Abstract
Although daunomycin and adriamycin are considered effective antitumor drugs and have been used in the clinic for over 40 years, their mechanism of action is still a matter of debate. We investigated the influence of daunomycin on interaction between linker or core histones and DNA in live HeLa cells in vitro, using image and flow cytometry. Exposure to daunomycin at clinically relevant concentrations (25–250 nM) caused dissociation of wild-type H1.1 as well as 4 H1 point mutants from DNA, followed by their accumulation in nucleoli and aggregation of chromatin. A detectable dissociation of H2B core histones occurred only at much higher concentrations of the drug (500 nM). Replication of DNA and synthesis of RNA were not halted by daunomycin (up to 2500 nM); however the characteristic subnuclear distribution of sites of transcription and replication was lost. Dissociation of the H1.1 linker histones and subsequent loss of higher order chromatin structures may constitute an important component of the mechanism of cytotoxicity of daunomycin.
Introduction
Although daunomycin and adriamycin are considered effective antitumor drugs and have been used in the clinic for over 40 years, their mechanism of action is still a matter of debate. Several mechanisms of cytotoxicity of these drugs have been proposed including inhibition of DNA replication and transcription, generation of free radicals, generation of DNA crosslinks, and creation of DNA breaks following stalling of topoisomerase II.Citation1-Citation4 However, some of these mechanisms were observed in the presence of daunomycin that exceed the concentrations occurring in plasma of treated patients.Citation1 Thus, contributions of these effects to cytotoxicity exerted on tumor and untransformed cells in patients remains unclear.
Histone H1 is a principal factor responsible for stabilization of chromatin higher order structures in cell nucleus.Citation5 It is also known that chromatin higher order structures are a factor involved in control of gene expression.Citation6 Thus, we tested a hypothesis based on the assumption that cytotoxicity of daunomycin is mediated by exerting adverse effects on interaction between histone H1 and DNA.
We demonstrate that daunomycin at clinically achievable concentrations causes dissociation of H1.1 histones from DNA and subsequent disruption of spatial organization of chromatin and nuclear structure. Replication and transcription are not halted; however, the characteristic subnuclear distribution of transcription and replication sites in the nucleus is lost. These observations suggest that daunomycin-induced dissociation of histones H1 and a subsequent loss of higher order chromatin structures are important components of the mechanism of cytotoxicity of this drug.
Results
Daunomycin-induced dissociation of H1.1 histones from DNA and aggregation of chromatin
The influence of daunomycin on interaction between histones and DNA was studied in live cells exposed to clinically relevant as well as higher drug concentrations. When added to culture medium, daunomycin readily crossed the plasma membrane and entered the nucleus (). The fluorescence of daunomycin which entered cell interiors became detectable in cytoplasmic vesicles, the Golgi apparatus, and chromatin within minutes after adding the drug to culture medium ().Citation7,Citation8 Although daunomycin is known to readily intercalate into DNA, however, the intensity of fluorescence of daunomycin in the nuclei of live cells was much lower than in the cytoplasm (). This unexpectedly low intensity of nuclear fluorescence is likely to reflect a limited access and a reduced ability of daunomycin to bind DNA in situ, as well as quenching of daunomycin fluorescence upon intercalation into DNA.Citation9,Citation10 Following formaldehyde fixation, the drug was released from the Golgi apparatus and was bound by nuclear DNA (). Unlike in the nucleus, the drug accumulated in the Golgi was not tightly bound as demonstrated by its release during a fixation procedure.
Figure 1. Entry and subcellular localization of daunomycin in live HeLa cells. (A and B) Transmitted light and fluorescence images showing a cell in medium supplemented with daunomycin (500 nM; 30 min); scale bar 10 μm. (C) The intensities of fluorescence of daunomycin accumulating in the region of the Golgi apparatus and in chromatin. (D−G) Images of a live cell exposed to daunomycin for 30 min and the same cell after formaldehyde fixation. Daunomycin is released from intracellular vesicles and Golgi apparatus upon fixation; thus flow cytometry of fixed cells detects only the DNA-bound drug (see below). (D and E) Transmitted light images; (F and G) Fluorescence (the detection level in [F and G] is the same), scale bar 5 μm.
