Abstract
Transmembrane protease/serine 4 (TMPRSS4) is a member of the type II transmembrane serine protease (TTSP) family and it was found highly expressed in several cancers. This study aims to evaluate the expression of TMPRSS4 in colorectal cancer (CRC) and investigate its role in proliferation and self-renewal of colon cancer cells. qRT-PCR and immunohistochemistry were used to detect the mRNA and protein expression level of TMRPSS4 in CRC samples respectively. Loss of function assay was conducted with RNAi technique. Cell proliferation was done with WST-8 assay; cell apoptosis and cell cycle analysis were performed with flow cytometry; invasion and migration were done with transwell assay. Plate and soft agarose clonogenic assays were used to detect clone-formation ability. CD44 and CD133 expressions were analyzed by flow cytometry and western blot. We found that TMPRSS4 was highly expressed in CRC tissues both at mRNA and protein level and correlated with pathological stage. Knockdown of TMPRSS4 in highly expressed colon cancer cell line HCT116 resulted in inhibition of cell proliferation, induction of cell apoptosis and suppression of invasion and migration; moreover, knockdown of TMPRSS4 suppressed the in vitro clone-formation ability of HCT116 and reduced the expressions of CD44 and CD133. The findings in this research showed that TMPRSS4 was associated with CRC stage and regulated the proliferation and self-renewal ability of colon cancer cells; TMRPSS4 was involved in the development and progression of CRC.
Introduction
Colorectal cancer (CRC) is the third most common cancer and the second leading cause of cancer-related deaths for both sexes in the US.Citation1 Although the 5-y survival rate for early stage CRCs is fairly high, the prognosis of late stage CRCs, i.e., high risk stage II, stage III, and stage IV, remains to be improved.Citation2 Of note, the major cause of CRC related deaths is metastases to distant organs, such as liver and lung.Citation3 Despite efforts in investigating comprehensive therapy including surgery, chemotherapy, and radiotherapy for CRCs, molecular targets which play crucial roles in the tumorigenesis and metastasis of CRCs need to be explored.
During the past decade, numerous studies had focused on the epithelial–mesenchymal transition (EMT) and cancer stem cell (CSC) concepts. By undergoing the EMT process, the epithelial cancer cells could acquire mesenchymal features and get enhancement of the invasion and migration ability, thus promoting cancer progression.Citation4 However, it would not be lethal if only the cancer cells metastasized from primary tumors to distant organs; the outgrowth of micrometastases into macro ones would ultimately result in cancer-related deaths, and self-renewal ability, a well-established trait of CSC, is a must for this process.Citation5 At the late stage of metastatic cascade, the CSC state associated self-renewal ability is indispensable for metastasized cancer cells to form new colonies. Interestingly, recent studies have found that the transcription factors (TFs) involved in EMT are functionally pleiotropic and they constitute the molecular links between EMT and CSC states.Citation6 Specially, EMT program has been found to be able to generate cancer cells with features of CSCs and it has also been proposed in the migrating CSC concept that cancer stem cells that have undergone EMT are facilitated to disseminate. In this way, cancer cells could acquire part of the CSC traits, mainly the self-renewal potential, via EMT and in turn, the activation of TFs during induction of the CSC state would further promote EMT, representing as one great challenge for therapy.
Previous studies have demonstrated that dysregulation of proteases plays an important role in the development and progression of cancer. The proteolytic activities of proteases are crucial for a variety of cellular activities including the digestion of the extracellular matrix, proliferation, survival, migration, and invasion of cancer cells.Citation7 The serine protease is the oldest and one of the largest protease families and type II transmembrane serine protease (TTSP) is such a member.Citation8 Several TTSPs have been found to play critical roles in normal organ developmentCitation9-Citation12 and some TTSPs like hepsin, corin, and matriptase were found dysregulated in tumors, implicating their possible roles in tumorigenesis and/or progression.Citation13 Among them, TMPRSS4 (transmembrane protease/serine 4) has been described to be able to regulate the invasive and metastatic ability of CRC cells by inducing EMT via integrin α5 upregulation and thus facilitate CRC progression.Citation14 Inhibition of TMPRSS4 with 2-hydroxydiarylamide derivatives could abolish its invasion promotion activity in TMRPSS4-overexpressed CRC cells.Citation15 Moreover, TMPRSS4 was found to be associated with tumor stage, lymph node metastasis, and tumor size, and independently predict the overall survival and disease free survival of breast cancer patients.Citation16,Citation17 Given all that, TMPRSS4 appears to play important roles in carcinogenesis and may represent as a potential therapeutic target. However, it’s still unknown whether TMPRSS4 is involved in the regulation of cell proliferation and self-renewal ability of colon cancer cells. Hereby, we reported that TMPRSS4 could regulate in vitro cell growth and self-renewal ability of CRC cells; its functions were exerted at least partially through regulation of cell apoptosis and CSC related state.
