Abstract
Small conditional RNAs were used to kill cells selectively in a prior report. The method utilized the cellular innate immune response to dsRNA, causing PKR activation and cell death. We designed small conditional RNAs specific to a highly restricted transcript, that of the mesothelin gene expressed in various cancer lines and specific to a tpc/hpr fusion transcript expressed in a published “control” line. Hairpins of small conditional RNAs were functionally active in cell-free conditions. Hairpins were transfected into 6 types of cells. We observed non-specific killing of cells after transfection of the hairpins targeting the reported fusion transcript or of the mesothelin transcript. Thus when attempting to use this system for a special purpose, to target cancer mutations, the results were not satisfactory. Specifically when repeating the work described in the publication, we could not replicate the results using the methods described. Recently the publication was retracted but without comment on the validity of the reported method. Here we provide scientific basis to consider the method impaired or invalid.
Introduction
Protein kinase R (PKR) plays an important role in the antiviral innate immune response within cells.Citation1 Double-stranded viral RNA (dsRNA) activates PKR, leading to inhibition of protein synthesis and induction of apoptosis.
Hairpins of small conditional RNAs can be designed to target a specific RNA sequence. When mixed, an RNA template initiates strand-displacement opening of a first RNA hairpin, which in turn opens a second RNA hairpin, propagating a chain reaction (hybridization chain reaction, HCR) and forming long nicked double-stranded RNA (dsRNA), details having been reviewed.Citation2 As proposed, HCR may be elicited in cells by targeting a specific mRNA species.Citation3 dsRNA formed inside the cells leads to PKR activation and apoptosis.
We attempted to replicate and extend the principle because of its novel implementation of cancer-specific discrimination. The protocol is unambiguous and should be extensible to other cellular targets. We were familiar with a common cancer-selective transcript (mesothelin),Citation4 which we tested along with the described positive controls. The results of our study contrasted with the claimed “selectivity” of HCR-mediated cell death, and our designed oligos were not effective in cell killing.
Results
Cell-free polymerization
Hairpins are functional
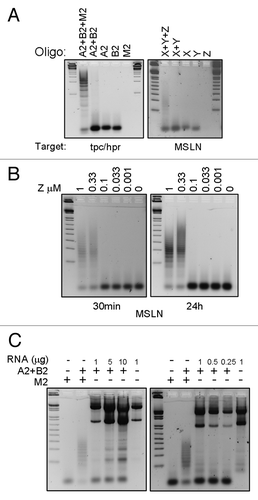
Cell-free polymerization reactions confirmed the functionality of the RNA hairpins. Fusion-targeting hairpins A2 and B2 polymerized in the presence of initiator M2 (). Mesothelin-targeting hairpins X and Y polymerized in the presence of initiator Z (). Polymerization was absent when initiators were omitted.
Figure 1. Cell-free hybridization chain reaction (HCR). Samples from HCR were separated on a 1% agarose gel. (A) A2 and B2 are tpc/hpr-targeting hairpins; X and Y are mesothelin-targeting hairpins; M2 is the initiator oligo for tpc/hpr HCR; Z is the initiator for mesothelin HCR. One or more oligos were added to the reaction, as indicated. The first and last lanes have a DNA ladder. (B) HCR using different concentrations of mesothelin initiator oligos. First lane has a DNA ladder. (C) Hairpins (A2 and B2) targeting tpc/hpr were incubated for 30 min with various quantities of RNA from LNCaP cells. The first lane has a DNA ladder.
Using different concentrations of initiator sequences
We used mesothelin hairpins with various concentrations of initiator Z to determine the minimum quantity of initiator required for polymerization. X and Y hairpins polymerized in the presence of 1 and 0.33 µM of initiator Z (). Polymerization was not observed using 0.1 µM or less of initiator, thus high concentrations of initiator were required for polymerization. Longer incubation (16 h) of the reaction did not induce polymerization when using lower concentrations of initiator.
Cell-free polymerization using LNCaP RNA as initiator sequence
We examined whether unfractionated RNA could serve as the initiator for HCR. Unexpectedly, A2 and B2 hairpins did not polymerize in presence of various quantities (0.25, 0.5, 1, 5, or 10 µg) of LNCaP RNA at 30 min or 16 h of incubation at 37 °C (). Possibly, unrelated RNA blocked the reaction and/or the transcript concentration specific to the initiator was too low to generate detectable polymerization.