![Figure 1. Entry and subcellular localization of daunomycin in live HeLa cells. (A and B) Transmitted light and fluorescence images showing a cell in medium supplemented with daunomycin (500 nM; 30 min); scale bar 10 μm. (C) The intensities of fluorescence of daunomycin accumulating in the region of the Golgi apparatus and in chromatin. (D−G) Images of a live cell exposed to daunomycin for 30 min and the same cell after formaldehyde fixation. Daunomycin is released from intracellular vesicles and Golgi apparatus upon fixation; thus flow cytometry of fixed cells detects only the DNA-bound drug (see below). (D and E) Transmitted light images; (F and G) Fluorescence (the detection level in [F and G] is the same), scale bar 5 μm.](/cms/asset/39e7ec9c-5ce7-4b6f-85bf-cb9817eff635/kcbt_a_10925328_f0001.gif)
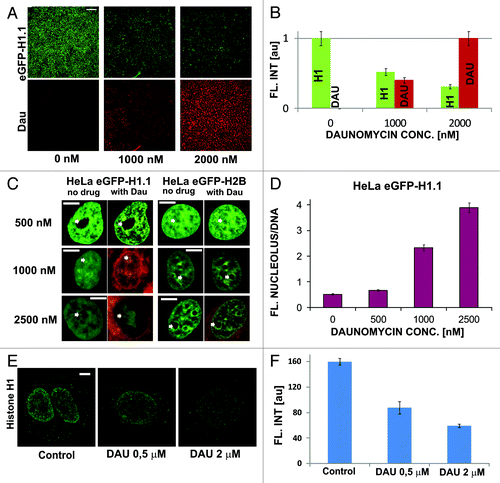
In order to investigate the influence of DNA-intercalated daunomycin on chromatin structure, we studied cells expressing H1.1 histone tagged with GFP. First, cells were exposed to a high concentration of daunomycin (1000, 2000, or 2500 nM) for 120 min. Accumulation of daunomycin in cells caused a decrease of the intensity of fluorescence of GFP-H1.1 (). At the same time, histone GFP-H1.1 accumulated in nucleoli (). Subsequently chromatin appeared to aggregate in numerous small foci (). No accumulation of H2B core histones in nucleoli was detected. This decrease of GFP-H1.1 fluorescence in chromatin may have arisen due to two reasons—the dissociation of linker histones from DNA and quenching of fluorescence of GFP-H1.1 bound to DNA by the intercalating daunomycin.Citation11 The drug-driven dissociation of endogenous H1.1 histones from DNA () was also detected by immunofluorescence. However, nucleolar binding of the H1.1 histones which had been dissociated from DNA was not detected by immunofluorescence since the dissociated H1.1 histones were not fixed by formaldehyde. The translocated histones were washed out from nucleoli during the subsequent steps of the immunofluorescence procedure (data not shown).
Figure 2. Dissociation of H1.1 linker histones from DNA and translocation to nucleoli upon exposure to daunomycin. (A and B) Images and a graph showing a decrease of the intensity of green fluorescence of GFP-H1.1 after a 2 h exposure of live HeLa cells to daunomycin (1 or 2 μM); scale bar 100 μm. (C) Images of histone GFP-H1.1, GFP-H2B, and daunomycin (0.5, 1, and 2 μM) in live cells. As the drug binds to DNA, chromatin aggregates and histones H1.1 but not H2B accumulate in nucleoli (arrows). (D) A ratio between the intensities fluorescence of GFP-H1.1 histones bound in nucleoli and to DNA outside of the nucleoli. At higher drug concentrations more histone H1.1 is translocated to nucleoli and the intensity of the nucleolar fluorescence of daunomycin exceeds that of chromatin by a factor of almost 4. (E and F) Dissociation of histone H1 from DNA in cells exposed to daunomycin, detected by immunofluorescence. Images and a graph showing the average fluorescence intensity of immunolabeled H1.1 histone detected in nuclei of untreated cells and in the treated cells, following fixation. The H1 histone, which dissociated from DNA, is washed out during the immunofluorescence procedure and escapes detection by this method, but the process can be observed directly under a microscope (data not shown); scale bar 5 μm.
In order to assess the contributions of fluorescence quenching and histone dissociation from DNA to the observed decrease of fluorescence we first studied the process of quenching in formaldehyde fixed cells (). Quenching of GFP-H1.1 was dependent on the concentration of daunomycin, whereas quenching of the GFP tag on the H2B core histones was only very weak (). As expected, no accumulation of histones in the nucleoli of fixed cells was detected ().