Results
TMPRSS4 was highly expressed in CRC tissues and correlated with pathological stage
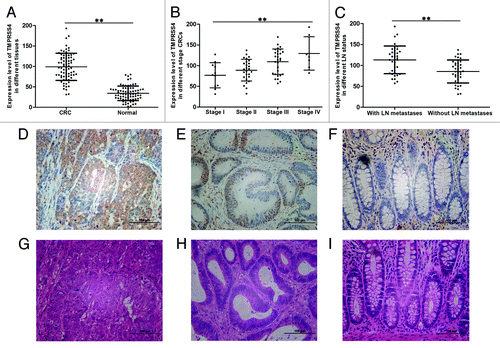
First, we evaluated the expression levels of TMPRSS4 in CRC tissues and the paired normal mucosa by qRT-PCR. The clinicopathological parameters of included patients were shown in . No significant differences were seen between TMPRSS4 mRNA level and clinicopathological parameters like age, gender, tumor site, and tumor histology. When compared with its level in normal tissues, the expression of TMPRSS4 in CRC samples was approximately 3-folds higher (P < 0.01) (); furthermore, the expression of TMPRSS4 increased as the stage became advanced, with stage IV CRCs being the highest and stage I CRCs the lowest (P < 0.01) (); the same trend was also observed when comparing CRCs containing lymph nodes metastases (stage III and IV) with non-metastases ones (stage I and stage II) (P < 0.01) (). We then made immunohistochemistry staining to validate these findings: the staining intensity declined from stage IV () to stage I CRC tissue (), and to normal mucosa (). The matched H&E images of above samples were shown as .
Table 1. Clinicopathological parameters
Figure 1. TMPRSS4 was highly expressed in CRC tissues. The mRNA expression level of TMPRSS4 in: (A) CRC tissues and normal mucosa; (B) different stage CRC tissues; and (C) CRCs with and without LN metastases. Immunohistochemical staining of TMPRSS4 (in brown) in: (D) stage IV CRC; (E) stage I CRC; and (F) normal mucosa. (G–I) are the matched HE staining of (D–F). (Scale bars: 100 μm. **P < 0.01)
Knockdown of TMPRSS4 suppressed cell proliferation rate and induced cell apoptosis
Given that TMPRSS4 was highly expressed in CRC tissues, we wanted to know whether it was also highly expressed in colon cancer cells. qRT-PCR and western blot showed that TMPRSS4 was differentially expressed in seven CRC cell lines and HCT116 had the highest expression (). Then we asked whether TMPRSS4 was involved in the regulation of CRC cell growth in vitro. Loss of function assay with RNAi was performed and transfection efficiency was evaluated by western blot (data not shown). By transfecting HCT116 with one validated siRNA, we found that the proliferation rate was significantly decreased in WST-8 assay (; P = 0.02). To exclude cell line dependency possibility, we used SW1116 and observed similar change (; P = 0.04).
Figure 2. Knockdown of TMPRSS4 suppressed cell proliferation. (A) mRNA (upper) and protein (lower) expression of TMPRSS4 in different CRC cell lines. (B and C) The proliferation rates of HCT116 and SW1116 cells were reduced after TMPRSS4 knockdown. (D and E) Induction of cell apoptosis after TMPRSS4 knockdown in HCT116 and SW1116 cells. (F and G) No significant changes of cell cycle were seen in HCT116 and SW1116 cells. (*P < 0.05)
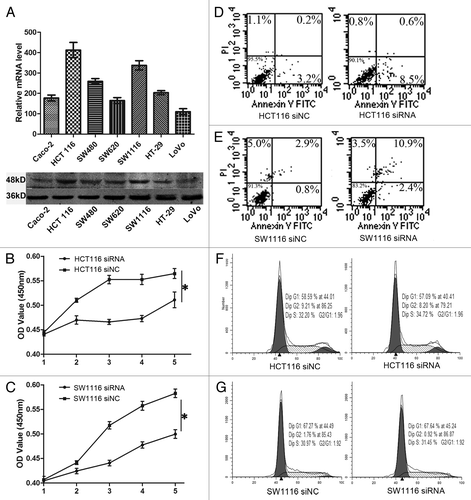
Next, we investigated the potential mechanisms through which TMPRSS4 was involved in cell proliferation. Interestingly, we found that HCT116 cells in the siRNA group showed a higher ratio of apoptosis than that of the siNC group (), but the cell cycle did not show any differences (), which was in conflict with previous study.Citation18 Then we used SW1116 cells to further confirm these results. Treatment with siRNA in SW1116 cells induced a higher cell apoptosis proportion (), but still, we did not observe any changes in cell cycle distribution (). Hence, TMPRSS4 may regulate in vitro cell growth through induction of apoptosis rather than regulating cell cycle distribution.
