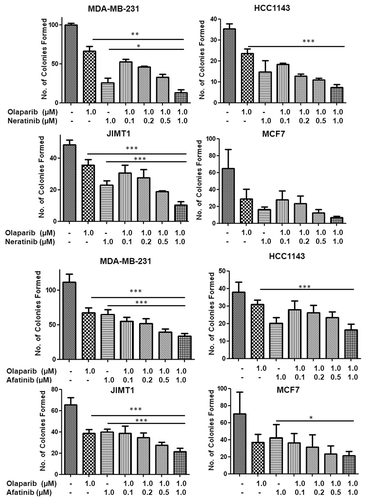
The combined effects of olaparib and neratinib as well as olaparib and afatinib on colony formation were investigated in four cell lines. As shown in , the effects of the dual treatment were cell-line dependent. Thus, in both the MDA-MB-231 and JIMT1 cells, the anti-growth effects of 1 µM of olaparib and either neratinib or afatinib were significantly more potent than either agent alone. In HCC1143 cells on the other hand, the combined treatment was more potent than olaparib alone but not significantly stronger than that of neratinib or afatinib alone. In contrast, using the MCF7 cells, combined treatment with olaparib and neratinib did not enhance cytotoxicity over either drug alone. The reason for this may relate to sensitivity of this cell line to neratinib. In the MCF7 cells, combined treatment with olaparib and afatinib gave significantly stronger inhibition of colony formation than afatinib alone but not compared with olaparib alone ().
Figure 3. Barcharts illustrating the effect of olaparib (constant 1 µM) alone or in combination with the pan-HER inhibitors neratinib (upper four plots) and afatinib (lower four plots) at varying concentrations on clonogenic survival of MDA-MB-231, HCC1143, JIMT1 and MCF7 breast cancer cells. *p < 0.05, **p < 0.01 and ***p < 0.005; Student’s paired t-test.
Discussion
Olaparib and iniparib are among two of the PARP/putative PARP inhibitors most widely investigated in clinical trials in patients with breast cancer.Citation4-Citation7,Citation13,Citation14 To our knowledge, this is one of the first in vitro studies to compare cytotoxicity of these agents in a broad panel of breast cancer cell lines, including TNBC, HER2-positive and ER-positive cells. Our findings suggest that response to both olaparib and iniparib is variable from cell line to cell line and independent of molecular subtype. Despite this variability, and in agreement with recent reports,Citation20,Citation21 we found that olaparib was a more potent inhibitor of cell growth than iniparib in almost all the cell lines investigated. Our results are thus consistent with recent studies showing that iniparib was a weak inhibitor of PARP1.Citation20-Citation22
Although MTT and colony formation assays are widely used to evaluate in vitro antiproliferative activities of investigational drugs, we showed that the IC50 values obtained with the colony formation assay were consistently lower than those obtained with the MTT assays. Furthermore, no significant correlation was found between the IC50 values obtained with the two assays across the panel cell lines investigated. Thus, the specific IC50 values determined, depended not only on the cell line but also on the specific viability assay used. It remains to be shown which assays better predict anti-tumor activity in vivo.
Because of the similarities between triple-negative and BRCA1/2 associated tumors,Citation10-Citation12 it might be expected that PARP inhibitors would be more effective in breast cancer cells, negative for ER, PR and HER2. Indeed, Hastak et al.Citation23 reported that TN breast cancer cell lines were more sensitive than luminal cell lines to the experimental PARP inhibitor, PJ34 (EMD Biosciences). In contrast, our preclinical study using a larger panel of breast cancer cell lines than investigated by Hastak et al.,Citation23 found no significant relationship between response to olaparib or iniparib and TN status. Indeed, of all the cell lines investigated in the current study, the most sensitive was the HER2-positive cell line, SKBR3 with the colony formation assay. Consistent with this finding, Nowsheen et al.Citation24 recently reported that HER2 overexpression conferred sensitivity to two different PARP inhibitors.
As mentioned in the Introduction above, the main success to date with PARP inhibitor monotherapy has been in BRCA1/2 associated breast and ovarian cancer.Citation4-Citation7 In these malignancies, defective BRCA1/2 impairs homologous recombination (HR) which in turn results in synthetic lethality in the presence of a PARP inhibitor.Citation8,Citation9 Although mutations in BRCA1 and 2 are rare in sporadic breast cancer, decreased expression of these proteins, possibly mediated by promoter methylation, gene loss or increased levels of negatively activating transcriptional factors, may occur. If such a decrease occurs, it might be expected, like that of mutation, to impair HR and thus confer sensitivity to PARP inhibitors.