Figure 3. Dissociation of H1.1 histones and changes in architecture of chromatin in cells exposed to slowly increasing concentrations of daunomycin. (A) Images of fluorescence of GFP-H1.1, GFP-H2B, and daunomycin in formaldehyde-fixed cells incubated with increasing concentrations of the drug. A loss of the green signal is due entirely to fluorescence quenching; scale bar 2 μm. (B) A graph illustrating quenching of GFP on H1.1 and H2B histones by daunomycin. (C) A ratio between fluorescence intensities of histones GFP-H1.1 in nucleoli vs nuclear DNA, in fixed cells. (D) Fluorescence intensity of histones GFP-H1.1 and GFP-H2B in live cells exposed to growing concentrations of daunomycin. A loss of the green signal is due primarily to H1.1 histone dissociation and subsequent degradation. (E) A graph illustrating a decrease of fluorescence of GFP-H1 and GFP-H2B histones in live cells exposed to daunomycin. (F) A ratio between fluorescence intensities of histones GFP-H1.1 in nucleoli vs nuclear DNA, in live cells, demonstrating accumulation of histone H1.1 in nucleoli at drug concentrations above 1 μM
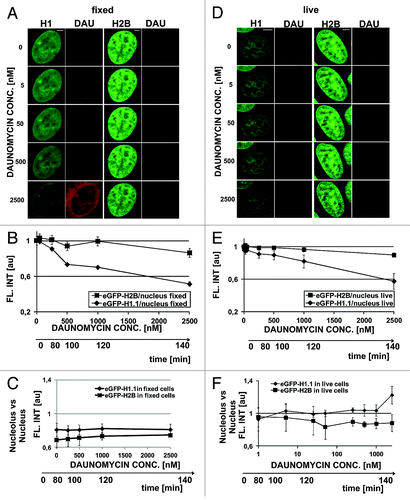
In order to further elucidate the influence of dissociation of H1.1 histones from DNA on the structure of the nucleus, we incubated live cells with stepwise increasing concentrations of daunomycin (20–30 min for each concentration) and analyzed subsequent images of the same selected cells (). The nuclear structure was seen via H1 or H2B histone distribution. Chromatin aggregation was more conspicuous in cells expressing GFP-H2B than GFP-H1.1. In the GFP-H1.1 cells changes in subnuclear chromatin distribution were obfuscated by a gradual loss of the green signal resulting from GFP-H1.1 histones fluorescence quenching and dissociation from DNA (). Aggregation was first detected in perinucleolar heterochromatin, even at a drug concentration as low as 5 nM. A step-wise increase of daunomycin concentration up to 2500 nM in this cell culture caused further, more pronounced chromatin aggregation () and dissociation of histone H1. Only a weak accumulation of H1.1 in nucleoli, similar to the process shown in , was observed during this prolonged incubation with daunomycin (1000 nM; ). It is possible that under conditions of this experiment, where the concentration of the drug was increasing quite slowly, efficient degradation of the dissociated histones occurred and the pool of the H1.1 detached histones available for translocation and binding in nucleoli was lower than in the case of a one-time rapid exposure to a high drug concentration, which is shown in .
Daunomycin-induced H1.1 histone dissociation measured by flow cytometry
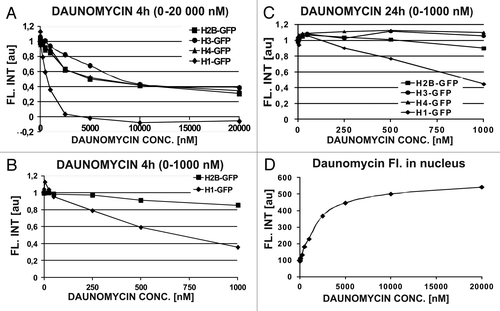
The experiments described above demonstrate the dissociation of H1.1 histones from DNA in individual cells assessed by image cytometry. We asked whether the specific H1 histone dissociation can be measured in the whole cell population by flow cytometry, at clinically relevant concentrations of daunomycin (25–250 nM). We addressed this question by studying the effects of a wide range of daunomycin concentrations and 4 h as well as 24 h exposures, on the H1.1 linker and three core histones. At the low concentrations, only the fluorescence of H1.1 histones, but not the core histones, was significantly reduced at DNA, suggesting a selective dissociation of the linker histone from DNA and subsequent degradation (). Such a dissociation of H1.1 but not the core histones suggests that a loss of higher order chromatin structures was not accompanied by dismantling of nucleosomes. The core histones were not dissociated until daunomycin reached a high concentration of 1 μM (), suggesting that nucleosomes can be dismantled by daunomycin only at high drug concentrations. Interestingly, a large proportion of nucleosomes seem resistant to daunomycin, since even at 10–20 μM daunomycin the fluorescence intensity of H2B, H3, or H4 core histones did not diminish below the level of 40% of the initial value. Following a 24 h long incubation at low drug concentrations (25−50 nM) the amount of core H2B, H3, and H4 histones transiently increased () probably as a result of endoreplication of DNA (see below). demonstrates that the amount of daunomycin bound to DNA in the nucleus increased as a function of extracellular concentration of the drug. The binding sites appear to be close to saturation at 10 000 nM, a concentration above which the remaining core histones could no longer be dissociated from DNA.
Figure 4. Daunomycin-induced dissociation of histones from DNA measured by flow cytometer. (A–C) Concentrations of linker (H1) and core (H2B, H3, and H4) histones in daunomycin-treated cells after 4 h or 24 h incubation with the daunomycin. (D) Fluorescence of daunomycin bound to DNA, in fixed cells, as function of the drug concentration (4 h incubation)
H1.1 histone dynamics in cells treated with daunomycin
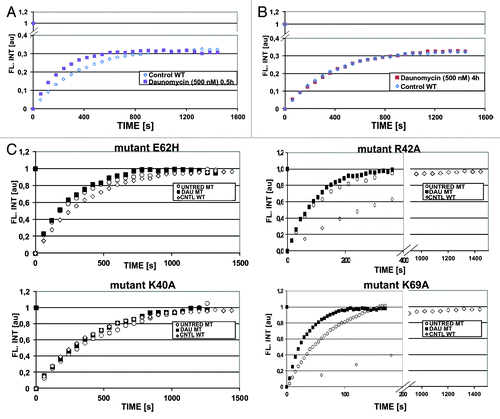
Despite exposure to daunomycin at 500 nM and subsequent aggregation of chromatin, a subpopulation of H1.1 histones (~60%, ) remained bound to DNA. FRAP experiments performed on the GFP-H1.1 histones, which remained associated with the aggregated chromatin 30 and 120 min after administration of the drug showed that H1.1 histones were still dynamic, i.e., exchanging with a mobile pool (). The recovery of fluorescence was faster than in untreated cells (), indicating a faster exchange rate and possibly a larger soluble pool. However, no such fast fluorescence recovery was observed after a 4 h incubation with daunomycin (). Thus, the postulated increased pool of soluble H1 histones appears to be created shortly after the cells are exposed to daunomycin (at a high concentration), but it is reduced later, possibly due to degradation of the unbound histones. The fact that the fluorescence recovery rate of H1 bound within daunomycin-aggregated chromatin is the same as in untreated cells suggests that there may exist some chromatin regions that are resistant to daunomycin intercalation.