Knockdown of TMPRSS4 inhibited in vitro clone-formation ability
Previous studies have demonstrated that TMPRSS4 was involved in the EMT program of CRC cellsCitation18 and consistently, we validated this result independently as knockdown of TMPRSS4 suppressed the invasion and migration of HCT116 cells (, P < 0.01, both). Accordingly, we also detected the expression of EMT markers after treatment with siRNA and observed an increase of E-cadherin and a decrease of vimentin (, upper panel), implicating a suppression of EMT. Unexpectedly, we found that Claudin-1 was also downregulated after TMPRSS4 knockdown (, upper panel). We also detected the key transcriptional factors of EMT and found a slight decrease of Snail, but expression of Slug did not show significant change and ZEB1 was not detectable in HCT116 cells (, lower panel).
Figure 3. Knockdown of TMPRSS4 inhibited self-renewal ability. (A) Cell invasion and migration ability were decreased in TMPRSS4 knockdown HCT116 cells. (B) Upper panel: EMT markers of E-cadherin and vimentin were downregulated and upregulated respectively, indicating a suppression of EMT; expressions of CD44 and CD133 were both suppressed after TMPRSS knockdown. Lower panel: Expression of key EMT transcriptional factors in wild type (wt), siNC, and siRNA group. Snail was downregulated and expression of Slug did not show significant change. ZEB1 was not detectable. GAPDH was used as loading control. (C) Plate clonogenic (upper) and soft agarose clonogenic (lower) assay of HCT116 cells. (D) Ppercentages of CD44 (upper) and CD133 (lower) positive HCT116 cells in the siRNA group (right) were lower than that of siNC group (left). (E) Percentages of CD44 (upper) and CD133 (lower) positive SW1116 cells in the siRNA group (right) were lower than that of siNC group (left). (Scale bars: 100μm; *P < 0.05; **P < 0.01)
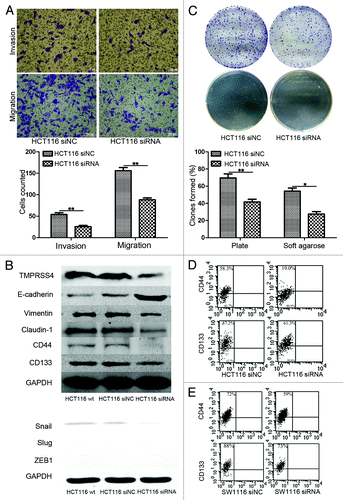
As described above, EMT program had been suggested to be linked with cancer stem cell state, which could be reflected by cell self-renewal ability and the expression pattern of CSC markers, such as CD44 and CD133. Consequently, we conducted plate and soft agarose clonogenic assay to detect self-renewal ability change of HCT116 cells after TMPRSS4 knockdown. As show in , the colonies formed by shNC treated cells were more than those formed by shRNA treated ones on both clonogenic assays (plate clonogenic assay, P < 0.01; soft agarose clonogenic assay, P = 0.016). We then asked whether knockdown of TMPRSS4 could influence the expression of cancer stem cell surface markers. Flow cytometry showed that the percentages of CD44 and CD133 positive cells decreased in siRNA treated cells (). Comparable results were observed in western blot (, upper panel).
Discussion
TMPRSS4, a member of the TTSP family, was first found in pancreatic cancer and played a role in the invasion and migration of cancer cells and promoted the spread of influenza virus.Citation18-Citation20 Hereby, we reported that TMPRSS4 was highly expressed in CRCs and correlated with TNM stage. Knockdown of TMPRSS4 in highly expressed colon cancer cells could inhibit cell proliferation by inducing apoptosis but without interfering with cell cycle. Moreover, knockdown of TMPRSS4 suppressed self-renewal ability and reduced the percentages of CD44 and CD133 positive cells.