Evidence for this was recently obtained when Moskwa et al.Citation19 reported that overexpression of miR-182 in the breast cancer cell line, MDA-MB-231 suppressed expression of BRCA1 and conferred sensitivity to two different PARP inhibitors. Conversely, antagonizing miR-182 increased BRCA1 levels and resulted in resistance to PARP1 inhibition. Based on these findings, either increased miR-182 levels or decreased BRCA1 protein might be expected to confer sensitivity to PARP inhibitors. In our study however, using a panel of cell rather than just the MDA-MB-231 cell line, neither miR-182 nor BRCA1 baseline levels correlated with response to olaparib or iniparib. A previous study using a larger panel of cell lines showed a non-significant trend for a correlation between low BRCA1 mRNA levels and sensitivity to olaparib.Citation19
One of the cell lines investigated in this study, i.e., HCC 1937 is known to harbor a BRCA1 mutation and thus might have been expected to be highly sensitive to the PARP inhibitors used. However, similar to previous reports (Drew et al.Citation28; Lehmann et al.Citation29), we also found that this cell line was poorly sensitive to olaparib. Thus, while BRCA1 mutations may be necessary for high sensitivity to PARP inhibitors, it alone is insufficient. Consistent with this in vitro finding, early clinical trial data show that only some patients with BRCA mutated tumors responded to PARP inhibitors.Citation5,Citation6
Since PARP1 is the primary target of PARP inhibitors, we also investigated if a relationship existed between PARP1 activity levels and response to olaparib and iniparib. As with, miR-182 and BRCA1, no significant relationship was found between the basal levels of PARP1 activity and response to either of the inhibitors. A previous study showed a lack of correlation between PARP1 mRNA and response to olaparib.Citation25 PARP1 catalytic activity, as measured in the current investigation might however, be expected to be a more meaningful measure of active PARP1 than PARP1 mRNA levels.
Although several preclinical studies have investigated combined treatment with PARP inhibitors and specific cytotoxic agents, until recently, few had analyzed combinations of PARP inhibitors and non-cytotoxic agents. Here we show that that combined treatment with olaparib and either of the panHER inhibitors, neratinib or afatinib enhanced growth inhibition. Indeed, combined treatment with olaparib and either neratinib or afatinib augmented growth inhibition over either agents alone in two of the four cell lines studied. While this work was in preparation, Nowsheen et al.Citation26 reported a synthetic lethal interaction between PARP inhibitors and lapatinib in TN breast cancer, while Ibrahim et al.Citation27 showed that PI3K inhibition sensitized TN breast cancer cells to PARP inhibition. All of these findings when taken together suggest that combinations of PARP inhibitors and specific tyrosine kinase inhibitors may have efficacy in TN breast cancer.
Since reduced CDK1 activity was previously shown to result in increased anti-tumor response to PARP inhibitors in a mouse model of lung cancer,Citation18 we investigated combined treatment with the CDK1 inhibitor, RO-3306 and olaparib. In agreement with the previously reported results from the lung cancer model,Citation18 we found that combined treatment with olaparib and RO-3306 was superior to either drug alone in a cell-line dependent manner. Based on this finding and previous data,Citation18 combinations of PARP inhibitors and CDK1 inhibitors might now be considered for clinical trials in sporadic breast cancer.
In addition to our study, a number of other preclinical studies have investigated PARP inhibitors in a panel of breast cancer cell lines. Drew et al.Citation28 evaluated AG-014699 (rucaparib) (Pfizer GRD, La Jolla, CA) in 9 human cell lines (most of which were breast cancer derived) and concluded that this agent was cytotoxic in cells with mutated BRCA1/2 genes, epigenetically silenced BRCA1 cells as well as in cells with XRCC3 mutations. In that study, unlike ours, no distinction was made between TN and non-TN breast cancer cell lines. Furthermore, unlike the present study, AG-014699 was not investigated in combination with other agents.
In a second in vitro study, Hastak et al.Citation23 compared the effect of the PARP inhibitor, PJ34 (EMD Biosciences) on the in vitro growth of 4 TN and 3 luminal breast cancer cell lines. As mentioned above, the TN cell lines were found to be more sensitive to PJ34 than the luminal cell lines investigated. Furthermore, synergy was observed between PJ34 and gemcitabine and cisplatinum in the TN cell lines.