Figure 5. Dynamics of histone GFP-H1.1 by FRAP. (A) FRAP of GFP-H1.1 in live HeLa cells exposed to 500 nM daunomycin for 0.5 h, indicating the presence of a mobile pool (open symbols, untreated; closed symbols, daunomycin-treated). (B) Dynamics of histone GFP-H1.1 returns to the level of untreated cells after 4 h incubation with daunomycin (500 nM). (C) Dynamics of four point mutants of GFP-H1.1, in 3T3 fibroblasts exposed to daunomycin (500 nM, for 4 h; open symbols, untreated; closed symbols, daunomycin-treated). Note a different time scale in graphs representing mutants R42A and K69A characterized by a rapid rate of exchange.
Dynamics of point mutated histones H1
We further investigated the dynamics of H1 histones altered by single point mutations.Citation12 Dynamics of three H1 histone mutants (K40A, R42A, and E62H) showed no significant difference between the treated and untreated cells (). However, the mutant K69A, which is characterized by the shortest residence time, showed a significantly faster fluorescence recovery in cells exposed to daunomycin. In this mutant alanine in position 69 is replaced with lysine. This change affects the region of histone H1 molecule that binds to DNA. Clearly this substitution strongly influences the DNA binding properties of the whole protein, while the other three mutations do not show such a strong effect.Citation12 This observation is in agreement with the notion that intercalation of daunomycin into DNA influences interaction between H1 linker histone and DNA in this particular region of the H1 molecule.
Daunomycin and DNA replication
Inhibition of DNA synthesis has been suggested as a possible mechanism of daunomycin cytotoxicity.Citation1,Citation13,Citation14 However, we found that DNA synthesis was only slightly decreased by daunomycin at 50 nM and significantly reduced but not entirely abolished even at high concentrations of the drug (2500 nM) (). These observations suggest that DNA synthesis slowed down but was not halted, while the cell divisions seized (Fig. S1). Although replication was still occurring in cells treated with the drug, changes in spatial distribution of this process were apparent (). In cells treated with daunomycin at concentrations exceeding 50 nM the patterns of replication, which are characteristic for different stages of S phase, were missing. The DNA replication seemed to occur throughout the nucleus, resembling the pattern characteristic for early S phase. This could be due to the arrest in early S phase as well as a loss of higher order chromatin structures.
Figure 6. Replication and transcription in cells treated with daunomycin. (A and B) Images of immunolabeled BrdU (A) and BrU (B) incorporated into DNA and RNA respectively, in daunomycin-treated cells; scale bar 5 μm (A) and 20 μm (B).(C) Relative amounts of RNA in daunomycin-treated cells. (D) Cell viability after treatment with daunomycin, measured by the MTT assay.
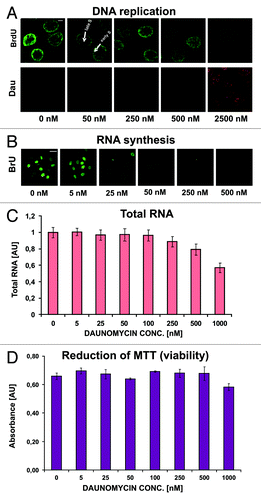
Daunomycin and RNA synthesis
Microscopy studies using BrU incorporation showed that RNA synthesis decreased in daunomycin-treated cells, but was still present even at high drug concentrations (). A significant decrease of incorporation of BrU was observed in cells exposed for 24 h to daunomycin at 250 nM or higher. A “viability” test (MTT reduction assay) showed that the drug treatment (0−1000 nM, 24 h) did not cause immediate loss of cell metabolic activity ().Citation15 Cell death occurred at later times, as indicated in Table S1. These observations suggest that RNA synthesis was not abolished, thus halting RNA production by daunomycin is unlikely to be a principal cause of cytotoxicity of this drug.