Previous studies had demonstrated that TMPRSS4 was highly expressed in different types of cancer, such as pancreatic cancer,Citation21 thyroid cancer,Citation22 lung cancer,Citation23 etc. In breast cancer, higher expression of TMPRSS4 was associated with positive lymph node metastasis, larger tumor size, advanced pathological grade, and shorter overall and disease-free survival;Citation16 in non-small cell lung cancer, overexpression of TMPRSS4 in patients with squamous histology was associated with poor prognosis;Citation24 in malignant thyroid neoplasm, TMPRSS4 was identified to be a diagnostic marker.Citation25 Of note, Jung et al. had reported TMPRSS4 was upregulated in colon cancer.Citation14 Hence, TMPRSS4 seems to be a factor regulating tumor development and progression. In this study, we confirmed that TMPRSS4 was highly expressed in CRCs and we went a step further in finding the association between TMPRSS4 expression and tumor pathological stage. Expression of TMPRSS4 increased as the colon mucosa transformed from normal to malignant phenotype and from stage I to stage IV CRCs, suggesting a positive correlation between TMPRSS4 and CRC progression. Moreover, we found that metastatic tumors showed higher TMPRSS4 level than non-metastatic ones, indicating its role in promoting invasion and migration. Considering the reported roles of TMPRSS4 in breast and thyroid cancer, these findings strongly suggested the importance of TMPRSS4 in the tumorigenesis and aggressiveness of CRC. However, the prognostic value of TMPRSS4 in CRC needs to be further evaluated.
The role of TMPRSS4 in colon cancer cell proliferation has not been thoroughly discussed before. In our study, we found that TMPRSS4 affected cell proliferation in a cell apoptosis dependent manner. Transfection of targeted siRNA into HCT116 inhibited cell proliferation rate in vitro. This inhibition effect was accompanied with an induction of cell apoptosis; but no cell cycle changes were seen. Apoptosis and cell cycle arrest are two well-established tumor suppressive mechanisms which could be induced to inhibit tumor development. In previous studies, phospho-ERK1/2 and phospho-p38 MAPK were reported to be inactivated after TMPRSS4 suppression.Citation16 Importantly, ERK and p38 MAPK are regulators of the MAPK pathwaysCitation26 and they are essential in some apoptotic responses.Citation27 Hence, we propose that knockdown of TMRPSS4 might induce apoptosis via reduced expression of activated ERK and p38 MAPK and further studies are warranted to validate this pathway. Interestingly, we did not observe any cell cycle changes; this contradicted with former study which found suppression of TMPRSS4 reduced cyclin D1 and cyclin A to arrest cell cycle progression and led to cell growth inhibition.Citation16 Whether TMPRSS4 could modulate cell cycle remains to be determined in the future.
TMPRSS4 has been demonstrated to regulate the EMT program and invasion ability of colon cancer cell SW480: TMPRSS4 could upregulate integrin α5 along with its signaling pathway to facilitate EMT and invasiveness.Citation14 In this study, we confirmed the pro-invasion and -migration function of TMPRSS4 and this may explain its higher expression level in CRCs with lymph nodes metastases. Unexpectedly, we found claudin-1 expression was decreased after TMPRSS4 knockdown. In human liver cancer cells, claudin-1 was reported to be able to induce EMT via c-Abl-ERK signaling pathwayCitation28 and contributed to the acquisition of invasive ability;Citation29 in lung cancer cells, claudin-1 was found to be a mediator of cell migration.Citation30 Furthermore, Snail, a key transcriptional factor in the EMT process, was also downregulated. Snail could directly modulate claudin-1 expressionCitation31 and its dystopic subcellular localizations with claudin-1 helped to form cellular morphology.Citation32 Hence, a new regulation pathway composed of cAbl/ERK/claudin-1/snail as the downstream of TMPRSS4 in EMT regulation of colon cancer cell deserves in-depth investigation. Additionally, EMT program has been suggested to generate cells with features of CSCsCitation33,Citation34 and we found that by inhibiting TMPRSS4, possibly suppressing EMT, the self-renewal ability of HCT116 could be reduced as reflected by clonogenic formation assays and CSC markers. CD44 and CD133 are two canonical cancer stem cell markers used in CRC research.Citation35 While CD133 is more widely expressed, the expression of CD44 confines to a smaller population, showing a higher selectivity. Consistently, we found percentages of CD44 and CD133 positive cells were decreased in TMPRSS4 knockdown group, with CD44 showing a higher reduction. The mechanisms underlying this shift deserves further exploration as some transcriptional factors of EMT, like Snail, Twist, and ZEB1, have crosstalk with the CSC related Wnt/β-catenin signaling pathway.
Conclusively, we demonstrated that TMPRSS4 correlated with CRC pathological stages and its downregulation could inhibit cell proliferation, induce cell apoptosis, suppress invasion, and reduce self-renewal ability. TMPRSS4 could be developed for its therapeutic potential and further studies are warranted to reveal the underlying mechanisms.
