Abstract
Bacteria are highly versatile and useful tools that could deliver short interfering RNA. In this study, a phoP/phoQ double-deleted Salmonella Typhimurium named VNP(PhoP/Q−) based on the genetic background of VNP20009. The biological safety and function of VNP(PhoP/Q−) were also analyzed. Our study revealed the following results: (1) VNP(PhoP/Q−) exhibited lower titers in tumor-free livers and spleens than VNP20009, (2) The survival of VNP(PhoP/Q−) in macrophages and 4T1 tumor cells was significantly reduced compared with that of VNP20009, (3) The tumor-targeting ability of VNP(PhoP/Q−) was significantly enhanced compared with that of VNP20009, and the anticancer effects of VNP(pPhoP/Q−) and VNP20009 on tumor-bearing mice were similar, (4) VNP(PhoP/Q−) could release an shRNA-expressing plasmid and express the EGFP reporter gene in tumor tissue. Therefore, VNP(PhoP/Q−) exhibited a better safety level in tumor-free mice and elicited an anti-tumor effect on tumor-bearing mice. Moreover, VNP(PhoP/Q−) could release an shRNA-expressing plasmid into the cytoplasm of host cells to silence targeted genes.
Introduction
RNA interference (RNAi) has emerged as a powerful research tool that can silence particular target genes.Citation1 Moreover, the immense therapeutic ability of RNAi to treat cancers has been proposed.Citation2 Studies have demonstrated that RNAi can possibly target the degradation of a specific harmful RNA (point mutation, translocation, or overexpression); as a result, the proliferation or apoptosis of cancer cells can be inhibited.Citation3-Citation5 However, advancements in RNAi technology for tumor therapy have been impeded by challenges involving the delivery of RNAi into the cytoplasm of cancer cells. Although viral vectors,Citation6,Citation7 nanoparticles,Citation8,Citation9 and liposomesCitation10 have been used to deliver RNAi, these delivery systems exhibit low security as well as target and silence the gene expressions of normal cells.
Facultative anaerobic Salmonella Typhimurium strains can preferentially accumulate in tumor tissues and elicit potential anti-tumor effects.Citation11,Citation12 As a protein drug delivery system and shRNA-expressing plasmid carrier, S. Typhimurium significantly inhibits tumor growth and metastasis.Citation13-Citation15 However, wild-type S. Typhimurium exhibits high toxicity. To reduce the possibility of lipopolysaccharide-induced septic shock in Salmonella-infected animals, researchers attenuated S. Typhimurium VNP20009 by genetic modification; as a result, purI and msbB genes are deleted, thereby eliciting toxicity in mice and swine.Citation16,Citation17 These mutants exhibit significantly reduced side effects, but tumor targeting, amplification, and growth suppression are still observed in mice. Clinical trials have also shown that VNP20009 can be used safely in humans.Citation18 As a drug delivery system, VNP20009 is used to express exogenous proteins (endostatin, TRAIL, and CCL21) to mediate anti-tumor effects because of its safety, replication, and high tumor specificity.Citation19-Citation21 VNP20009 can survive and proliferate in host cells and it cannot or rarely release interference plasmids into the cytoplasm of host cells. So, VNP20009 is not suitable for use as a shRNA-expressing plasmid carrier for the purpose of gene silencing in mammalian cells because S. Typhimurium PhoP/PhoQ operon is a typical bacterial two-component regulatory system composed of a membrane-associated sensor kinase (phoQ) and a cytoplasmic transcriptional regulator (phoP).Citation22,Citation23 Preliminary studies showed that the phoP/phoQ locus modulates important virulence functions, such as the survival of bacteria in macrophages and resistance to endogenous antimicrobial peptides.Citation24,Citation25 When deleted, the PhoP/PhoQ locus significantly reduces the survival of bacteria in macrophages and their virulence in BALB/c mice.Citation26,Citation27 Strains with deleted PhoP/PhoQ have been used as an effective vaccine and shRNA-expressing plasmid delivery vehicles.Citation28-Citation31 Considering the acceptable safety profile and anti-tumor effect of VNP20009, we aimed to delete PhoP/PhoQ based on the genetic background of VNP20009 to create a delivery carrier mediating shRNA plasmid expression with lower toxicity or better safety.
In our study, VNP(PhoP/Q−) was successfully engineered by red-mediated recombination mutagenesis and its biological characteristics were evaluated. Our results showed that VNP(PhoP/Q−) could be used as an RNAi delivery vector to target genes safely for cancer therapy.
Results
Toxic effect of VNP(PhoP/Q−) infection on normal mice
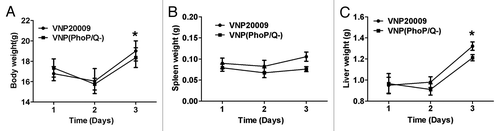
Body, liver, and spleen weights are major markers used to evaluate treatment-related toxicities in pre-clinical studies. Our results showed that the body, liver, and spleen weights of VNP(PhoP/Q−)- and VNP20009-infected mice at 2 d post-infection slightly decreased or remained unchanged compared with those at 1 d post-infection (P > 0.05). The body and liver weights of VNP(PhoP/Q−)- and VNP20009-infected mice at 3 d post-infection were significantly enhanced compared with those at 1 d post-infection (P < 0.05). However, VNP(PhoP/Q−) and VNP20009 infection did not result in any increase in spleen weight at 3 d post-infection (P > 0.05). Compared with VNP20009 infection, VNP(PhoP/Q−) infection did not induce a significant change at 2 d post-infection (P > 0.05). VNP(PhoP/Q−) infection significantly increased the liver and spleen weights compared with VNP20009 infection at 3 d post-infection (P < 0.05) (). The results suggested that phoP/phoQ deletion reduced the toxicity effect of VNP(PhoP/Q−) and further inhibited the swelling of the liver and the spleen.