Daunomycin and DNA damage (histone H2AX phosporylation)
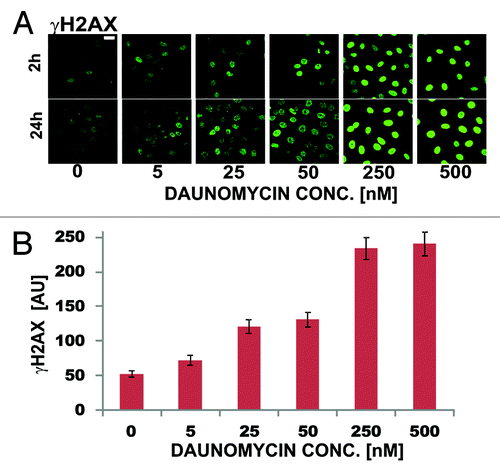
DNA strand brakes induced by daunomycin, due to stalling of topoisomerases, are thought to be involved in the mechanism of cytotoxicity of anthracycline drugs.Citation4 We employed histone H2AX phosphorylation assay to detect DNA breaks.Citation16 At drug concentration typically present in plasma of patients (25 nM) only a modest increase of the number of γH2AX foci was detected (2–3 times, ). Higher concentrations of daunomycin (250 nM) were needed to cause widespread histone H2AX phosphorylation (almost 5-fold increase of the number of γH2AX foci in comparison with the untreated cells). The level of phosphorylation of H2AX was similar after 2 and 24 h exposure to the drug ().
Figure 7. DNA damage assessed by means of immunodetection of histone H2AX phosphorylation in daunomycin-treated cells (2 h and 24 h). (A) images of immunolabeled histone γH2AX in cells treated with increasing concentrations of daunomycin for 2 and 24 h; scale bar 20 μm. (B) Average fluorescence intensity of immunolabeled histone γH2AX per cell, following 2 h treatment with daunomycin at different concentrations.
Long-term observation of cells treated with various concentrations of daunomycin
The importance of various proposed cytotoxicity mechanisms is likely to be dependent on the concentration of the drug vs. the cell number and duration of the treatment. In order to search for a correlation between cell death and drug-induced dissociation of H1.1 histones, we performed long-term observation of drug-treated cells (Fig. S1; Table S1). At clinical concentrations of daunomycin (25–50 nM) no obvious adverse effects were seen in the first 4 h. Even after 24 h, only a few cells showed morphological signs of apoptosis. Figure S1 reveals that 24 h after the beginning of drug exposure cells showed no obvious signs of death, even after a treatment with 1000 nM daunomycin. Longer drug exposure times were required in order to cause morphological changes. Cell death occurred a few days after the drug treatment, even when the drug was no longer present in the culture medium. After 48 h, a significant decrease of cell numbers occurred only in cultures exposed to high daunomycin concentrations (>500 nM). After 192 h a significant decrease in cell number was seen even in a culture originally exposed to 25 nM daunomycin. In a culture treated with 50 nM daunomycin, only single cells remained attached to substratum. These observations, combined with the data obtained by flow cytometry () demonstrate that the drug concentration leading to cell damage and death (250 nM) observed 8 d after the daunomycin treatment (Fig. S1) is similar to the concentration leading to dissociation of H1.1 histones from DNA and a loss of higher order chromatin structures.
Discussion
Daunomycin, chromatin structure, and mechanism of cytotoxicity
The results presented here demonstrate that daunomycin causes dissociation of H1.1 histones from chromatin and induces changes in higher order chromatin organization, which ultimately lead to chromatin aggregation. A loss of chromatin structure due to dissociation of H1 histones from the DNA is consistent with the known role of the linker histone in maintaining higher order chromatin structures.Citation5,Citation17,Citation18 Interestingly, daunomycin does not appear to lead to eviction of the core histones from DNA. This implies that in cells challenged with clinically relevant concentrations of daunomycin the nucleosomes are not dismantled. Since higher order chromatin structures are involved in control of gene expression, we postulate that the mechanism of cytotoxic action of daunomycin may be initiated by altering interactions between the linker histones and DNA.
The fate of the dissociated H1.1 histones
When the linker histones are expelled from DNA, a transient soluble pool of H1.1 is created (). It is also possible that the detached histones are modified by daunomycin. It has been shown that daunomycin can bind directly to histones, with a preference for the linker histone (although this was observed ex vivo, at very high drug concentrations).Citation19,Citation20 Such histone alteration is likely to be recognized by the cellular protein quality control machinery and may lead to H1.1 degradation.Citation21 We cannot exclude the possibility that the exciting light, which we have to use in the imaging experiments, accelerates degradation of the modified histones as well. Interestingly, however, we did not observe H1.1 histone degradation in cells exposed to DRAQ5—a different anthracycline derivative. DRAQ5 causes dissociation of H1.1 histone from DNA and, to a lesser degree, dissociation of the core histones as well.Citation11 The molecules of H1.1, which are expelled from DNA by daunomycin appear to accumulate in nucleoli readily (). Interestingly, the H1 mutants with reduced affinity to DNA were also reported to accumulate in nucleoli.Citation12 Thus, nucleolus appears to be the default localization for H1 histones that were dissociated from DNA following drug intercalation or due to a lower binding constant resulting from a mutation.