Materials and Methods
Cell culture
Seven human colon cancer cell lines, Caco-2, HT-29, HCT116, SW480, SW620, SW1116, and LoVo were purchased from American Type Culture Collection (ATCC) and preserved by Shanghai Institute of Digestive Surgery. HT-29 and HCT116 were cultured in McCoy’s 5A medium (10-050-CV, Corning) with 10% fetal bovine serum (FBS) (35-076-CV, Corning); SW480, SW620, and SW1116 were cultured in L-15 medium (GNM41300, Genom) containing 10% FBS; Caco-2 and LoVo were cultured in RPMI 1640 (10-043-CV, Corning) containing 10% FBS and F-12K containing 10% FBS (GNM21127, Genom), respectively. Cells were placed in a humiliated incubator at 37 °C with 5% CO2.
Clinical specimens and immunohistochemistry
Seventy-four pairs of matched CRC tissues and normal mucosa located at least 5 cm away from the tumor were collected from Shanghai Minimally Invasive Surgery Center between Apr 2011 and Sept 2012. All included patients gave their informed consents prior to surgeries and all the processes were conducted according to the protocol approved by the Ethical Committee of Ruijin Hospital, Shanghai Jiao Tong University School of Medicine. Samples were obtained by sharp dissection immediately after surgical specimens were removed during the operation, and transported in liquid nitrogen, preserved in –80 °C refrigerator and processed within 3 mo. Tissues for immunohistochemistry were fixed with formalin, embedded with paraffin and stained with hematoxylin–eosin and primary antibody to TMPRSS4 (PA5-18871, Pierce Antibodies), according to established protocols.Citation36
RNA isolation and qRT-PCR
TRIZOL reagent (15596026, Invitrogen) was used to extract total RNAs and cDNA was synthesized from 1 μg RNA using PrimeScript RT Master Mix (RR036A, TAKARA). qRT-PCR was performed using SYBR Green PCR Master Mix (4309155, Invitrogen) on the Applied Biosystems 7900HT sequence detection system (Applied Biosystems). Primers of TMPRSS4 were: 5′-TCCAAGGACC GATCCACACT-3′ (forward), and 5′-AAGTTGTCGA AACAGGCAGA G-3′ (reverse); primers of GAPDH were: 5′-TTGGCATCGT TGAGGGTCT-3′ (forward) and 5′-CAGTGGGAAC ACGGAAAGC-3′ (reverse). The mRNA level of GAPDH was used for normalization.
Whole cell protein extraction and western blot
Cells were lysed with RIPA (R0010, Solarbio) at a confluence about 90% and protein concentration was determined by BCA Protein Assay Kit (23225, Thermo Scientific). Western blot was performed according to established protocolsCitation37 and primary antibodies were: TMPRSS4 (Pierce Antibodies), CD44 (sc-65412, Santa Cruz), CD133 (130-092-395, Miltenyi Biotec); E-cadherin, vimentin, claudin-1, Snail, Slug, and ZEB1 from Epithelial Mesenchymal Transition (EMT) Antibody Sampler Kit (9782S, Cell Signaling Technology); GAPDH (KC-5G4, Kangchen) was used as loading control.
Transient transfection
Commercial siRNAs and shRNAs targeting TMPRSS4 were purchased from GenePharma, Shanghai. Cells were seeded in 6-well culture plates (Corning) at a density of 3 × 105 cells/well and transfection of siRNAs, shRNA, and negative controls (NCs) were performed 24 h later with lipofectamine 2000 (12566014, Invitrogen). qRT-PCR was used to examine transfection efficiency. Cell function assays, RNA isolation and whole cell protein extraction were conducted 48 h after transfection.
Cell proliferation assay
Cells were plated in 96-well cell culture plates 24 h after transfection, at a density of 1000 cells/well in sixplicate and incubated for 4 d. Cell Counting Kit-8 (CCK8) (CK04-13, Dojindo) was used to assess cell proliferation. Percentages of viable cells were evaluated by OD values at 450 nm, using microplate reader (Epoch, BioTek) at the same time point each day (day 1, 2, 3, 4, and 5), in accordance with the manufacturer’s instruction.
Flow cytometry
Cells were seeded in 6-well plates and harvested 48 h after transfection. For cell cycle analysis, cells were fixed with ice-cold 75% ethanol and then stored at 4 °C for 24 h; before analysis, cells were washed with PBS, treated with RNase at 37 °C for 20 min, and then stained with propidium iodide for 30 min in dark; for apoptosis analysis, an Annexin V-FITC Apoptosis Detection Kit I (556547, BD Bioscience) was used in accordance with manufacturer’s instructions. To analyze CD44 and CD133, cells were harvested, rinsed with phosphate-buffered saline (PBS), incubated in 1% PBS-BSA (A2058-100G, Sigma) for 30 min, washed with PBS again and stained with PE conjugated CD44 (130-095-180, Miltenyi Biotec) and CD133 (130-080-801, Miltenyi Biotec) for another 30 min before flow cytometry scan. PE conjugated Mouse IgG1 (130-092-212, Miltenyi Biotec) was used as isotype control.