Using hairpin transfection and assessing cell viability
We transfected cells with the hairpins to determine whether hairpin transfection selectively killed cells as described.Citation3 Transfection with A2 and B2 using three different conditions resulted in killing of LNCaP cells,Citation3 but unexpectedly it killed negative control BXPC3 cells in a similar fashion (). Cell death in LNCaP and BXPC3 was equal, ~90% for condition 1, ~60% for condition 2, and ~40–50% for condition 3 (condition 1 had the highest hairpin concentration). H2126 and AsPC1 cells were also non-selectively killed (20–40%) using condition 1. Viability of 293 cells was not affected by transfection. We found the reported HCR hairpins not to be “selective” as claimed.Citation3
Figure 2. Cell killing by RNA hairpins is not selective. Six cell types were transfected with RNA hairpins. Normalized cell viability at 96 h is depicted. Transfections were in duplicates and averaged, data shown are representative of two experiments. The various transfection conditions are described in the text.
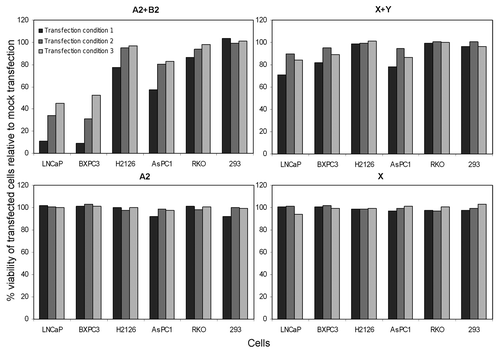
Transfection with X and Y resulted in ~20% of BXPC3 and AsPC1 cells being killed by only transfection condition 1, but unexpectedly ~20% of LNCaP cells were also killed by the same condition (). H2126, RKO, and 293 cells were unaffected by the hairpin transfections. Mesothelin-expressing (H2126) cells were thus resistant to mesothelin hairpins. Thus, HCR was not selective or highly effective using novel target transcripts.
The viability of every cell type was unaffected by transfection with only one hairpin or transfection with only HiPerFect, which were procedural negative controls. 293 and RKO cells, which were cellular negative controls lacking the targeting transcripts of tpc/hpr or mesothelin, were not affected by any hairpin transfection.
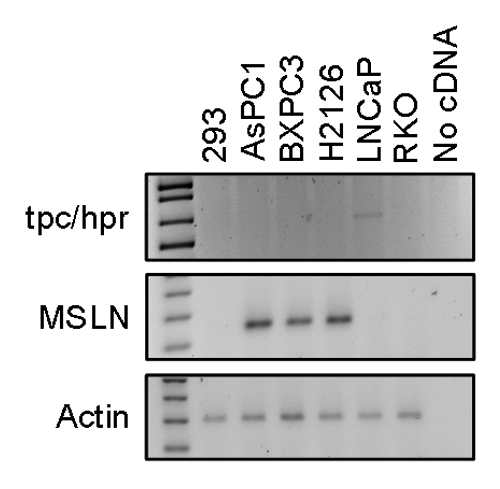
RT-PCR confirmed the selective presence of target transcripts in cells
Because we observed non-selective killing of cells by hairpins, we re-examined whether the “non-selectivity” of the HCR method was due to the presence of tpc/hpr fusion transcript in BXPC3 cells and which of the cells had a mesothelin transcript. By RT-PCR, only LNCaP cells, of the six cell lines, had a tpc/hpr fusion transcript (), as expected. Thus, selective cell death using the reported hairpins should have occurred only in LNCaP cells and not in the other cell lines. AsPC1, BXPC3, and H2126 cells contained the mesothelin transcript (), as expected. Thus, using mesothelin hairpins, selective cell death should have occurred only in AsPC1, BXPC3, and H2126 cells. Yet, we observed non-selective cell killing by both sets of hairpins (tpc/hpr and mesothelin, see above). Thus, the presence of aberrant transcripts was not the reason for non-selective killing of cells when using the HCR method.
FITC-tagged control siRNA transfection
We examined whether a variable transfectability of cells was the reason for non-selective killing. Using FITC-tagged control siRNA, visual comparison revealed that all cell lines had >80% transfectability. Differential cell transfectability was thus not the reason for unexpected patterns of cell killing in our experiments.
Discussion
Venkatraman et al.Citation3 proposed that by use of specifically designed RNA hairpin oligos targeting a unique cellular RNA transcript, a hybridization chain reaction (HCR) can be generated in cells. dsRNA produced in the process would then be recognized by PKR, leading to apoptosis. This theoretically compelling model could be used to kill specific cells by targeting selective RNA species. In the publication, the authors described killing more than 90% of treated cells by use of the above model. The proposal further aimed to treat cancers by targeting mutated genes.
We observed non-selective killing of cells using transfection employing either the tpc/hpr fusion-targeting or MSLN-targeting HCR oligos. Also, our transfection of the recommended amounts of RNA oligos was not sufficient to kill cells as claimed, nor were higher or lower amounts. Reasons for these failures could include the chosen experimental conditions, which differed between in-tube and in-cell HCR. Only one species of target RNA oligo is present during in-tube HCR, but there are multiple sequences of RNA in cells that may initiate the reaction non-specifically. While the oligo design potentially could be improvedCitation5 for better results, we experienced nonselective killing even when using the published oligos.