Figure 1. The toxicity effect of VNP(PhoP/Q−) infection in normal mice. Tumor-free mice were infected with VNP(PhoP/Q−) and VNP20009. Body weight, spleen weight and liver weight were determined at 1, 2 and 3 d post infection (n = 6). (A) Body weight, *P < 0.05, body weight in mice of VNP(PhoP/Q−) or VNP20009 infection at 3 d post infection vs. 1 and 2 d post infection. (B) Spleen weight. (C) Liver weight, *P < 0.05, liver weight in mice of VNP(PhoP/Q−) or VNP20009 infection at 3 d post infection vs. 1 and 2 d post infection.
VNP(PhoP/Q−) biodistribution in normal mice and tumor-bearing mice
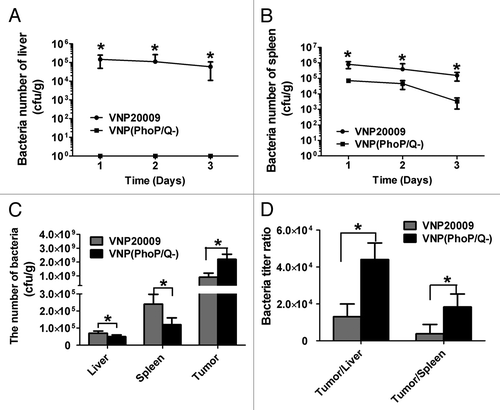
To determine bacterial biodistribution, we inoculated VNP20009 and VNP(PhoP/Q−) in tumor-free and tumor-bearing mice. Tumor-free mice were sacrificed at 1, 2, and 3 d post-injection. The liver and the spleen were removed and then homogenized. VNP20009 and VNP(PhoP/Q−) were isolated. The results showed that the bacterial titers in the livers and the spleens of mice infected with VNP20009 were significantly enhanced compared with those of mice infected with VNP(PhoP/Q−) at 1, 2, and 3 d post-injection (P < 0.05) (). Moreover, bacteria were observed in VNP(PhoP/Q−)-infected mice at 1, 2, and 3 d post-injection. In tumor-bearing mice, VNP20009 and VNP(PhoP/Q−) preferentially accumulated in tumors and reached 1.0 × 109 cfu/g. Furthermore, the bacteria titers in the tumor tissues of mice infected with VNP(PhoP/Q−) were significantly enhanced compared with those of mice infected with VNP20009 (P < 0.05) (). In addition to tumor tissues, normal tissues, such as the spleen and the liver, were infiltrated with VNP20009 and VNP(PhoP/Q−) (). However, the bacteria titers in the livers and the spleens of mice infected with VNP20009 were significantly lower than those of mice infected with VNP(PhoP/Q−) at 6 d post-injection (P < 0.05) (). The ratio of tumors in the liver to the tumors in the spleen of mice infected with VNP(PhoP/Q−) was significantly enhanced compared with that of tumor mice infected with VNP20009 (P < 0.05) (). In summary, VNP(PhoP/Q−) exhibited an excellent safety profile and a higher degree of tumor specificity than VNP20009.
Figure 2. VNP(PhoP/Q−) biodistribution in normal mice and tumor bearing mice. Tumor-free mice or tumor bearing mice were inoculated with VNP20009 and VNP(PhoP/Q−) by intraperitoneal injection. Bacteria titer in liver and spleen of tumor-free mice were determined at 1, 2, and 3 d post infection. Bacteria titer in liver, spleen, and tumor of tumor-free mice were determined at 6 d post infection. (A and B) Bacteria titer in liver and spleen of tumor-free mice, *P < 0.05, VNP20009 vs. VNP(PhoP/Q−). (C) Bacteria titer of liver, spleen, and tumor in tumor-free mice with VNP20009 and VNP(PhoP/Q−) infection, *P < 0.05, VNP20009 vs. VNP(PhoP/Q−). (D) Bacteria titer ratio in in tumor free mice with VNP20009 and VNP(PhoP/Q−) infection, *P < 0.05, VNP20009 vs. VNP(PhoP/Q−).
Immune cells of VNP(PhoP/Q−) infection in the liver and the spleen of normal mice
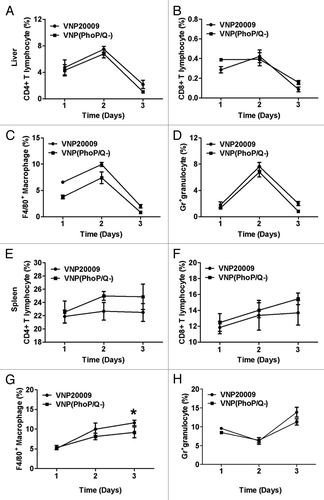
The above study showed that VNP20009 or VNP(PhoP/Q−) can multiply in the spleen and the liver of normal mice. To analyze the changes in immune cells during VNP20009 or VNP(PhoP/Q−) infection in the liver and the spleen of normal mice, we determined the percentage of CD4+ T cells, CD8+ T cells, macrophages, and granulocytes in the spleens and the livers at 1, 2, and 3 d post-injection by flow cytometry assay. The results showed that VNP20009 and VNP(PhoP/Q−) infection induced the infiltration of CD4+ T cells, CD8+ T cells, macrophages, and granulocytes; the peaks were reached at 2 d post-infection and the baseline levels were reduced at 3 d post-infection in the livers (). The percentages of CD4+ T cells, CD8+ T cells, macrophages, and granulocytes in the livers of mice infected with VNP20009 or VNP(PhoP/Q−) did not statistically differ (P > 0.05) (). In the spleen, the percentages of CD4+ T cells, CD8+ T cells, and macrophages increased with the duration of VNP20009 or VNP(phoP/Q−) infection in mice. Furthermore, the percentages of CD4+ T cells, CD8+ T cells, and granulocytes in the spleens of mice infected with VNP20009 or VNP(PhoP/Q−) did not statistically differ at 1, 2, and 3 d post-injection (P > 0.05) (). The percentage of macrophages in the spleens of mice infected with VNP20009 or VNP(PhoP/Q−) did not statistically differ at 1 and 2 d post-injection (P > 0.05) (). By contrast, the percentage of macrophages in the spleens of mice infected with VNP(PhoP/Q−) was significantly reduced than that of mice infected with VNP20009 at 3 d post-injection (P < 0.05) (). In general, VNP20009 and VNP(PhoP/Q−) exhibited a similar level of biological safety, although the safety indexes of the latter were superior to those of the former.