Invasion and migration assay
For invasion assay, the inner side of the membrane of cell culture inserts (Millipore) were coated with 100 μl (dilution, 1:9) ice-cold basement membrane matrix (356234, BD Bioscience) and inserts were incubated at 37 °C for 4 h before use; for migration assay, cell culture inserts without coating matrix were used. All inserts were put in 24-well plates. Then, 1 × 105 single cell suspension in 200 μl serum free medium was gently pipetted into the chambers of cell culture inserts and medium with 10% FBS placed outside the chamber was used as chemoattractant. After incubation at 37 °C for 24 (in migration assay) or 48 h (in invasion assay), inserts were taken out and cells attached inside the chambers were rubbed off softly with cotton swabs, and cells attached at the outside of culture inserts were stained with 1% crystal violet for 30 min at room temperature. Inserts were washed and photographed and stained cells of 5 random fields were counted.
Plate and soft agarose clonogenic assay
To perform plate clonogenic assay, cells were resuspended and filtered with a strainer (BD Falcon) to get the single cell suspension; then cells were seeded in 6-well plates at a density of 1000 cells/well. After macro clones (larger than 5 mm) formed, cells were fixed with methanol and stained with 1% crystal violet. For soft agarose clonogenic assay, 1.2% and 0.6% PBS-agarose (A9045-5G, Sigma) and RMPI 1640 medium containing 20% FBS were prepared. To make the base layer, melted 1.2% PBS–agarose at 40 °C was mixed with the above medium at a ratio of 1:1 and 4 ml mixture was pipetted gently into each well. After the base layer solidified, plates were stored at 4 °C for future use. To make upper layer, melted 0.6% PBS-agarose at 40 °C was mixed with 37 °C medium at a ratio of 1:1 and then 100 cells/10 μl single cell suspension was added to 2 ml of the mixture, which was then pipetted onto the base gel in each well. After upper gel became solidified, plates were incubated at 37 °C, in a humiliated atmosphere for 2 wk.
Statistical analysis
A Student t test was used to analyze differences between two groups and one-way ANOVA was employed in case of data consisted of more than two groups. Data represents means ± SD from 3 independent experiments. All statistical analyses were performed using the SPSS 15.0 software. A two-tailed value of P < 0.05 was considered statistically significant.
| Abbreviations: | ||
| CRC | = | colorectal cancer |
| EMT | = | epithelial- mesenchymal transition |
| CSC | = | cancer stem cell |
| TF | = | transcription factors |
| TTSP | = | type II transmembrane serine protease |
| TMPRSS4 | = | transmembrane protease/serine 4 |
| NC | = | negative control |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin 2013; 63:11 - 30; http://dx.doi.org/10.3322/caac.21166; PMID: 23335087
- Gao P, Song YX, Wang ZN, Xu YY, Tong LL, Sun JX, Yu M, Xu HM. Is the prediction of prognosis not improved by the seventh edition of the TNM classification for colorectal cancer? Analysis of the surveilla006Ece, epidemiology, and end results (SEER) database. BMC Cancer 2013; 13:123; http://dx.doi.org/10.1186/1471-2407-13-123; PMID: 23496812
- Sadahiro S, Suzuki T, Ishikawa K, Nakamura T, Tanaka Y, Masuda T, Mukoyama S, Yasuda S, Tajima T, Makuuchi H, et al. Recurrence patterns after curative resection of colorectal cancer in patients followed for a minimum of ten years. Hepatogastroenterology 2003; 50:1362 - 6; PMID: 14571738
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest 2009; 119:1420 - 8; http://dx.doi.org/10.1172/JCI39104; PMID: 19487818
- Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell 2011; 147:275 - 92; http://dx.doi.org/10.1016/j.cell.2011.09.024; PMID: 22000009
- Scheel C, Weinberg RA. Cancer stem cells and epithelial-mesenchymal transition: concepts and molecular links. Semin Cancer Biol 2012; 22:396 - 403; http://dx.doi.org/10.1016/j.semcancer.2012.04.001; PMID: 22554795
