Abstract
Pancreatic cancer is the fourth leading cause of cancer-related death in the world. The etiology of pancreatic cancer is heterogeneous with a wide range of alterations that have already been reported at the level of the genome, transcriptome, and proteome. The past decade has witnessed a large number of experimental studies using high-throughput technology platforms to identify genes whose expression at the transcript or protein levels is altered in pancreatic cancer. Based on expression studies, a number of molecules have also been proposed as potential biomarkers for diagnosis and prognosis of this deadly cancer. Currently, there are no repositories which provide an integrative view of multiple Omics data sets from published research on pancreatic cancer. Here, we describe the development of a web-based resource, Pancreatic Cancer Database (http://www.pancreaticcancerdatabase.org), as a unified platform for pancreatic cancer research. PCD contains manually curated information pertaining to quantitative alterations in miRNA, mRNA, and proteins obtained from small-scale as well as high-throughput studies of pancreatic cancer tissues and cell lines. We believe that PCD will serve as an integrative platform for scientific community involved in pancreatic cancer research.
Keywords: :
Introduction
Pancreatic cancer is the fourth leading cause of cancer related deaths with an estimated 227 000 deaths reported globally per year.Citation1 In the United States alone, it is estimated that 39 590 pancreatic cancer related deaths will occur in the year 2014.Citation2 The prognosis of patients with pancreatic cancer is extremely poor with a 5-y relative survival rate of ~6%.Citation2,Citation3 Chemotherapy and radiotherapy have not been effective in improving the survival rate of these patients and carbohydrate antigen 19-9 (CA19-9) has significant limitations as a biomarker.Citation3-Citation6 Thus, there is an urgent need for identification and evaluation of new biomarkers for this cancer for translation into clinical practice.
Recent advances in high-throughput technology platforms have enabled several Omics types of studies, which have led to identification of a large number of transcripts and proteins that are differentially expressed in pancreatic cancer tissues or cell lines when compared with their non-tumor counterparts. A vast amount of such Omics data are scattered across the literature, which makes it difficult for biologists to make the most effective use of such data in generating new hypotheses or in identifying candidate markers to pursue. A central repository that integrates information regarding molecules that have been observed to be differentially expressed in pancreatic cancer will accelerate clinical research as well as basic science. To achieve this goal, we had previously cataloged a list of potential biomarkers for pancreatic cancer (RNA and protein) that were overexpressed in pancreatic cancer.Citation7 However, we felt that there was a need to make this resource more accessible to the biologists. Thus, we decided to develop a web-based resource to provide easy access to the data related to pancreatic cancer. Pancreatic Cancer Database (PCD) is a web-based (http://www.pancreaticcancerdatabase.org/) compendium of molecules that are differentially expressed in pancreatic cancer along with the corresponding fold-change, citation(s) in PubMed, and details of the tumor subtype or cell lines used in the studies.
PCD Design and Architecture
PCD was developed using PHP (http://www.php.net) as an application server. MySQL (http://www.mysql.com) was used as the data storage system at the backend. Data in PCD can be queried using gene symbol, protein name, molecular alterations, cancer types, cell lines and experimental methods. To provide a quick and simple access to the information, an autocomplete option has been enabled in the database. The “browse” option can be used to navigate through the molecular alterations reported at RNA and protein levels alphabetically and miRNA levels.
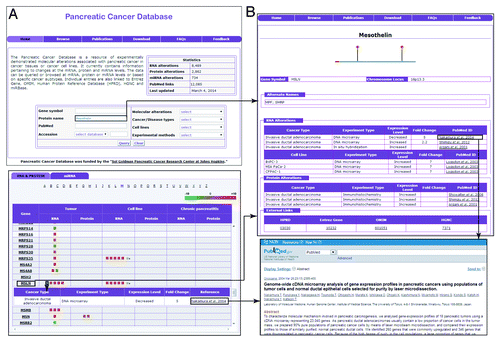
For every molecule cataloged in PCD, the precise pathologic subtype of pancreatic cancer is annotated (e.g., invasive ductal adenocarcinoma). We have also documented the status of the cataloged molecules in chronic pancreatitis (whenever available from literature) since it is an inflammatory condition with symptoms similar to pancreatic cancer. We have also provided information whether these molecules are found in body fluids or known to be present on the plasma membrane. In order to provide a ready reference to the user, PubMed citations are made available with each data entry. External links to various resources available in the public domain including Human Protein Reference Database (HPRD),Citation8,Citation9 Entrez Gene,Citation10,Citation11 Online Mendelian Inheritance in Man (OMIM),Citation12 Swiss-Prot,Citation13 and HUGO Gene Nomenclature Committee (HGNC)Citation14 have also been provided for all the molecules. For miRNAs, an external link to miRBaseCitation15 is also provided. As community participation is vital for updating and improving a database, we have also incorporated an option for submitting new articles and comments to the support team. A screenshot of the molecule page for mesothelin, for an instance, is explored as shown in .