In a third study, Chung et al.,Citation21 investigated 4 PARP inhibitors in three triple-negative cell lines. Of the four examined, AG-014699 (rucaparib) was the most potent, followed by olaparib, ABT-888 (veliparib) and iniparib. Potency was related to their effects on G2/M arrest and extent of DNA damage. Of potential significance was the finding that AG-014699 suppressed STAT3 phosphorylation, suggesting that this anti-PARP agent may also act as a potential inhibitor of cell signaling.
A fourth study identified a seven-gene panel whose mRNA levels were associated with response to olaparib.Citation25 This panel included five genes, i.e., BRCA1, XPA, TDG, NBS1 and MRE11A, whose mRNAs were associated with resistance and two genes, i.e., MK2 and CHEK2, whose mRNAs correlated with sensitivity. However, as mentioned above, levels of BRCA1 mRNA, when treated as a single variable, did not show a statistically significant relation (i.e, at the 0.05 level) with sensitivity to olaparib.
Our findings when combined with results from the recent reports indicating that iniparib does not appear to act as a competitive PARP inhibitorCitation19 and indeed has little inhibitory impact on PARP1,Citation19-Citation21 may explain the lack of efficacy observed in patients with advanced triple-negative breast cancer.Citation14 These negative results reported with iniparibCitation14 however, may not apply to competitive-type PARP inhibitors, which constitute the vast majority of PARP inhibitors currently undergoing clinical trials. We therefore suggest that trials of bona-fide PARP inhibitors in triple-negative breast cancer should not be abandoned at this point in time because of the negative phase III trial results with iniparib. Furthermore, our results suggest that bona fide PARP inhibitors such as olaparib may have anti-tumor activity in HER2-positive and ER-positive breast cancers.
In conclusion, we showed that olaparib is more potent than iniparib in the inhibition of breast cancer cell line growth in vitro. Our results also show that sensitivity to olaparib is cell line-dependent and that this anti-cancer agent inhibits growth of both TN and non-TN cell lines. Olaparib may thus have benefit in at least some sporadic breast cancers. Furthermore, combined treatment involving olaparib and RO-3306 or olaparib and a pan HER inhibitor (neratinib or afatinib) was more potent than either agent alone in at least some of the cell lines investigated. These results should now be confirmed in appropriate animal models of breast cancer. Pending the outcome of these preclinical experiments, clinical trials involving olaparib and selective CDK1 inhibitors or olaparib and a pan HER inhibitor, may be warranted in breast cancer.
Materials and Methods
Cell culture
The following panel of breast cancer cell lines were used: MDA-MB-231, MDA-MB-468, BT20, HCC1143, HCC1937, Hs578t, Hs578t(si) (all TN), BT474, JIMT1, SKBR3, SUM159 (all HER2-positive), CAMA1, MCF7 and T47D (ER-positive/luminal). Further details relating to these cell lines are listed in in supplementary data. All were obtained from the American Tissue Culture Collection, apart from Hs578t(si) cells which were supplied by Dr Susan McDonnell, University College, Dublin. This cell line was derived from the parental Hs578t cell line by sequential selection through in vitro invasive chambers.Citation30 Cell lines were maintained in RPMI 1640, supplemented with 10% fetal bovine serum (FBS) (Invitrogen Life Technologies), 1% penicillin/streptomycin (Invitrogen Life Technologies) and 1% Fungizone (Invitrogen Life Technologies) and maintained in a 37°C CO2-humidified incubator.
Olaparib and iniparib were obtained from Selleck Chemicals; RO-3306 from Merck and neratinib and afatinib from Sequoia Research Products.
Cell viability assays
Cell proliferation was assessed using the MTT Cell Proliferation Kit I (Roche Applied Science) as previously described.Citation31 To test the effect of iniparib or olaparib on proliferation, cells were plated at a density of 1 × 103/well in 96-well flat-bottomed plates (Corning® Costar®, Sigma-Aldrich). Following overnight incubation, quadruplicate wells were treated with varying concentrations of compounds alone or in combination, for 5 d.
Clonogenic Assays
Cells were seeded in six-well plates (Corning® Costar®, Sigma-Aldrich) at a density of 1 × 103 cells/well in quadruplicate and treated with compound for 14 d. Cells were fixed in 1% glutaraldehyde (Sigma) and stained with 0.1% crystal violet (Pro-Lab Diagnostics). The mean colony count and standard error of the mean were calculated.