- Netzel-Arnett S, Hooper JD, Szabo R, Madison EL, Quigley JP, Bugge TH, Antalis TM. Membrane anchored serine proteases: a rapidly expanding group of cell surface proteolytic enzymes with potential roles in cancer. Cancer Metastasis Rev 2003; 22:237 - 58; http://dx.doi.org/10.1023/A:1023003616848; PMID: 12784999
- Szabo R, Bugge TH. Type II transmembrane serine proteases in development and disease. Int J Biochem Cell Biol 2008; 40:1297 - 316; http://dx.doi.org/10.1016/j.biocel.2007.11.013; PMID: 18191610
- Yuan X, Zheng X, Lu D, Rubin DC, Pung CY, Sadler JE. Structure of murine enterokinase (enteropeptidase) and expression in small intestine during development. Am J Physiol 1998; 274:G342 - 9; PMID: 9486188
- Guipponi M, Tan J, Cannon PZ, Donley L, Crewther P, Clarke M, Wu Q, Shepherd RK, Scott HS. Mice deficient for the type II transmembrane serine protease, TMPRSS1/hepsin, exhibit profound hearing loss. Am J Pathol 2007; 171:608 - 16; http://dx.doi.org/10.2353/ajpath.2007.070068; PMID: 17620368
- Scott HS, Kudoh J, Wattenhofer M, Shibuya K, Berry A, Chrast R, Guipponi M, Wang J, Kawasaki K, Asakawa S, et al. Insertion of beta-satellite repeats identifies a transmembrane protease causing both congenital and childhood onset autosomal recessive deafness. Nat Genet 2001; 27:59 - 63; http://dx.doi.org/10.1038/83768; PMID: 11137999
- Ohler A, Becker-Pauly C. Morpholino knockdown of the ubiquitously expressed transmembrane serine protease TMPRSS4a in zebrafish embryos exhibits severe defects in organogenesis and cell adhesion. Biol Chem 2011; 392:653 - 64; http://dx.doi.org/10.1515/bc.2011.070; PMID: 21657981
- Choi SY, Bertram S, Glowacka I, Park YW, Pöhlmann S. Type II transmembrane serine proteases in cancer and viral infections. Trends Mol Med 2009; 15:303 - 12; http://dx.doi.org/10.1016/j.molmed.2009.05.003; PMID: 19581128
- Kim S, Kang HY, Nam EH, Choi MS, Zhao XF, Hong CS, Lee JW, Lee JH, Park YK. TMPRSS4 induces invasion and epithelial-mesenchymal transition through upregulation of integrin alpha5 and its signaling pathways. Carcinogenesis 2010; 31:597 - 606; http://dx.doi.org/10.1093/carcin/bgq024; PMID: 20118200
- Kang S, Min HJ, Kang MS, Jung MG, Kim S. Discovery of novel 2-hydroxydiarylamide derivatives as TMPRSS4 inhibitors. Bioorg Med Chem Lett 2013; 23:1748 - 51; http://dx.doi.org/10.1016/j.bmcl.2013.01.055; PMID: 23414802
- Liang B, Wu M, Bu Y, Zhao A, Xie F. Prognostic value of TMPRSS4 expression in patients with breast cancer. Med Oncol 2013; 30:497; http://dx.doi.org/10.1007/s12032-013-0497-8; PMID: 23420063
- Cheng D, Kong H, Li Y. TMPRSS4 as a poor prognostic factor for triple-negative breast cancer. Int J Mol Sci 2013; 14:14659 - 68; http://dx.doi.org/10.3390/ijms140714659; PMID: 23857060
- Jung H, Lee KP, Park SJ, Park JH, Jang YS, Choi SY, Jung JG, Jo K, Park DY, Yoon JH, et al. TMPRSS4 promotes invasion, migration and metastasis of human tumor cells by facilitating an epithelial-mesenchymal transition. Oncogene 2008; 27:2635 - 47; http://dx.doi.org/10.1038/sj.onc.1210914; PMID: 17968309
- Chaipan C, Kobasa D, Bertram S, Glowacka I, Steffen I, Tsegaye TS, Takeda M, Bugge TH, Kim S, Park Y, et al. Proteolytic activation of the 1918 influenza virus hemagglutinin. J Virol 2009; 83:3200 - 11; http://dx.doi.org/10.1128/JVI.02205-08; PMID: 10825129
- Bahgat MM, Błazejewska P, Schughart K. Inhibition of lung serine proteases in mice: a potentially new approach to control influenza infection. Virol J 2011; 8:27; http://dx.doi.org/10.1128/JVI.02205-08; PMID: 19158246
- Wallrapp C, Hähnel S, Müller-Pillasch F, Burghardt B, Iwamura T, Ruthenbürger M, Lerch MM, Adler G, Gress TM. A novel transmembrane serine protease (TMPRSS3) overexpressed in pancreatic cancer. Cancer Res 2000; 60:2602 - 6; http://dx.doi.org/10.1186/1743-422X-8-27; PMID: 21251300