Figure 1. A screenshot of the primary information page for mesothelin in Pancreatic Cancer Database. (A) The query, browse and comments page for mesothelin are shown. (B) The molecule page for mesothelin with the mRNA and protein level alterations along with the cancer subtype, level of regulation, experimental assay used, PubMed citation, and external links to publicly available resources.
Annotation Strategy
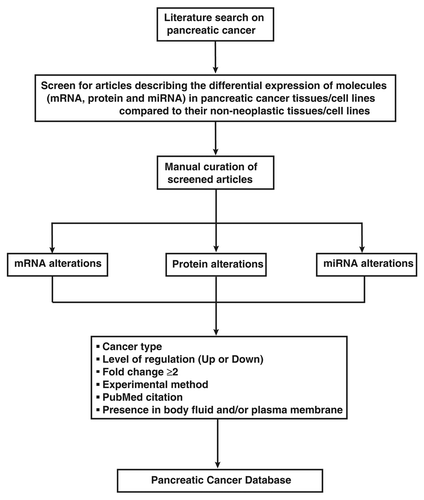
PCD contains manually curated alterations reported at mRNA, miRNA, and protein levels from the published literature. Searches using keywords and Medical Subject Headings (MeSH) in NCBI retrieved around 5000 articles related to expression alterations in pancreatic cancer. These articles were then screened to capture molecules reported to be up or downregulated at the RNA, protein, and miRNA levels in pancreatic cancer tissues/cell lines compared with the normal tissues/cell lines. A ≥2-fold change was used to consider a molecule as upregulated or downregulated in pancreatic cancer. We have not considered unpublished data for inclusion into PCD. The detailed criteria used to annotate the molecular alterations are shown in .
Figure 2. Strategy for annotation of molecular alterations in PCD. Articles published on pancreatic cancer are screened to identify the molecules (mRNA, miRNA, and protein) reported to be differentially expressed by ≥2-fold in pancreatic cancer tissues/cell lines when compared with their normal counterparts. The screened articles are curated manually to catalog the mRNA, protein, and miRNA level alterations. Information pertaining to the pancreatic cancer subtype, level of regulation (up or downregulation), the experimental method used, and the PubMed citation are provided for each molecule. The presence of the molecules in any body fluids and/or plasma membrane is also included, whenever available.
mRNA Alterations
mRNA alterations were annotated from both microarray and non-microarray data. An mRNA molecule, identified from a microarray study was considered for inclusion into PCD if it was upregulated or downregulated by ≥2-fold in neoplastic pancreatic tissues/cell lines as compared with non-neoplastic pancreatic tissue/cell lines. However, if an mRNA molecule was reported to be over/under expressed by multiple methods, it was included in the database even if no fold-change information was available. For non-microarray data, the data was included if ≥2-fold change was present or if evidence at the protein level was present.
Protein Alterations
Both mass spectrometry and other proteomics-based studies were considered for curation of protein level alterations. A protein identified from quantitative proteomic methodologies (e.g., ICAT, SILAC,Citation16 or iTRAQCitation17 methods) was included in PCD if it was reported to be up- or down- regulated by ≥2-fold in pancreatic cancer tissues/cell lines when compared with their normal counterparts. However, proteins identified using non-quantitative proteomic methods (e.g., 2D gel electrophoresis) were included only if it was validated by other techniques such as western blot, immunohistochemistry, or ELISA.
miRNA Alterations
miRNAs have shown promise as prognostic markers for cancers. Their stability in body fluids and tissues make them as suitable markers for early detection of cancers.Citation18 Individual miRNAs and miRNA signatures reported to be associated with pancreatic cancer have been cataloged according to the same criteria mentioned above.