Figure 3. Subpopulation analysis of immune cells in liver and spleen of tumor-free mice. Tumor-free mice or tumor-bearing mice were inoculated with VNP20009 and VNP(PhoP/Q−) by intraperitoneal injection. Immune cells in liver and spleen of tumor free mice were determined at 1, 2, and 3 d post infection. (A) CD4+ T cells in liver. (B) CD8+ T cells in liver. (C) F4/80+ macrophages in liver. (D) Gr-1+ granulocytes in liver. (E) CD4+ T cells in spleen. (F) CD8+ T cells in spleen. (G) F4/80+ macrophages in spleen. *P < 0.05, macrophage in mice of VNP20009 infection at 3 d post infection vs. VNP(phoP/Q−)infection at 3 d post infection. (H) Gr-1+ granulocytes in spleen.
Intracellular survival of VNP(PhoP/Q−) in vitro
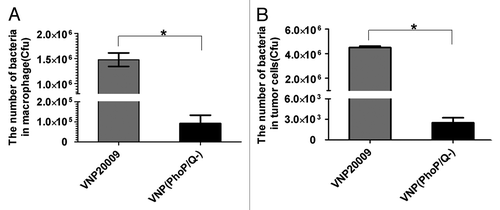
PhoP/PhoQ participate in the intracellular survival of S. Typhimurium. We analyzed the intracellular survival ability of S. Typhimurium in vitro after PhoP and PhoQ genes were deleted. The results showed that the intracellular survival ability of VNP(PhoP/Q−) was significantly reduced than that of VNP20009 in RAW 264.7 macrophage and 4T1 tumor cells (P < 0.05) ().
Figure 4. Intracellular survival of VNP(PhoP/Q−) in vitro. Raw 264.7 macrophages and tumor cells were incubated with VNP(PhoP/Q−) or VNP20009 for 1 h at 37 °C to permit phagocytosis. One hour after infection, gentamicin (25 μg/mL) was added to kill extracellular bacteria. After 3 h of culture at 37 °C, cells were lysed and the titer of bacteria was determined by counting colonies. (A) The number of bacteria in macrophage, *P < 0.05, VNP20009 vs. VNP(PhoP/Q−). (B) The number of bacteria in tumor, *P < 0.05, VNP20009 vs. VNP(PhoP/Q−).
Antitumor effect of VNP(PhoP/Q−)
Our previous study showed that VNP20009 could significantly inhibit Lewis lung carcinoma model and B16F10 melanoma model by intraperitoneal injection. We further analyzed whether PhoP/PhoQ deletion could influence anti-tumor effect of VNP20009 in 4T1 breast cancer model. The results showed that VNP20009 or VNP(PhoP/Q−) significantly inhibited growth of 4T1 breast cancer than that of PBS (P < 0.05) (). However, VNP20009 or VNP(PhoP/Q−) exhibited a similar anticancer effect (P > 0.05) (). The tumor doubling time was significantly prolonged from 5.30 d (CI, 4.59 d to 6.25 d) in the PBS control group to 7.13 d (CI, 6.64 to 7.70 d) in VNP20009 group and 7.33 d (CI, 6.52 d to 8.38 d) in VNP(PhoP/Q−) group (P < 0.05) (; ). Tumor growth delay also significantly increased from 15.62 d (CI, 15.41 d to 15.91 d) in PBS control group to 22.07 d (CI, 21.51 d to 22.72 d) in VNP20009 group and 23.27 d (CI, 22.24 d to 24.60 d) in VNP(PhoP/Q−) group (P < 0.05) (; ).
Figure 5. Antitumor effect of VNP(PhoP/Q−). 4T1 breast cancer mice per group were injected i.p. with 1 × 104 cfu of VNP20009 and VNP(PhoP/Q−), or with PBS at day 6 and 12. Tumor volumes among different groups were compared. Data are presented as mean ± SD. (A) Tumor growth curves. *P < 0.05 for PBS vs. VNP20009 and VNP(PhoP/Q−). (B) Tumor doubling time, *P < 0.05 for PBS vs. VNP20009 and VNP(PhoP/Q−). (C) Growth delay time,*P < 0.05 for PBS vs. VNP20009 and VNP(PhoP/Q−).