We verified our methods by cellular transcript analysis, transfectability testing, and by use of in vitro and cellular control experiments. Due to the early observation of unpromising results in cells, we did not look for the PKR activation, DNA double-stranded breaks, or markers for cell apoptosis.
When we searched for non-specific polymerization in vitro, however, we failed to observe it. Why, then, might we have observed non-specific killing? Perhaps only a small number of dsRNA molecules are required to be recognized by PKR, so as to induce the apoptotic response. Presumably, a small number of dsRNA hybrids produced by leaky non-specific polymerization in the presence of a pool of cellular RNA could remain undetected in the gel assay we used and yet induce non-specific cell killing. The prior authors might have overlooked these possibilities due to the contrived, but perhaps misleading, clarity in the in vitro polymerization reaction they used, which employed only artificial target oligos.
Of the 34 publications citing the original report (Web of Science), none provided a replication of the work. There are other studies using dsRNA-PKR model for specific cell killing,Citation6-Citation8 but the techniques were different than described here, and our critique does not apply to them. Recently, the HCR publication was retracted from the Proceedings of the National Academy of Sciences, and the retraction letter failed to comment on the technique’s validity. Here, we offer the basis for doubts. All of our work was done before the retraction of the Proceedings of the National Academy of Sciences paper.
Materials and Methods
Cell lines
Cell lines 293, AsPC1, BXPC3, H2126, LNCaP, and RKO were from ATCC. AsPC1, BXPC3, H2126, and LNCaP cells were cultured in RPMI medium and 293 and RKO cells in DMEM medium, supplemented with 10% FBS and antibiotics. LNCaP cells have a known tpc/hpr translocation and hence express the fusion transcript. AsPC1, BXPC3, and H2126 cells were mesothelin-expressing cell lines. 293 and RKO cells lacked tpc/hpr and mesothelin transcripts.
Design of HCR oligonucleotides
We used two target (initiator) sequences; they were 18 bp long and represented on the targeted mRNA. One spanned the fusion joint in tpc/hpr, target sequence M2: 5′-AAGAAGAAAU AUGUGGGA-3′.Citation3 The second was a part of mesothelin transcript, target sequence Z: 5′-UAAACUGGAU GAGCUCUA-3′. These sequences were targeted in cell-free and cellular HCR. Hairpins targeting the fusion transcript derived from the reported reaction “HCR2”;Citation3 hairpin A2: 5′-UCCCACAUAU UUCUUCUUGA GUAAGAAGAA AUAUGU-3′ and hairpin B2: 5′-AAGAAGAAAU AUGUGGGAACA UAUUUCUUCU UACUC-3′. Hairpins targeting the mesothelin transcript were designed based on rules of the prior reportCitation3 as follows. The map of hairpin X would be: (18 bp reverse compliment sequence of Z)-(4bp loop)-(first 14 bp of Z), which is 5′-UAGAGCUCAU CCAGUUUAGU GGUAAACUGG AUGAGC-3′. The map of hairpin Y would be: (18 bp Z sequence)-(reverse compliment sequence of first 14 bp of Z)-(reverse compliment sequence of 4 bp loop), which is 5′-UAAACUGGAUG AGCUCUAGCU CAUCCAGUUU ACCAC-3′. RNA oligonucleotides were synthesized and HPLC-purified (Integrated DNA Technologies).
Oligonucleotides were diluted in 1× PKR buffer (20 mM HEPES, pH7.4, 4 mM MgCl2, 100 mM KCl), heated at 90 °C for 90 sec, cooled rapidly on ice and incubated at RT for 30 min.
Cell-free polymerization
Using oligonucleotides as initiator sequence
Paired hairpins were mixed with or without initiator oligonucleotide in 1× PKR buffer. Hairpins A2 and B2 were mixed with initiator M2 and hairpins X and Y were mixed with initiator Z. In this reaction, hairpins were 0.8 µM each and initiator 0.4 µM. In the other reaction, hairpins X and Y were mixed with different concentrations of initiator sequence Z. Hairpins were 1.0 µM each and initiator Z was 0.01, 0.033, 1, 0.33, or 1 µM.
Using unfractionated LNCaP RNA to provide the initiator sequence
Hairpins A2 and B2 (0.66 µm each) were mixed with or without various quantities (0.25, 0.5, 1, 5, or 10 µg/reaction) of RNA from LNCaP cells or of target oligonucleotide M2.
All reactions were performed in 13–15 µl final volume. Reaction mixes were incubated at 37 °C for 30 min or for 16 h, resolved on a 1% agarose gelCitation9 containing SYBR Safe (Life Technologies) and visualized under UV light.