Salient Features of PCD
All molecular level alterations reported from different studies are displayed on a single page, thus providing the users an easy access to the molecule information at a glance. The “Browse Page” allows visualization of the level of regulation at the mRNA, protein, or miRNA as a heat map. This heat map is provided for each molecule in the browse page. A red box indicates upregulation and a green box represents downregulation. A gradient scale is shown from red-to-green color that reflects the fold change of the molecules from a scale of +10 to −10, representing upregulation or downregulation, respectively. A mouse-over option allows display of the fold-change values. Upon clicking the box, the corresponding cancer type and level of regulation with fold-change values along with PubMed citation are displayed. This feature in PCD permits the users to obtain an easy overview of the level of regulation of the molecules.
PCD Statistics
PCD currently contains a total of 3481 unique genes reported to be altered at the expression level in pancreatic cancer. Of these, 703 genes have altered expression at both mRNA and protein levels, 570 genes only at the protein level, and 1982 genes only at the mRNA level. Apart from these, 226 miRNAs that have been shown to be associated with pancreatic cancer have been included in PCD. provides the overall statistics of the annotations in PCD.
Table 1. Summary of molecules in Pancreatic Cancer Databases
Future Outlook
PCD will be updated on a regular basis as new data become available in the published literature. In the future, we will also incorporate genomic, epigenetic, and metabolomic alterations in PCD. We expect PCD to ultimately become a comprehensive and integrate resource that will allow users to navigate through multiple levels of various Omics data pertaining to pancreatic cancer.
Conclusions
Although a few databases for pancreatic cancer research have recently become available, it is still difficult to explore a single gene of interest across different data sets/publications.Citation19,Citation20 Pancreatic Cancer Database was developed with the major goal of providing users an integrated view of genes that are observed to be altered in pancreatic cancer. This centralized data portal should not only help design of future experiments but also reduce the time spent in searching for published literature by biologists. In addition to guiding and facilitating future studies, we anticipate that PCD will be invaluable to those investigating biomarkers and prognosis factors in pancreatic cancer. Over time, we anticipate that this database could become a model database for development of future databases on other cancers.
| Abbreviations: | ||
| PCD | = | Pancreatic Cancer Database |
| HPRD | = | Human Protein Reference Database |
| OMIM | = | Online Mendelian Inheritance in Man |
| HGNC | = | HUGO Gene Nomenclature Committee |
| MeSH | = | Medical Subject Headings |
| ICAT | = | isotope-coded affinity tag |
| SILAC | = | stable isotope labeling with amino acids in cell culture |
| iTRAQ | = | isobaric tags for relative and absolute quantitation |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
Pancreatic Cancer Database was funded by the Sol Goldman Pancreatic Cancer Research Center at Johns Hopkins University School of Medicine, Baltimore, MD USA.
Acknowledgments
We thank Department of Biotechnology, Government of India for Research Support to Institute of Bioinformatics. Aneesha Radhakrishnan is a recipient of Senior Research Fellowship from Council of Scientific and Industrial Research (CSIR), Government of India. We acknowledge Praveen Kumar, Renu Goel and Sandhya Rani, Institute of Bioinformatics, Bangalore, India for their curation support and database development. We also thank Adwait Sodani, Aniruddha Sahu, Ankit Chawla, Ankit Kumar, Mahashweta Dash, and Satish Kumar from Armed Forces Medical College (AFMC), Pune, India, for critical reading of the manuscript.