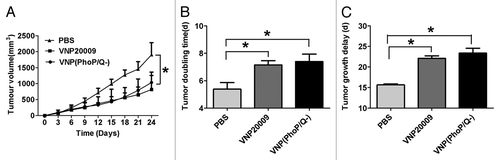
Table 1. Regression analysis for treatment effects on tumor growth for treatment effects on survival
Effect of VNP(PhoP/Q−) on the released plasmid
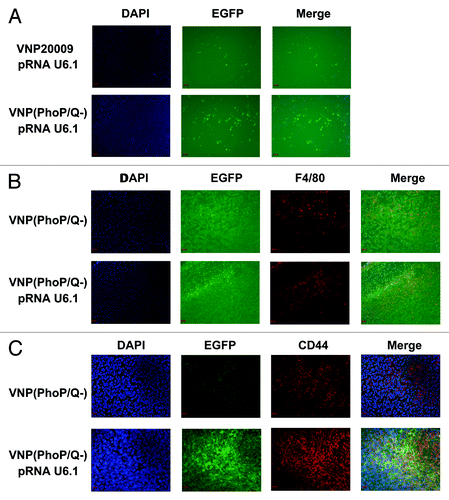
We analyzed whether or not VNP(PhoP/Q−) can release a shRNA-expressing plasmid carrier into the cytoplasm of host cells to produce shRNA. The interference vector pRNA U6.1 RNA (expresses EGFP) was targeted into the tumor tissue by VNP20009 and VNP(PhoP/Q−). EGFP expression indicated that the interference plasmid expression of shRNA was observed in the tumor cells. Immunofluorescence results at low-power lens also showed that the EGFP expression level in the tumor tissue of mice with VNP(phoP/Q−)-pRNA U6.1 RNA infection was significantly enhanced compared with that of VNP20009-pRNA U6.1 RNA infection (). We further determined the cell that expresses the EGFP protein by co-localization. The results showed that EGFP protein was expressed by 4T1 tumor cells and macrophages (). In summary, VNP(PhoP/Q−) could release shRNA-expressing plasmid into the cytoplasm of 4T1 tumor cells and macrophages to express shRNA.
Figure 6. Releasing plasmid effect of VNP(PhoP/Q−). 4T1 breast cancer mice per group were injected i.p. with 1 × 104 cfu of VNP20009 and VNP(PhoP/Q−) bearing pRNAU6.1 RNA interference vector. Six days post-treatment, frozen tumor sections were prepared and EGFP expression in tumor tissues was analyzed. EGFP expression indicated the shRNA expression ability of interference plasmid in tumor cells. (A) EGFP expression in tumor tissue. (B) EGFP expression in macrophages of tumor tissues. (C) EGFP expression in 4T1 tumor cells of tumor tissues.
Discussion
RNA interference (RNAi) is a biological process in which RNA molecules inhibit gene expression, typically by causing the destruction of specific mRNA molecules.Citation32,Citation33 As a therapeutic methods, RNAi has several advantages over traditional pharmaceutical approaches including high specificity and long gene silencing times (several days in vivo).Citation34 Although RNAi have enormous potential therapeutics funnction, to date, few clinical studies use this technology.Citation35,Citation36 The reason is that the application of RNAi-based therapies in vivo has been hampered due to difficulties with delivery.Citation37 Recent, serveral strategies such as chemically modified siRNA, liposomes, nanoparticles and viral vectors were used to overcome delivery problem.Citation38 However, these methods were limited for expensive price and potential danger.
In the study, we engineered VNP(PhoP/Q−), a genetically modified PhoP/PhoQ-deleted S. Typhimurium VNP20009, could release shRNA-expressing plasmid into the cytoplasm of host cells to produce shRNA. In contrast to viral vectors, VNP(PhoP/Q−) does not integrate the genetic material into the host genome and can be controlled by antibiotics.Citation39,Citation40 Moreover, facultative anaerobic S. Typhimurium strains can preferentially accumulate and replicate in the hypoxic regions of solid tumors.Citation12,Citation41,Citation42 This property provides the pre-conditions for VNP(PhoP/Q−) to deliver the fragment of RNAi to mediate anti-tumor effects.
As a RNAi delivery vector, VNP20009 should release RNAi plasmid into host cells to express shRNA. As an intracellular bacterial pathogen, VNP20009 can survive and proliferate in host cells. Therefore, VNP20009 cannot or rarely release interference plasmids into the cytoplasm of host cells. A previous study showed that PhoP/PhoQ is required for virulence; if phoP/phoQ is deleted, S. Typhimurium poorly survives in macrophages.Citation26,Citation43 Therefore, we deleted the phoP/phoQ genes of VNP20009 to construct VNP(phoP/Q−). Our study showed that VNP(phoP/Q−) strain not only has reduced survival rate in macrophages but also had decreased survival rate in tumor cells.
Studies have focused on the safety of VNP(PhoP/Q−). Although VNP20009 has shown efficient biological safety, whether or not PhoP/PhoQ knockout likely affects safety should be verified. Our study showed that VNP(PhoP/Q−) yielded lower titers in the livers and the spleens of tumor-free mice than VNP20009 at 1, 2, and 3 d post-injection. Moreover, the body weight, liver weight, spleen weight, and immune cell infiltration of mice infected with VNP(PhoP/Q−) and VNP20009 did not statistically differ at 1 and 2 d post-injection. Hence, the lysis of VNP(PhoP/Q−) in host cells induced inflammation and caused immune cell infiltration in the livers and the spleens. With the duration of infection time, inflammation in mice infected with VNP(PhoP/Q−) gradually decreased. In the spleen, immune cells, such as macrophages, were significantly reduced than those in mice infected with VNP20009.
In addition, our study showed that the bacterial titers in the tumor tissues of mice infected with VNP(PhoP/Q−) were significantly enhanced compared with those of mice infected with VNP20009. However, the study found that VNP20009 and VNP(PhoP/Q−) exhibited similar anti-cancer effects. This phenomenon was also observed in other gene knockout bacteria (unpublished data). Therefore, anti-tumor effect was possibly not correlated with bacterial tumor targeting. However, a higher tumor specificity of VNP(PhoP/Q−) possibly induced numerous bacteria to infect host cells and release interference plasmids into host cells.