Some H1 histones remain associated with DNA and are still dynamic
The weaker binding of an H1 point mutant to DNA in the presence of daunomycin is consistent with the notion that daunomycin alters the binding sites for this histone. However, daunomycin does not expel all H1 histone molecules from DNA and leaves the core histones in place (). The dynamics of these bound H1 histones is unaffected by daunomycin. It is thus justified to postulate that in live cells intercalation of daunomycin into DNA pushes bases apart and causes a change of the DNA structure which alters the binding sites for histones H1. Alteration of the binding sites could, in turn, decrease the affinity of binding of H1 histones to the daunomycin-decorated DNA and result in shifting the balance toward the unbound pool of H1. Thus, an interesting question remains—whether the molecules of H1 histone that remain in contact with DNA and are characterized by an unaltered dynamic behavior, are bound in short DNA stretches between the intercalated molecules of daunomycin or whether some large areas of DNA are resistant to daunomycin intercalation and maintain a normal association with H1. This could happen if some shielding chromatin proteins were bound exclusively in discrete areas of DNA. Even if the size of such intercalator-resistant DNA regions were relatively large, i.e., many thousands of bases, they would still be unresolvable by standard confocal microscopy which we used in this study.
Daunomycin perturbs but does not halt replication and transcription
Replication and transcription are not halted by daunomycin at clinically relevant concentrations. High daunomycin concentrations (above 2 μM) are needed to stop replication and transcription. In this respect daunomycin differs from supravital DNA binding agents like DRAQ5, Hoechst, and Syto17 which significantly reduce replication and transcription (Wójcik and Dobrucki, unpublished data). Thus, we postulate that daunomycin-induced dissociation of H1.1 histone that leads to a collapse of higher order chromatin structures, perturbs the control of DNA and RNA synthesis.
DNA damage in daunomycin-treated cells
We observed slightly elevated levels of phosphorylation of histone γH2AX after exposure of cells to clinically relevant concentrations of daunomycin (). Since phosphorylation of histone H2AX is a generally accepted marker of DNA strand brakes, it is possible to interpret our data as evidence of some DNA breakage resulting from daunomycin intercalation.Citation22 Phosphorylation of H2AX in cells exposed to another anthracycline, DRAQ5, was also reported.Citation23 However we also note that the absence of a clear evidence in support of DNA double strand breaks in cells treated with anthracycline antibiotics under therapeutic conditions was reported.Citation1 On the other hand it has been shown that histone phosphorylation may result from changes of chromatin structure which are unlikely to be associated with DNA breaks and cause no major loss of cell viability.Citation24 Since our data demonstrate chromatin aggregation even at very low daunomycin concentrations (5 nM, ), we suggest that changes of chromatin structure caused by daunomycin via H1 histone depletion may lead to phosphorylation of H2AX, without any major increase in the number of DNA breaks.
Mechanism of daunomycin cytotoxicity
We demonstrate that at clinically relevant concentrations of daunomycin no immediate cell death, and no complete inhibition of replication, transcription and metabolic activity occur. Lethal drug effects become visible at a later time, days after the drug removal from the surrounding environment. Thus, based on the existing body of knowledge and the observations described above we postulate the following sequence of events: daunomycin enters a cell interior, becomes partly sequestered by endosomes and the Golgi; some molecules of the drug are pumped out by MDR mechanisms. The molecules that reach the nucleus compete with H1.1 histone for binding sites on a DNA double helix. Once a molecule of daunomycin has intercalated into DNA, the structure of the double helix changes so that the affinity of H1 histone for the altered polymer is decreased. The pool of mobile H1.1 transiently increases, but a part of the pool of soluble H1.1 is degraded, whereas the remaining H1.1 molecules reach the nucleolus and bind to RNA. Dissociation of H1.1 from DNA is dependent on the concentration of daunomycin, leads to a loss of higher order chromatin structures and ultimately interferes with transcription and replication.
Materials and Methods
Daunomycin and cells
Daunomycin (Sigma) stock solutions were kept frozen and thawed prior to adding to cell cultures. HeLa cells were cultured according to standard procedures. HeLa cells expressing histones tagged with eGFP were kindly provided by Dr T Kanda, Dr H Kimura, and Prof PR Cook.Citation25,Citation26 3T3 cells with histone H1 point mutants were kindly provided by Dr DT Brown.Citation12
Microscopy and live cell imaging
Bio-Rad MRC1024 confocal system (Bio-Rad Microscience) was used.Citation27 GFP and daunomycin were excited simultaneously by 488 nm light. Optical filters used were: primary dichroic 510DCLP, secondary 565DRLP, emission 540/30, and 585 LP. Live cells were prepared and imaged as described previously.Citation11,Citation27
Immunolabeling of BrdU, BrU, H1, and H2AX
BrU or BrdU was added to culture medium for 2 h or 30 min respectively; subsequently the cells were washed with PBS and fixed with formaldehyde.Citation28 0,5% Triton X-100 was used to permeate cell membranes. DNA in cells exposed to BrdU was denatured with 2 M HCl. Incubation in 3% BSA before addition of the primary antibody was performed in order to prevent nonspecific binding of the antibody. After 1 h incubation with the primary antibody (anti BrdU/Bru or anti H1 or anti-H2AX) cells were washed with PBS, and goat anti-mouse secondary antibody labeled with AlexaFluor 488 was added for the next 1 h incubation. Cells were washed with PBS and confocal images were recorded.