Detection of miR-182
Following RNA isolation,Citation32 reverse transcription (RT) was performed using the TaqMan microRNA reverse transcription kit (Applied Biosystems). Briefly, the RT reaction consisted of 1.5 μL 10 × RT Buffer, 0.15 μL dNTPs 100 mM, 0.19 μL RNase Inhibitor 20 U/μL, 1.0 MultiScribe reverse transcriptase, 3 μL of RT primer and total RNA (10 ng from cells or 5 µl of RNA from conditioned media), in a final volume of 15 μL. Real-time PCR was performed, using a Mastercycler ep realplex 2S system (Eppendorf) for 30 min at 16°C, 30 min at 42°C, 5 min at 85°C and then held at 4°C. The RT products were subsequently amplified with sequence-specific primers. The 20 μL PCR mix contained 1.33 μL RT product, 1 μL TaqMan® Universal PCR Master Mix (20 × ), 1 μL TaqMan® probe. The reaction mix was incubated in a 96-well plate at 95°C for 10 min followed by 40 cycles of 95°C for 15 s and at 60°C for 1 min.
Expression of miR-182 (002334, Applied Biosystems) was normalized to the endogenous control, RNU44 (001094, Applied Biosystems) for RNA from both the cells and conditioned medium. The comparative CT method was used for data analysis. For miR-182 in conditioned medium, fold changes were calculated using their respective cells CT values as calibrator.
Western blotting
CDK1 and BRCA1 levels were determined using western blotting analysis. Total protein was extracted from cell lines using 50 mmol/L TRIS-HCl (pH 7.4) containing a cocktail of protease and phosphatase inhibitors (Roche Applied Science) and Triton X-100 (1%), under agitation at 4°C for 1 h. Equal amounts of protein were separated on 10% SDS-PAGE and transferred to PVDF membranes (Millipore). Membranes were blocked in 5% low-fat dry milk (Marvel instant dried skimmed milk) in PBS-T and then stained for CDK1 with a monoclonal mouse antibody (1:200) (MAB8878, Millipore) for 1 h at room temperature or with the anti-BRCA1 (Ab-1) mouse monoclonal antibody (1:100) (MS110, Calbiochem) overnight at 4°C.
Following three washes for 10 min in PBS-T, the membrane was incubated with 1:2,000 horseradish peroxidase-conjugated anti-mouse secondary antibody (Cell Signaling) for 1 h at room temperature before incubation with chemiluminescence reagent (Luminol, Santa Cruz Biotechnology) for 1 min. Membranes were then exposed to X-ray film (Fujifilm). The intensity of the protein bands observed was semiquantified using the Autochemi UVP Bioimaging System, with normalization against β-actin (Sigma).
PAR ELISA
PARP1 catalytic activity was measured by monitoring the formation of PAR polymer by ELISA (Trevigen®, R&D Systems). Manufacturer’s instructions were followed for all steps, including protein extraction.
Statistical analysis
The Student’s paired t test was used to compare the effects of the PARP inhibitors alone vs. their combinations with other agents. Data was analyzed using PASW Statistics Version 18 (SPSS Inc.) and Prism version 5.0b software (GraphPad Software). The IC50 (concentration required to inhibit growth by 50%) for each inhibitor was determined using CalcuSyn software (Biosoft).
Additional material
Download Zip (316 KB)Acknowledgments
The authors wish to thank Science Foundation Ireland, Strategic Research Cluster Award (08/SRC/B1410) to Molecular Therapeutics for Cancer Ireland, the Health Research Board Clinician Scientist Award (CSA/2007/11) and the Cancer Clinical Research Trust, for funding this work.
Disclosure of Potential Conflicts of Interest
A. Pierce, PM McGowan, M. Cotter, M. Mullooly, N. O’Donovan, S. Rani, L. O’Driscoll and MJ Duffy have no conflict of interest to report. J. Crown has received speaker’s fees and research support from GlaxoSmithKline, Roche, Novartis, Sanofi-Aventis and Pfizer.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/nucleus/article/24349.