VNP(PhoP/Q−) as RNAi delivery vehicle has several key advantages. First, VNP(PhoP/Q−) can be delivered by intravenous (iv), intraperitoneal (ip), or intratumoral (it) injection as VNP20009. Second, VNP(PhoP/Q−) preferentially accumulate to five times greater within the tumor as compared with VNP20009. This allows for the potential of delivering high levels of RNA interference plasmids and avoid systemic toxicity. Third, VNP(PhoP/Q−) as Salmonella mutation, it can target a broad range of solid tumors such as melanoma, breast and lung cancer. Fourth, VNP(PhoP/Q−) can continuously replicate and produce RNAi plasmids. Fifth, VNP(PhoP/Q−) exhibit better safety than VNP20009. Sixth, VNP(PhoP/Q−) does not integrate the genetic material into the host genome and can be allow cessation of treatment or post-treatment elimination by antibiotics.
In summary, VNP(PhoP/Q−) is a safe delivery system of RNA interference vector. Moreover, this delivery system could be used to target shRNA in tumor cells and TAMs to inhibit tumor directly or indirectly.
Materials and Methods
Animals, cell lines, gene, plasmids, and bacterial strains
Six-week-old male Balb/c mice were obtained from the Comparative Medicine Center of Yangzhou University and maintained in pathogen-free conditions for one week before the start of the experiment. Raw 264.7 macrophage cells were purchased from American Type Culture Collection (ATCC). Cell lines were cultured at 37 °C in 5% CO2 in a humidified atmosphere in Dulbecco’s modified Eagle’s media (DMEM) supplemented with 10% fetal bovine serum (FBS). 4T1-Luciferase murine breast cancer cells were purchased from Xenogen. Cell lines were cultured at 37 °C in 5% CO2 in a humidified atmosphere in RPMI medium 1640 supplemented with 10% fetal bovine serum (FBS). pRNA U6.1 RNA interference vetor were purchased from GenScript Corporation. S. Typhimurium VNP20009 (msbB−/purI−) (strain YS1646) was purchased from ATCC and cultured in modified Luria-Bertani (LB) media at 37 °C. Restriction enzyme and DNA polymerases were purchased TAKARA (China).
Construction of VNP(PhoP/Q−)
Construction of VNP(PhoP/Q−) were described by Datsenko et al.Citation44 Briefly, the phoP/phoQ genes on the S. Typhimurium chromosome were replaced with PCR generated kanamycin resistance cassettes. The antibiotic resistance cassette was amplified by PC-R from pkd4 plasmid with primers:
PhoP-F: 5′-CACCATAATC AACGCTAGAC TGTTCTTATT GTTAACACAA GGGAGAAGAG GTGTAGGCT GGAGCTGCTT C-3′,
phoQ-R: 5′-CGATTATAAC GGATGCTTAA CGAGATGCGT GGAAGAACGC ACAGAAATGT CATATGAATA TCCTCCTTAG-3′. At the 5′ end of either primer contains a 40–50 base-long extension with homology to the PhoP/PhoQ gene. The PCR products were used to replace the coding sequence of the PhoP/PhoQ genes on chromosome using the Lambda Red recombination system.
Construction of tumor model
4T1 breast cancer cells, which had been grown in RPMI medium 1640 plus 10% FBS, were collected and suspended in PBS (pH 7.4). To obtain primary tumor model, 2 × 105 4T1 cells dispersed in 100 μL of PBS was injected subcutaneously into the hind flank region of the Balb/c mice. The tumors were allowed to grow. Animal studies designed to maintain a high standard of animal welfare were approved by the Nanjing University Animal Care and Use Committee.
Bacteria biodistribution in tumor free and tumor bearing mice
To determine bacterial biodistribution, tumor free mice or tumor bearing mice were inoculated with VNP20009 and VNP(PhoP/Q−) by intraperitoneal injection. Tumor free mice were sacrificed after 1, 2 and 3 d and tumor bearing mice were sacrificed after 6 d. The tissue samples (liver and spleen or tumor) were weighed and homogenized in a known volume of PBS and plated on LB agar (without salt) by serial dilution. After 16 h of culture at 37 °C, the titer of bacteria was determined by counting colonies and dividing them by the weight of the tissue (colony forming unit [cfu]/g tissue).
Flow cytometry analysis
Tumor-free mice were inoculated with VNP20009 and VNP(PhoP/Q−) by intraperitoneal injection. Mice were sacrificed after 1, 2, and 3 d. Liver and spleens were collected post-infection for subpopulation analysis of immune cells. Briefly, the livers were digested by collagenase IV and the spleens were ground. The single cell suspension were obtained using a 40-mL cell strainer (BD Falcon), deleted red blood cells by Red Blood Cell Lysing Buffer (Sigma), and followed by washing twice in incomplete RPMI 1640 medium. To determine the percentage of immune cells in liver and spleen tissue, 106 cells resuspended in 100 μL phosphate-buffered saline (PBS) containing 1% FCS and stained for 30 min at 4 °C by using appropriate isotype controls and phycoerythrin-conjugated anti-CD4, anti-CD8, anti-F4/80, FITC-conjugated anti-Gr (BD Biosciences/PharMingen). Samples were incubated in dark for 45 min. After washing in PBS, the cells were analyzed on a FACSCalibur using CELLQUEST software (BD Biosciences).