FRAP studies of histone dynamics
Bleaching of eGFP-H1.1 in HeLa cells was performed in a square 8.65 μm × 8.65 μm, ND filter 10%, 4 scans. When recording fluorescence recovery, 512 × 512 pixel images were collected in 60 sec intervals for 1440 sec. In 3T3 cells with histone H1 point mutations bleached square was 2 μm × 2 μm, ND filter 30%, 1 scan, intervals between collected recovery images were adjusted to the mutant's dynamics.Citation12 Image processing and manual measurements of fluorescence recovery were done using ImagePro Plus software.
Flow cytometry
Cells were seeded on 40 mm Petri dish, various amounts of daunomycin were added to culture medium and cells were incubated for 4 or 24 h. Then cells were washed with PBS, trypsinized and 4% formaldehyde was added for 15 min. Subsequently cells were washed with PBS and flow cytometry measurements were performed, using a CyFlow ML flow cytometer (Partec GmbH). A red laser diode (635 nm) was used for excitation of daunomycin, a solid-state blue laser (488 nm) was used for excitation of GFP.
RNA content
RNA in cells treated with daunomycin was measured using a spectrophotometric method. RNA was extracted using fenozol (A&A Biotechnology) as described previously.Citation29 Concentration of the RNA in each probe was assessed spectrophotometrically using NanoDrop ND-1000 Spectrophotometer (absorption at 260 and 280 nm).
MTT test
General cell metabolic activity was assessed using MTT reduction assay.Citation15,Citation30 The dissolved formazan was collected from each well and the absorbance was measured at 560 nm using microplate reader Tecan GENios Plus.
Additional material
Download Zip (4.4 MB)Acknowledgments
We are indebted to Dr DT Brown for the H1 point mutants. The authors thank Dr S Gołda and Dr G Szewczyk for assistance with MTT assay and RNA measurements. This work was supported by a research grant 2011/01/B/NZ3/00609 from the National Centre for Science, Kraków. Confocal microscopes were acquired through generous grants awarded by Foundation for Polish–German Cooperation in Warsaw and European Union Structural Funds (POIG.02.01.00-12-064/08; BMZ).
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
References
- Gewirtz DA. A critical evaluation of the mechanisms of action proposed for the antitumor effects of the anthracycline antibiotics adriamycin and daunorubicin. Biochem Pharmacol 1999; 57:727 - 41; http://dx.doi.org/10.1016/S0006-2952(98)00307-4; PMID: 10075079
- Sinha BK, Katki AG, Batist G, Cowan KH, Myers CE. Differential formation of hydroxyl radicals by adriamycin in sensitive and resistant MCF-7 human breast tumor cells: implications for the mechanism of action. Biochemistry 1987; 26:3776 - 81; http://dx.doi.org/10.1021/bi00387a006; PMID: 2820475
- Sawyer DB, Fukazawa R, Arstall MA, Kelly RA. Daunorubicin-induced apoptosis in rat cardiac myocytes is inhibited by dexrazoxane. Circ Res 1999; 84:257 - 65; http://dx.doi.org/10.1161/01.RES.84.3.257; PMID: 10024299
- Tewey KM, Rowe TC, Yang L, Halligan BD, Liu LF. Adriamycin-induced DNA damage mediated by mammalian DNA topoisomerase II. Science 1984; 226:466 - 8; http://dx.doi.org/10.1126/science.6093249; PMID: 6093249
- Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol 1979; 83:403 - 27; http://dx.doi.org/10.1083/jcb.83.2.403; PMID: 387806
- Pliss A, Malyavantham K, Bhattacharya S, Zeitz M, Berezney R. Chromatin dynamics is correlated with replication timing. Chromosoma 2009; 118:459 - 70; http://dx.doi.org/10.1007/s00412-009-0208-6; PMID: 19296120
- Moriyama Y, Manabe T, Yoshimori T, Tashiro Y, Futai M. ATP-dependent uptake of anti-neoplastic agents by acidic organelles. J Biochem 1994; 115:213 - 8; PMID: 8206870
- Ouar Z, Bens M, Vignes C, Paulais M, Pringel C, Fleury J, et al. Inhibitors of vacuolar H+-ATPase impair the preferential accumulation of daunomycin in lysosomes and reverse the resistance to anthracyclines in drug-resistant renal epithelial cells. Biochem J 2003; 370:185 - 93; http://dx.doi.org/10.1042/BJ20021411; PMID: 12435274
- Darzynkiewicz Z, Traganos F, Kapuscinski J, Staiano-Coico L, Melamed MR. Accessibility of DNA in situ to various fluorochromes: relationship to chromatin changes during erythroid differentiation of Friend leukemia cells. Cytometry 1984; 5:355 - 63; http://dx.doi.org/10.1002/cyto.990050411; PMID: 6468176