References
- Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell 2010; 39:8 - 24; http://dx.doi.org/10.1016/j.molcel.2010.06.017; PMID: 20603072
- Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer 2010; 10:293 - 301; http://dx.doi.org/10.1038/nrc2812; PMID: 20200537
- Yap TA, Sandhu SK, Carden CP, de Bono JS. Poly(ADP-ribose) polymerase (PARP) inhibitors: Exploiting a synthetic lethal strategy in the clinic. CA Cancer J Clin 2011; 61:31 - 49; http://dx.doi.org/10.3322/caac.20095; PMID: 21205831
- Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 2009; 361:123 - 34; http://dx.doi.org/10.1056/NEJMoa0900212; PMID: 19553641
- Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet 2010; 376:235 - 44; http://dx.doi.org/10.1016/S0140-6736(10)60892-6; PMID: 20609467
- Audeh MW, Carmichael J, Penson RT, Friedlander M, Powell B, Bell-McGuinn KM, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet 2010; 376:245 - 51; http://dx.doi.org/10.1016/S0140-6736(10)60893-8; PMID: 20609468
- Gelmon KA, Tischkowitz M, Mackay H, Swenerton K, Robidoux A, Tonkin K, et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol 2011; 12:852 - 61; http://dx.doi.org/10.1016/S1470-2045(11)70214-5; PMID: 21862407
- Ashworth A. Drug resistance caused by reversion mutation. [Review] Cancer Res 2008; 68:10021 - 3; http://dx.doi.org/10.1158/0008-5472.CAN-08-2287; PMID: 19074863
- Banerjee S, Kaye SB, Ashworth A. Making the best of PARP inhibitors in ovarian cancer. Nat Rev Clin Oncol 2010; 7:508 - 19; http://dx.doi.org/10.1038/nrclinonc.2010.116; PMID: 20700108
- Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer 2004; 4:814 - 9; http://dx.doi.org/10.1038/nrc1457; PMID: 15510162
- Anders CK, Winer EP, Ford JM, Dent R, Silver DP, Sledge GW, et al. Poly(ADP-Ribose) polymerase inhibition: “targeted” therapy for triple-negative breast cancer. Clin Cancer Res 2010; 16:4702 - 10; http://dx.doi.org/10.1158/1078-0432.CCR-10-0939; PMID: 20858840
- Hartman AR, Kaldate RR, Sailer LM, Painter L, Grier CE, Endsley RR, et al. Prevalence of BRCA mutations in an unselected population of triple-negative breast cancer. Cancer 2012; 118:2787 - 95; http://dx.doi.org/10.1002/cncr.26576; PMID: 22614657
- O’Shaughnessy J, Osborne C, Pippen JE, Yoffe M, Patt D, Rocha C, et al. Iniparib plus chemotherapy in metastatic triple-negative breast cancer. N Engl J Med 2011; 364:205 - 14; http://dx.doi.org/10.1056/NEJMoa1011418; PMID: 21208101
- O'Shaughnessy JS, Schwartzberg LS, Danso MA, et al. A randomized phase III study of iniparib (BSI-201) in combination with gemcitabine/carboplatin (G/C) in metastatic triple-negative breast cancer (TNBC. J Clin Oncol 2011; 29 107
- Ossovskaya V, Li L, Broude E, Lim CU, Roninson I, Bradley C, et al. (2009) BSI-201 enhances the activity of multiple classes of cytotoxic agents and irridation in triple negative breast cancer [abstract]. In: Proceedings of the 100th Annual Meeting of the American Association for Cancer Research; 2009 Apr 18-22, Denver, CO. Philadelphia: AACR. Abstract nr 5552.
- Ji J, Lee MP, Kadota M, et al. (2011) Pharmacodynamic and pathway analysis of three presumed inhibitors of poly (ADP-ribose) polymerase: ABT-888, AZD2281, and BSI201 [abstract]. In: Proceedings of the 101st Annual Meeting of the American Association for Cancer Research; Apr 17–21; Washington, DC. Philadelphia (PA): AACR; 2011. Abstract nr 4527.
- Maegley KA, Bingham P, Dalvie D, Bergqvist S, Tatlock J, Kania R. (2010) An in vitro mechanistic comparison of clinical PARP inhibitors. PARP 2010. In: Proceedings of the 18th International Conference on ADP-ribose metabolism; Aug 18–21; Zurich, Switzerland. Abstract nr 72.