Survival ability assay in macrophage and tumor cell
Intracellular survival of bacteria was measured by the method of Buchmeier and Heffron. Briefly, 1 × 106 Raw 264.7 macrophages and tumor cells in RPMI plus 5% FCS containing no antibiotics were allowed to adhere to each well of a 6-well plate. VNP20009 and VNP(PhoP/Q−) were added to each well at a ratio of 10 bacteria per macrophage and tumor cells. The cells were incubated for 1 h at 37 °C to permit phagocytosis, and the free bacteria were removed by four washes with PBS. RPMI plus 5% FCS plus gentamicin (25 μg/mL) (control extracellular bacteria) was added, and the cells were incubated at 37 °C. Wells were analyzed at 3 h after infection by removing the medium, lysing the macrophages and tumor cells with 0.5 mL PBS containing 0.1% Triton X-100, rinsing each well with 0.5 mL of PBS, and plated on LB agar (without salt) by serial dilution. After 16 h of culture at 37 °C, the titer of bacteria was determined by counting colonies.
Anti-tumor assay in vivo
4T1 breast cancer mice with a tumor volume of 150 mm3 were randomly divided into three groups: PBS, VNP20009 and VNP(PhoP/Q−). The mice in the PBS group were injected intraperitoneally with 100 μL of PBS per mouse, whereas the mice in the VNP20009 and VNP(PhoP/Q−) were injected intraperitoneally with 1 × 105 cfu VNP20009 and VNP(PhoP/Q−) per mouse, respectively. After 6 d, the mice of the four groups were repeatedly treated with same treatment regime. The length and width of the tumor were measured every two days using a Vernier caliper (Mytutoyo Co.) across its two perpendicular diameters. Tumor volume was calculated using the following formula: tumor volume = length × width2 × 0.52. The numbers and dates of death of mice were recorded to calculate the survival rate.
Immunofluorescence
4T1 breast cancer mice with a tumor volume of 150 mm3 were randomly divided into two groups. One group mice were injected intraperitoneally with 100 μL of VNP20009 bearing pRNA U6.1 RNA interference vector. Other group mice were injected intraperitoneally with 100 μL of VNP(PhoP/Q−) bearing pRNA U6.1 RNA interference vetor. After treatment for 6 d, mice in two groups were sacrificed. Frozen tumor sections were prepared using standardized procedure. To analyzed EGFP expression of 4T1 tumor cells, 4T1 tumor cells were stained with rat anti-mouse CD44 (BD PharmingenTM). The secondary antibodies were goat anti-rat IgG labeled with Cy3 (Jackson ImmunoResearch). The nuclear of cells were stained by DAPI. EGFP expression indicated interference plasmid expression shRNA ability in tumor cells.
Statistical analysis
The results are presented as mean ± SD. Data were assessed using the SPSS 13.0 software. Statistical significance for comparisons among two or three groups was analyzed using the Student t test or one way ANOVA, respectively. The level of significance was set at P < 0.05.
| Abbreviations: | ||
| VNP20009 | = | S. Typhimurium ATCC14028 (msbB−/purI−) |
| VNP(PhoP/Q−) | = | VNP20009 deleted phoP/phoQ gene |
| RNAi | = | RNA interference |
Disclosure of Potential Conflicts of Interest
The authors declare that they have no conflict of interest.
Acknowledgments
The authors are grateful to grants from the Ministry of Science and Technology (2012CB967004, 2014CB744501, 2012AA020809, and 2012ZX09401012), the Doctoral Station Science Foundation from the Chinese Ministry of Education (20130091130003), the Chinese National Natural Sciences Foundation (81121062), the Jiangsu Provincial Nature Science Foundation (BZ2012050 and BE2013630), and the Bureau of Science and Technology of Changzhou (CZ20130011, CE20135013, CZ20120004, CM20122003, and WF201207).
References
- Lieberman J, Song E, Lee SK, Shankar P. Interfering with disease: opportunities and roadblocks to harnessing RNA interference. Trends Mol Med 2003; 9:397 - 403; http://dx.doi.org/10.1016/S1471-4914(03)00143-6; PMID: 13129706
- Chakraborty C. Potentiality of small interfering RNAs (siRNA) as recent therapeutic targets for gene-silencing. Curr Drug Targets 2007; 8:469 - 82; http://dx.doi.org/10.2174/138945007780058988; PMID: 17348839
- Lakka SS, Gondi CS, Yanamandra N, Olivero WC, Dinh DH, Gujrati M, Rao JS. Inhibition of cathepsin B and MMP-9 gene expression in glioblastoma cell line via RNA interference reduces tumor cell invasion, tumor growth and angiogenesis. Oncogene 2004; 23:4681 - 9; http://dx.doi.org/10.1038/sj.onc.1207616; PMID: 15122332
- Gondi CS, Lakka SS, Yanamandra N, Olivero WC, Dinh DH, Gujrati M, Tung CH, Weissleder R, Rao JS. Adenovirus-mediated expression of antisense urokinase plasminogen activator receptor and antisense cathepsin B inhibits tumor growth, invasion, and angiogenesis in gliomas. Cancer Res 2004; 64:4069 - 77; http://dx.doi.org/10.1158/0008-5472.CAN-04-1243; PMID: 15205313