- Kebebew E, Peng M, Reiff E, Duh QY, Clark OH, McMillan A. ECM1 and TMPRSS4 are diagnostic markers of malignant thyroid neoplasms and improve the accuracy of fine needle aspiration biopsy. Ann Surg 2005; 242:353 - 61, discussion 361-3; PMID: 16135921
- Nguyen TH, Weber W, Havari E, Connors T, Bagley RG, McLaren R, Nambiar PR, Madden SL, Teicher BA, Roberts B, et al. Expression of TMPRSS4 in non-small cell lung cancer and its modulation by hypoxia. Int J Oncol 2012; 41:829 - 38; PMID: 22692880
- Larzabal L, Nguewa PA, Pio R, Blanco D, Sanchez B, Rodríguez MJ, Pajares MJ, Catena R, Montuenga LM, Calvo A. Overexpression of TMPRSS4 in non-small cell lung cancer is associated with poor prognosis in patients with squamous histology. Br J Cancer 2011; 105:1608 - 14; http://dx.doi.org/10.1038/bjc.2011.432; PMID: 22067904
- Kebebew E, Peng M, Reiff E, McMillan A. Diagnostic and extent of disease multigene assay for malignant thyroid neoplasms. Cancer 2006; 106:2592 - 7; http://dx.doi.org/10.1002/cncr.21922; PMID: 16688775
- Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 2002; 298:1911 - 2; http://dx.doi.org/10.1126/science.1072682; PMID: 12471242
- Bacus SS, Gudkov AV, Lowe M, Lyass L, Yung Y, Komarov AP, Keyomarsi K, Yarden Y, Seger R. Taxol-induced apoptosis depends on MAP kinase pathways (ERK and p38) and is independent of p53. Oncogene 2001; 20:147 - 55; http://dx.doi.org/10.1038/sj.onc.1204062; PMID: 11313944
- Suh Y, Yoon CH, Kim RK, Lim EJ, Oh YS, Hwang SG, An S, Yoon G, Gye MC, Yi JM, et al. Claudin-1 induces epithelial-mesenchymal transition through activation of the c-Abl-ERK signaling pathway in human liver cells. Oncogene 2013; 32:4873 - 82; http://dx.doi.org/10.1038/onc.2012.505; PMID: 23160379
- Yoon CH, Kim MJ, Park MJ, Park IC, Hwang SG, An S, Choi YH, Yoon G, Lee SJ. Claudin-1 acts through c-Abl-protein kinase Cdelta (PKCdelta) signaling and has a causal role in the acquisition of invasive capacity in human liver cells. J Biol Chem 2010; 285:226 - 33; http://dx.doi.org/10.1074/jbc.M109.054189; PMID: 19897486
- Shiozaki A, Bai XH, Shen-Tu G, Moodley S, Takeshita H, Fung SY, Wang Y, Keshavjee S, Liu M. Claudin 1 mediates TNFα-induced gene expression and cell migration in human lung carcinoma cells. PLoS One 2012; 7:e38049; http://dx.doi.org/10.1371/journal.pone.0038049; PMID: 22675434
- Ikenouchi J, Matsuda M, Furuse M, Tsukita S. Regulation of tight junctions during the epithelium-mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. J Cell Sci 2003; 116:1959 - 67; http://dx.doi.org/10.1242/jcs.00389; PMID: 12668723
- Bezdekova M, Brychtova S, Sedlakova E, Langova K, Brychta T, Belej K. Analysis of snail-1, e-cadherin and claudin-1 expression in colorectal adenomas and carcinomas. Int J Mol Sci 2012; 13:1632 - 43; http://dx.doi.org/10.3390/ijms13021632; PMID: 22408413
- Morel AP, Lièvre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One 2008; 3:e2888; http://dx.doi.org/10.1371/journal.pone.0002888; PMID: 18682804
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008; 133:704 - 15; http://dx.doi.org/10.1016/j.cell.2008.03.027; PMID: 18485877
- Sanders MA, Majumdar AP. Colon cancer stem cells: implications in carcinogenesis. Front Biosci (Landmark Ed) 2011; 16:1651 - 62; http://dx.doi.org/10.2741/3811; PMID: 21196254
- Mao Z, Sun J, Feng B, Ma J, Zang L, Dong F, Zhang D, Zheng M. The metastasis suppressor, N-myc downregulated gene 1 (NDRG1), is a prognostic biomarker for human colorectal cancer. PLoS One 2013; 8:e68206; http://dx.doi.org/10.1371/journal.pone.0068206; PMID: 23874544
- Dong TT, Zhou HM, Wang LL, Feng B, Lv B, Zheng MH. Salinomycin selectively targets ‘CD133+’ cell subpopulations and decreases malignant traits in colorectal cancer lines. Ann Surg Oncol 2011; 18:1797 - 804; http://dx.doi.org/10.1245/s10434-011-1561-2; PMID: 21267784