- Johnston FP, Jorgenson KF, Lin CC, van de Sande JH. Interaction of anthracyclines with DNA and chromosomes. Chromosoma 1978; 68:115 - 29; http://dx.doi.org/10.1007/BF00287144; PMID: 80312
- Wojcik K, Dobrucki JW. Interaction of a DNA intercalator DRAQ5, and a minor groove binder SYTO17, with chromatin in live cells--influence on chromatin organization and histone-DNA interactions. Cytometry A 2008; 73:555 - 62; http://dx.doi.org/10.1002/cyto.a.20573; PMID: 18459157
- Brown DT, Izard T, Misteli T. Mapping the interaction surface of linker histone H1(0) with the nucleosome of native chromatin in vivo. Nat Struct Mol Biol 2006; 13:250 - 5; http://dx.doi.org/10.1038/nsmb1050; PMID: 16462749
- Karukstis KK, Thompson EH, Whiles JA, Rosenfeld RJ. Deciphering the fluorescence signature of daunomycin and doxorubicin. Biophys Chem 1998; 73:249 - 63; http://dx.doi.org/10.1016/S0301-4622(98)00150-1; PMID: 9700924
- Bremerskov V, Linnemann R. Some effects of daunomycin on the nucleic acid synthesis in synchronized L-cells. Eur J Cancer 1969; 5:317 - 30; http://dx.doi.org/10.1016/0014-2964(69)90045-0; PMID: 5388282
- Bernas T, Dobrucki J. Mitochondrial and nonmitochondrial reduction of MTT: interaction of MTT with TMRE, JC-1, and NAO mitochondrial fluorescent probes. Cytometry 2002; 47:236 - 42; http://dx.doi.org/10.1002/cyto.10080; PMID: 11933013
- Olive PL. Detection of DNA damage in individual cells by analysis of histone H2AX phosphorylation. Methods Cell Biol 2004; 75:355 - 73; http://dx.doi.org/10.1016/S0091-679X(04)75014-1; PMID: 15603433
- Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. Dynamic binding of histone H1 to chromatin in living cells. Nature 2000; 408:877 - 81; http://dx.doi.org/10.1038/35048610; PMID: 11130729
- Thoma F, Koller T. Influence of histone H1 on chromatin structure. Cell 1977; 12:101 - 7; http://dx.doi.org/10.1016/0092-8674(77)90188-X; PMID: 561660
- Rabbani A, Finn RM, Thambirajah AA, Ausió J. Binding of antitumor antibiotic daunomycin to histones in chromatin and in solution. Biochemistry 2004; 43:16497 - 504; http://dx.doi.org/10.1021/bi048524p; PMID: 15610044
- Rabbani A, Abdosamadi S, Sari-Saraf N. Affinity of anticancer drug, daunomycin, to core histones in solution: comparison of free and cross-linked proteins. Acta Pharmacol Sin 2007; 28:731 - 7; http://dx.doi.org/10.1111/j.1745-7254.2007.00542.x; PMID: 17439730
- Ullrich O, Grune T. Proteasomal degradation of oxidatively damaged endogenous histones in K562 human leukemic cells. Free Radic Biol Med 2001; 31:887 - 93; http://dx.doi.org/10.1016/S0891-5849(01)00672-4; PMID: 11585707
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998; 273:5858 - 68; http://dx.doi.org/10.1074/jbc.273.10.5858; PMID: 9488723
- Zhao H, Traganos F, Dobrucki J, Wlodkowic D, Darzynkiewicz Z. Induction of DNA damage response by the supravital probes of nucleic acids. Cytometry A 2009; 75:510 - 9; http://dx.doi.org/10.1002/cyto.a.20727; PMID: 19373929
- Baure J, Izadi A, Suarez V, Giedzinski E, Cleaver JE, Fike JR, et al. Histone H2AX phosphorylation in response to changes in chromatin structure induced by altered osmolarity. Mutagenesis 2009; 24:161 - 7; http://dx.doi.org/10.1093/mutage/gen064; PMID: 19064695
- Kanda T, Sullivan KF, Wahl GM. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol 1998; 8:377 - 85; http://dx.doi.org/10.1016/S0960-9822(98)70156-3; PMID: 9545195
- Kimura H, Cook PR. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J Cell Biol 2001; 153:1341 - 53; http://dx.doi.org/10.1083/jcb.153.7.1341; PMID: 11425866
- Dobrucki JW, Feret D, Noatynska A. Scattering of exciting light by live cells in fluorescence confocal imaging: phototoxic effects and relevance for FRAP studies. Biophys J 2007; 93:1778 - 86; http://dx.doi.org/10.1529/biophysj.106.096636; PMID: 17416613
- Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol 1998; 140:1285 - 95; http://dx.doi.org/10.1083/jcb.140.6.1285; PMID: 9508763
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 1987; 162:156 - 9; http://dx.doi.org/10.1016/0003-2697(87)90021-2; PMID: 2440339
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 1983; 65:55 - 63; http://dx.doi.org/10.1016/0022-1759(83)90303-4; PMID: 6606682