- Johnson N, Li YC, Walton ZE, Cheng KA, Li D, Rodig SJ, et al. Compromised CDK1 activity sensitizes BRCA-proficient cancers to PARP inhibition. Nat Med 2011; 17:875 - 82; http://dx.doi.org/10.1038/nm.2377; PMID: 21706030
- Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ, et al. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell 2011; 41:210 - 20; http://dx.doi.org/10.1016/j.molcel.2010.12.005; PMID: 21195000
- Liu X, Shi Y, Maag DX, Palma JP, Patterson MJ, Ellis PA, et al. Iniparib nonselectively modifies cysteine-containing proteins in tumor cells and is not a bona fide PARP inhibitor. Clin Cancer Res 2012; 18:510 - 23; http://dx.doi.org/10.1158/1078-0432.CCR-11-1973; PMID: 22128301
- Patel AG, De Lorenzo SB, Flatten KS, Poirier GG, Kaufmann SH. Failure of iniparib to inhibit poly(ADP-Ribose) polymerase in vitro. Clin Cancer Res 2012; 18:1655 - 62; http://dx.doi.org/10.1158/1078-0432.CCR-11-2890; PMID: 22291137
- Chuang HC, Kapuriya N, Kulp SK, Chen CS, Shapiro CL. Differential anti-proliferative activities of poly(ADP-ribose) polymerase (PARP) inhibitors in triple-negative breast cancer cells. Breast Cancer Res Treat 2012; 134:649 - 59; http://dx.doi.org/10.1007/s10549-012-2106-5; PMID: 22678161
- Hastak K, Alli E, Ford JM. Synergistic chemosensitivity of triple-negative breast cancer cell lines to poly(ADP-Ribose) polymerase inhibition, gemcitabine, and cisplatin. Cancer Res 2010; 70:7970 - 80; http://dx.doi.org/10.1158/0008-5472.CAN-09-4521; PMID: 20798217
- Nowsheen S, Cooper T, Bonner JA, LoBuglio AF, Yang ES. HER2 overexpression renders human breast cancers sensitive to PARP inhibition independently of any defect in homologous recombination DNA repair. Cancer Res 2012; 72:4796 - 806; http://dx.doi.org/10.1158/0008-5472.CAN-12-1287; PMID: 22987487
- Daemen A, Wolf DM, Korkola JE, Griffith OL, Frankum JR, Brough R, et al. Cross-platform pathway-based analysis identifies markers of response to the PARP inhibitor olaparib. Breast Cancer Res Treat 2012; 135:505 - 17; http://dx.doi.org/10.1007/s10549-012-2188-0; PMID: 22875744
- Nowsheen S, Cooper T, Stanley JA, Yang ES. Synthetic lethal interactions between EGFR and PARP inhibition in human triple negative breast cancer cells. PLoS One 2012; 7:e46614; http://dx.doi.org/10.1371/journal.pone.0046614; PMID: 23071597
- Ibrahim YH, García-García C, Serra V, He L, Torres-Lockhart K, Prat A, et al. PI3K inhibition impairs BRCA1/2 expression and sensitizes BRCA-proficient triple-negative breast cancer to PARP inhibition. Cancer Discov 2012; 2:1036 - 47; http://dx.doi.org/10.1158/2159-8290.CD-11-0348; PMID: 22915752
- Drew Y, Mulligan EA, Vong W-T, Thomas HD, Kahn S, Kyle S, et al. Therapeutic potential of poly(ADP-ribose) polymerase inhibitor AG014699 in human cancers with mutated or methylated BRCA1 or BRCA2. J Natl Cancer Inst 2011; 103:334 - 46; http://dx.doi.org/10.1093/jnci/djq509; PMID: 21183737
- Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011; 121:2750 - 67; http://dx.doi.org/10.1172/JCI45014; PMID: 21633166
- Hughes L, Malone C, Chumsri S, Burger AM, McDonnell S. Characterisation of breast cancer cell lines and establishment of a novel isogenic subclone to study migration, invasion and tumourigenicity. Clin Exp Metastasis 2008; 25:549 - 57; http://dx.doi.org/10.1007/s10585-008-9169-z; PMID: 18386134
- McGowan PM, Mullooly M, Caiazza F, Sukor S, Madden SF, Maguire AA, et al. ADAM-17: a novel therapeutic target for triple negative breast cancer. Ann Oncol 2013; 24:362; http://dx.doi.org/10.1093/annonc/mds279; PMID: 22967992
- Rani S, Clynes M, O’Driscoll L. Detection of amplifiable mRNA extracellular to insulin-producing cells: potential for predicting beta cell mass and function. Clin Chem 2007; 53:1936 - 44; http://dx.doi.org/10.1373/clinchem.2007.087973; PMID: 17717131