References
- Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet 2011; 378:607 - 20; http://dx.doi.org/10.1016/S0140-6736(10)62307-0; PMID: 21620466
- Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin 2014; 64:9 - 29; http://dx.doi.org/10.3322/caac.21208; PMID: 24399786
- Vaccaro V, Melisi D, Bria E, Cuppone F, Ciuffreda L, Pino MS, Gelibter A, Tortora G, Cognetti F, Milella M. Emerging pathways and future targets for the molecular therapy of pancreatic cancer. Expert Opin Ther Targets 2011; 15:1183 - 96; http://dx.doi.org/10.1517/14728222.2011.607438; PMID: 21819318
- Chu D, Kohlmann W, Adler DG. Identification and screening of individuals at increased risk for pancreatic cancer with emphasis on known environmental and genetic factors and hereditary syndromes. JOP 2010; 11:203 - 12; PMID: 20442513
- Tempero MA, Uchida E, Takasaki H, Burnett DA, Steplewski Z, Pour PM. Relationship of carbohydrate antigen 19-9 and Lewis antigens in pancreatic cancer. Cancer Res 1987; 47:5501 - 3; PMID: 3308077
- Kim MS, Kuppireddy SV, Sakamuri S, Singal M, Getnet D, Harsha HC, Goel R, Balakrishnan L, Jacob HK, Kashyap MK, et al. Rapid characterization of candidate biomarkers for pancreatic cancer using cell microarrays (CMAs). J Proteome Res 2012; 11:5556 - 63; http://dx.doi.org/10.1021/pr300483r; PMID: 22985314
- Harsha HC, Kandasamy K, Ranganathan P, Rani S, Ramabadran S, Gollapudi S, Balakrishnan L, Dwivedi SB, Telikicherla D, Selvan LD, et al. A compendium of potential biomarkers of pancreatic cancer. PLoS Med 2009; 6:e1000046; http://dx.doi.org/10.1371/journal.pmed.1000046; PMID: 19360088
- Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, Bala P, Shivakumar K, Anuradha N, Reddy R, Raghavan TM, et al. Human protein reference database--2006 update. Nucleic Acids Res 2006; 34:D411 - 4; http://dx.doi.org/10.1093/nar/gkj141; PMID: 16381900
- Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human Protein Reference Database--2009 update. Nucleic Acids Res 2009; 37:D767 - 72; http://dx.doi.org/10.1093/nar/gkn892; PMID: 18988627
- Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res 2005; 33:D54 - 8; http://dx.doi.org/10.1093/nar/gki031; PMID: 15608257
- Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res 2007; 35:D26 - 31; http://dx.doi.org/10.1093/nar/gkl993; PMID: 17148475
- Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res 2005; 33:D514 - 7; http://dx.doi.org/10.1093/nar/gki033; PMID: 15608251
- Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O’Donovan C, Phan I, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res 2003; 31:365 - 70; http://dx.doi.org/10.1093/nar/gkg095; PMID: 12520024
- Bruford EA, Lush MJ, Wright MW, Sneddon TP, Povey S, Birney E. The HGNC Database in 2008: a resource for the human genome. Nucleic Acids Res 2008; 36:D445 - 8; http://dx.doi.org/10.1093/nar/gkm881; PMID: 17984084
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 2006; 34:D140 - 4; http://dx.doi.org/10.1093/nar/gkj112; PMID: 16381832
- Harsha HC, Molina H, Pandey A. Quantitative proteomics using stable isotope labeling with amino acids in cell culture. Nat Protoc 2008; 3:505 - 16; http://dx.doi.org/10.1038/nprot.2008.2; PMID: 18323819
- Venugopal A, Chaerkady R, Pandey A. Application of mass spectrometry-based proteomics for biomarker discovery in neurological disorders. Ann Indian Acad Neurol 2009; 12:3 - 11; PMID: 20151002
- Steele CW, Oien KA, McKay CJ, Jamieson NB. Clinical potential of microRNAs in pancreatic ductal adenocarcinoma. Pancreas 2011; 40:1165 - 71; http://dx.doi.org/10.1097/MPA.0b013e3182218ffb; PMID: 22001830
- Dayem Ullah AZ, Cutts RJ, Ghetia M, Gadaleta E, Hahn SA, Crnogorac-Jurcevic T, Lemoine NR, Chelala C. The pancreatic expression database: recent extensions and updates. Nucleic Acids Res 2014; 42:D944 - 9; http://dx.doi.org/10.1093/nar/gkt959; PMID: 24163255
- Nagpal G, Sharma M, Kumar S, Chaudhary K, Gupta S, Gautam A, Raghava GP. PCMdb: pancreatic cancer methylation database. Sci Rep 2014; 4:4197; http://dx.doi.org/10.1038/srep04197; PMID: 24569397