- Fleming JB, Shen GL, Holloway SE, Davis M, Brekken RA. Molecular consequences of silencing mutant K-ras in pancreatic cancer cells: justification for K-ras-directed therapy. Mol Cancer Res 2005; 3:413 - 23; http://dx.doi.org/10.1158/1541-7786.MCR-04-0206; PMID: 16046552
- Devroe E, Silver PA. Therapeutic potential of retroviral RNAi vectors. Expert Opin Biol Ther 2004; 4:319 - 27; http://dx.doi.org/10.1517/14712598.4.3.319; PMID: 15006726
- Kargiotis O, Chetty C, Gondi CS, Tsung AJ, Dinh DH, Gujrati M, Lakka SS, Kyritsis AP, Rao JS. Adenovirus-mediated transfer of siRNA against MMP-2 mRNA results in impaired invasion and tumor-induced angiogenesis, induces apoptosis in vitro and inhibits tumor growth in vivo in glioblastoma. Oncogene 2008; 27:4830 - 40; http://dx.doi.org/10.1038/onc.2008.122; PMID: 18438431
- Wang Y, Li Z, Han Y, Liang LH, Ji A. Nanoparticle-based delivery system for application of siRNA in vivo. Curr Drug Metab 2010; 11:182 - 96; http://dx.doi.org/10.2174/138920010791110863; PMID: 20359287
- Lee JM, Yoon TJ, Cho YS. Recent developments in nanoparticle-based siRNA delivery for cancer therapy. Biomed Res Int 2013; 2013:782041; http://dx.doi.org/10.1155/2013/782041; PMID: 23844368
- Ozpolat B, Sood AK, Lopez-Berestein G. Liposomal siRNA nanocarriers for cancer therapy. Adv Drug Deliv Rev 2014; 66:110 - 6; http://dx.doi.org/10.1016/j.addr.2013.12.008; PMID: 24384374
- Zhao M, Geller J, Ma H, Yang M, Penman S, Hoffman RM. Monotherapy with a tumor-targeting mutant of Salmonella typhimurium cures orthotopic metastatic mouse models of human prostate cancer. Proc Natl Acad Sci U S A 2007; 104:10170 - 4; http://dx.doi.org/10.1073/pnas.0703867104; PMID: 17548809
- Yu B, Yang M, Shi L, Yao Y, Jiang Q, Li X, Tang LH, Zheng BJ, Yuen KY, Smith DK, et al. Explicit hypoxia targeting with tumor suppression by creating an “obligate” anaerobic Salmonella Typhimurium strain. Sci Rep 2012; 2:436; http://dx.doi.org/10.1038/srep00436; PMID: 22666539
- Zhang L, Gao L, Zhao L, Guo B, Ji K, Tian Y, Wang J, Yu H, Hu J, Kalvakolanu DV, et al. Intratumoral delivery and suppression of prostate tumor growth by attenuated Salmonella enterica serovar typhimurium carrying plasmid-based small interfering RNAs. Cancer Res 2007; 67:5859 - 64; http://dx.doi.org/10.1158/0008-5472.CAN-07-0098; PMID: 17575154
- Ganai S, Arenas RB, Forbes NS. Tumour-targeted delivery of TRAIL using Salmonella typhimurium enhances breast cancer survival in mice. Br J Cancer 2009; 101:1683 - 91; http://dx.doi.org/10.1038/sj.bjc.6605403; PMID: 19861961
- Cao HD, Yang YX, Lü L, Liu SN, Wang PL, Tao XH, Wang LJ, Xiang TX. Attenuated Salmonella typhimurium carrying TRAIL and VP3 genes inhibits the growth of gastric cancer cells in vitro and in vivo. Tumori 2010; 96:296 - 303; PMID: 20572589
- Low KB, Ittensohn M, Le T, Platt J, Sodi S, Amoss M, Ash O, Carmichael E, Chakraborty A, Fischer J, et al. Lipid A mutant Salmonella with suppressed virulence and TNFalpha induction retain tumor-targeting in vivo. Nat Biotechnol 1999; 17:37 - 41; http://dx.doi.org/10.1038/5205; PMID: 9920266
- Clairmont C, Lee KC, Pike J, Ittensohn M, Low KB, Pawelek J, Bermudes D, Brecher SM, Margitich D, Turnier J, et al. Biodistribution and genetic stability of the novel antitumor agent VNP20009, a genetically modified strain of Salmonella typhimurium. J Infect Dis 2000; 181:1996 - 2002; http://dx.doi.org/10.1086/315497; PMID: 10837181
- Toso JF, Gill VJ, Hwu P, Marincola FM, Restifo NP, Schwartzentruber DJ, Sherry RM, Topalian SL, Yang JC, Stock F, et al. Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. J Clin Oncol 2002; 20:142 - 52; http://dx.doi.org/10.1200/JCO.20.1.142; PMID: 11773163
- Loeffler M, Le’Negrate G, Krajewska M, Reed JC. Salmonella typhimurium engineered to produce CCL21 inhibit tumor growth. Cancer Immunol Immunother 2009; 58:769 - 75; http://dx.doi.org/10.1007/s00262-008-0555-9; PMID: 18633610
- Chen J, Yang B, Cheng X, Qiao Y, Tang B, Chen G, Wei J, Liu X, Cheng W, Du P, et al. Salmonella-mediated tumor-targeting TRAIL gene therapy significantly suppresses melanoma growth in mouse model. Cancer Sci 2012; 103:325 - 33; http://dx.doi.org/10.1111/j.1349-7006.2011.02147.x; PMID: 22054098
- Chen J, Wei D, Zhuang H, Qiao Y, Tang B, Zhang X, Wei J, Fang S, Chen G, Du P, et al. Proteomic screening of anaerobically regulated promoters from Salmonella and its antitumor applications. Mol Cell Proteomics 2011; 10:009399; http://dx.doi.org/10.1074/mcp.M111.009399; PMID: 21474796
- Hohmann EL, Oletta CA, Miller SI. Evaluation of a phoP/phoQ-deleted, aroA-deleted live oral Salmonella typhi vaccine strain in human volunteers. Vaccine 1996; 14:19 - 24; http://dx.doi.org/10.1016/0264-410X(95)00173-X; PMID: 8821644