Transfection of HCR hairpins
Transfections of hairpins were done using HiPerFect (Qiagen) according to manufacturer’s Fast-Forward transfection protocol and using three conditions: Condition 1: HiPerFect 10 µl/ml in medium, each hairpins, 100 nM; Condition 2: HiPerFect 5 µl/ml, hairpins, 50 nM; and Condition 3: HiPerFect 5 µl/ml, hairpins, 25 nM. Transfections of LNCaP, BXPC3, H2126, AsPC1, RKO, and 293 cells were done in 96-well tissue culture plates at 10,000 cells/well in duplicate. Control transfections used single hairpins at equimolar concentrations or used HiPerFect without hairpins. We incubated cells at 37 °C for 96h, 5% CO2. Cell viability was measured.
Picogreen dsDNA quantification assay as a measure of viability in cell populations
We aspirated medium from wells of 96-well plates, added 50 µl of 0.3% SDS and incubated at RT for 20min. Picogreen was added (50 µl of 1:200 dilution, Invitrogen P7589) and incubated at RT for 50min. Fluorescence was read at excitation 480 ± 20 nm and emission 530 ± 25 nm in a fluorometer (Fusion HT, Packard). The percent of viable experimentally treated cells relative to control cells (treated only with HiPerFect) was calculated.
RT-PCR
RNA was extracted from cell lines using RNEasy (Qiagen). cDNA was synthesized (Superscript III cDNA synthesis kit, Life technologies) from RNA of each cell line. PCR-amplification of the fusion, mesothelin and actin transcripts used the following primer sets. Fusion primers:Citation3 5′-CCAATCTGGG AAACACTATC-3′ and 5′-TTAGACATGC CGACACAGAA-3′; mesothelin primers: 5′-ATGAAGCTGC GGACGGATGC-3′ and 5′-GAGGCAGGGC GTCCCCGAGA-3′; and actin primers: 5′-AGAAAATCTG GCACCACACC-3′ and 5′-AGAGGCGTAC AGGGATAGCA-3′. We resolved PCR products in gels as above.
Transfection of FITC-tagged control siRNA
Cells were transfected and seeded in four-chamber slides (Permanox, Sigma-Aldrich) with HiPerFect (10 µl/ml) and FITC-tagged control siRNA (50 nM, Santa Cruz, sc36869). We compared cellular fluorescence visually by fluorescence microscopy on the following day.
Acknowledgments
This work was supported by NIH grants CA62924 and CA134292 and the Everett and Marjorie Kovler Professorship in Pancreas Cancer Research.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
References
- García MA, Meurs EF, Esteban M. The dsRNA protein kinase PKR: virus and cell control. Biochimie 2007; 89:799 - 811; http://dx.doi.org/10.1016/j.biochi.2007.03.001; PMID: 17451862
- Dirks RM, Pierce NA. Triggered amplification by hybridization chain reaction. Proc Natl Acad Sci U S A 2004; 101:15275 - 8; http://dx.doi.org/10.1073/pnas.0407024101; PMID: 15492210
- Venkataraman S, Dirks RM, Ueda CT, Pierce NA. Selective cell death mediated by small conditional RNAs. Proc Natl Acad Sci U S A 2010; 107:16777 - 82; http://dx.doi.org/10.1073/pnas.1006377107; PMID: 20823260
- Ren YR, Patel K, Paun BC, Kern SE. Structural analysis of the cancer-specific promoter in mesothelin and in other genes overexpressed in cancers. J Biol Chem 2011; 286:11960 - 9; http://dx.doi.org/10.1074/jbc.M110.193458; PMID: 21288909
- Choi HM, Chang JY, Trinh A, Padilla JE, Fraser SE, Pierce NA. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat Biotechnol 2010; 28:1208 - 12; http://dx.doi.org/10.1038/nbt.1692; PMID: 21037591
- Shir A, Levitzki A. Inhibition of glioma growth by tumor-specific activation of double-stranded RNA-dependent protein kinase PKR. Nat Biotechnol 2002; 20:895 - 900; http://dx.doi.org/10.1038/nbt730; PMID: 12205508
- Shir A, Ogris M, Wagner E, Levitzki A. EGF receptor-targeted synthetic double-stranded RNA eliminates glioblastoma, breast cancer, and adenocarcinoma tumors in mice. PLoS Med 2006; 3:e6; http://dx.doi.org/10.1371/journal.pmed.0030006; PMID: 16318410
- Li YJ, Zeng JM, Huang SF, Wang XZ, Zhao SQ, Bai WJ, et al. Selective leukemia cell death by activation of the double-stranded RNA-dependent protein kinase PKR. Int J Mol Med 2011; 28:215 - 22; PMID: 21468538
- Brody JR, Kern SE. Sodium boric acid: a Tris-free, cooler conductive medium for DNA electrophoresis. Biotechniques 2004; 36:214 - 6; PMID: 14989083