- Galán JE, Curtiss R 3rd. Virulence and vaccine potential of phoP mutants of Salmonella typhimurium. Microb Pathog 1989; 6:433 - 43; http://dx.doi.org/10.1016/0882-4010(89)90085-5; PMID: 2671582
- Miller SI, Kukral AM, Mekalanos JJ. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci U S A 1989; 86:5054 - 8; http://dx.doi.org/10.1073/pnas.86.13.5054; PMID: 2544889
- Miller SI, Loomis WP, Alpuche-Aranda C, Behlau I, Hohmann E. The PhoP virulence regulon and live oral Salmonella vaccines. Vaccine 1993; 11:122 - 5; http://dx.doi.org/10.1016/0264-410X(93)90006-J; PMID: 8438611
- Miller SI, Mekalanos JJ. Constitutive expression of the phoP regulon attenuates Salmonella virulence and survival within macrophages. J Bacteriol 1990; 172:2485 - 90; PMID: 2185222
- Fields PI, Swanson RV, Haidaris CG, Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci U S A 1986; 83:5189 - 93; http://dx.doi.org/10.1073/pnas.83.14.5189; PMID: 3523484
- Hohmann EL, Oletta CA, Killeen KP, Miller SI. phoP/phoQ-deleted Salmonella typhi (Ty800) is a safe and immunogenic single-dose typhoid fever vaccine in volunteers. J Infect Dis 1996; 173:1408 - 14; http://dx.doi.org/10.1093/infdis/173.6.1408; PMID: 8648213
- Jia H, Li Y, Zhao T, Li X, Hu J, Yin D, Guo B, Kopecko DJ, Zhao X, Zhang L, et al. Antitumor effects of Stat3-siRNA and endostatin combined therapies, delivered by attenuated Salmonella, on orthotopically implanted hepatocarcinoma. Cancer Immunol Immunother 2012; 61:1977 - 87; http://dx.doi.org/10.1007/s00262-012-1256-y; PMID: 22527247
- Tian Y, Guo B, Jia H, Ji K, Sun Y, Li Y, Zhao T, Gao L, Meng Y, Kalvakolanu DV, et al. Targeted therapy via oral administration of attenuated Salmonella expression plasmid-vectored Stat3-shRNA cures orthotopically transplanted mouse HCC. Cancer Gene Ther 2012; 19:393 - 401; http://dx.doi.org/10.1038/cgt.2012.12; PMID: 22555509
- Li X, Li Y, Wang B, Ji K, Liang Z, Guo B, Hu J, Yin D, Du Y, Kopecko DJ, et al. Delivery of the co-expression plasmid pEndo-Si-Stat3 by attenuated Salmonella serovar typhimurium for prostate cancer treatment. J Cancer Res Clin Oncol 2013; 139:971 - 80; http://dx.doi.org/10.1007/s00432-013-1398-0; PMID: 23463096
- Hannon GJ, Conklin DS. RNA interference by short hairpin RNAs expressed in vertebrate cells. Methods Mol Biol 2004; 257:255 - 66; PMID: 14770011
- Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Rådmark O. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J 2002; 21:5864 - 74; http://dx.doi.org/10.1093/emboj/cdf578; PMID: 12411504
- Aagaard L, Rossi JJ. RNAi therapeutics: principles, prospects and challenges. Adv Drug Deliv Rev 2007; 59:75 - 86; http://dx.doi.org/10.1016/j.addr.2007.03.005; PMID: 17449137
- Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genet 2007; 8:173 - 84; http://dx.doi.org/10.1038/nrg2006; PMID: 17304245
- de Fougerolles AR. Delivery vehicles for small interfering RNA in vivo. Hum Gene Ther 2008; 19:125 - 32; http://dx.doi.org/10.1089/hum.2008.928; PMID: 18257677
- Shankar P, Manjunath N, Lieberman J. The prospect of silencing disease using RNA interference. JAMA 2005; 293:1367 - 73; http://dx.doi.org/10.1001/jama.293.11.1367; PMID: 15769970
- Li CX, Parker A, Menocal E, Xiang S, Borodyansky L, Fruehauf JH. Delivery of RNA interference. Cell Cycle 2006; 5:2103 - 9; http://dx.doi.org/10.4161/cc.5.18.3192; PMID: 16940756
- Keates AC, Fruehauf J, Xiang S, Li CJ. TransKingdom RNA interference: a bacterial approach to challenges in RNAi therapy and delivery. Biotechnol Genet Eng Rev 2008; 25:113 - 27; http://dx.doi.org/10.5661/bger-25-113; PMID: 21412352
- Xu DQ, Zhang L, Kopecko DJ, Gao L, Shao Y, Guo B, Zhao L. Bacterial delivery of siRNAs: a new approach to solid tumor therapy. Methods Mol Biol 2009; 487:161 - 87; http://dx.doi.org/10.1007/978-1-60327-547-7_8; PMID: 19301647
- Jia LJ, Xu HM, Ma DY, Hu QG, Huang XF, Jiang WH, Li SF, Jia KZ, Huang QL, Hua ZC. Enhanced therapeutic effect by combination of tumor-targeting Salmonella and endostatin in murine melanoma model. Cancer Biol Ther 2005; 4:840 - 5; http://dx.doi.org/10.4161/cbt.4.8.1891; PMID: 16210914
- Morrissey D, O’Sullivan GC, Tangney M. Tumour targeting with systemically administered bacteria. Curr Gene Ther 2010; 10:3 - 14; http://dx.doi.org/10.2174/156652310790945575; PMID: 20156191
- Groisman EA, Chiao E, Lipps CJ, Heffron F. Salmonella typhimurium phoP virulence gene is a transcriptional regulator. Proc Natl Acad Sci U S A 1989; 86:7077 - 81; http://dx.doi.org/10.1073/pnas.86.18.7077; PMID: 2674945
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 2000; 97:6640 - 5; http://dx.doi.org/10.1073/pnas.120163297; PMID: 10829079
