Abstract
Ran, a member of the RasGTPase family, has been showed to function in diverse cellular processes of cancer. In the present study, we examined the effects of Ran on the cell motility in pancreatic cancer cells and explored the possible mechanism of Ran’s function in the metastasis of pancreatic cancer. We demonstrated that the expression of Ran was remarkably higher in lymph lode metastases than in primary pancreatic cancer tissues. In the functional studies, stable knockdown of Ran by shRNA could efficiently inhibit the migration and invasion of pancreatic cancer cells both in vitro and in vivo. By PCR array, we analyzed the differences in the expression levels of metastasis-associated genes before and after the downregulation of Ran, and it was showed that the regulation of pancreatic cancer metastasis by Ran was partially mediated by AR and CXCR4. We further confirmed that AR and CXCR4 were significantly decreased following knockdown of Ran. These data indicated that Ran could regulate the invasion and metastasis of pancreatic cancer cells through AR and CXCR4.
Introduction
Pancreatic cancer is one of the most common malignant tumors of the digestive system. It was estimated that 277 000 new cases and 266 000 deaths occurred annually worldwide.Citation1 Due to the lack of specific symptoms, pancreatic cancer have progressed into advanced stage in 80–85% of the patients when first diagnosed, and local invasion and distant metastasis were the main causes to disease death.Citation2,Citation3 Therefore, the 5-y survival rate of pancreatic cancers was less than 5%.Citation4
The small GTPase Ran is a Ras-related GTPase that acts in diverse cellular processes. It could regulate the nucleocytoplasmic transport of molecules through the nuclear pore complexCitation5,Citation6 and modulate the cell cycle progression through the regulation of the mitotic spindle assembly and cell cycle-related proteins.Citation5,Citation7-Citation9 It has been reported that Ran has been found constitutively activated in a wide variety of human tumor tissues and cell lines of renal cell carcinoma, ovarian cancer and colon cancer.Citation10-Citation12 Ran could regulate a lot of oncogenes, tumor suppressors, and spindle assembly factors, and these facts suggested that Ran might play critical roles in the parthenogenesis of cancers.Citation13,Citation14
In the previous study, we found that knockdown of Ran in pancreatic cancer cell lines could inhibit cell proliferation and induced cell apoptosis through survivin and cell cycle proteins.Citation9 In addition, Ran was frequently reported to be involved in invasion and metastasis of tumor cells.Citation10,Citation15,Citation16To better elucidate the roles of Ran in pancreatic cancer, we herein established cells lines with stable transfection of Ran-shRNA. We found that knockdown of Ran could significantly suppress the invasive and metastatic potential of pancreatic cancer cells, and these effects of Ran were showed to be mediated through AR and CXCR4.
Results
Ran was upregulated in pancreatic cancer tissues with high metastatic potential
The expression of Ran was examined in pancreatic cancer tissues and the matched lymph lodes by immunohistochemistry (). Previously, we found that the expression level of Ran in pancreatic cancer tissues was associated with the histological grade, but not statistically related to gender, age, and cancer differentiation.Citation9 In the present study, further analysis showed that increased Ran expression was statistically correlated with the status of distant metastasis (, P = 0.006). Meanwhile, the expression level of Ran was significantly higher in metastatic lymph nodes than that in primary cancer tissues (, P = 0.042).
Figure 1. The expression of Ran in pancreatic cancer tissues. (A) Normal pancreatic tissues; (B) Well-differentiated; (C) Poorly-differentiated; (D) Metastatic site in the lymph node; (E and F) Negative controls of normal tissues and pancreatic cancer tissues using PBS instead of primary antibody.
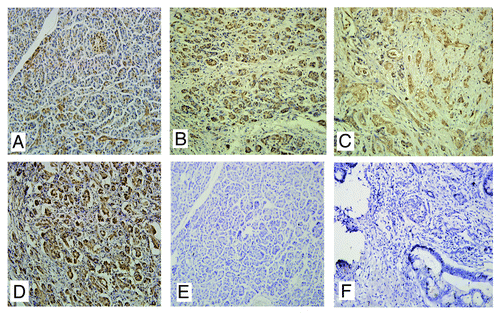
Table 1. Increased Ran expression was statistically correlated with the distant metastasis
Table 2. Expression of Ran in pancreatic cancer and matched lymph node metastases
Ran promoted the migratory, invasive, and metastatic capacities of pancreatic cancer cells
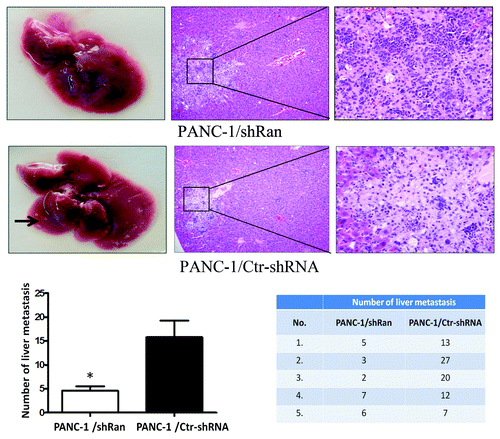
Previously, we constructed the lentivirus containing an Ran-shRNA and transfected it into PANC-1, ASPC-1, and HUVECs cells.Citation9 We next explored the effect of Ran on the invasive and migratory abilities of pancreatic cancer cells using an in vitro wound-healing and transwell assays. In wound-healing assay, the downregulation of Ran resulted in the significant inhibition of the wound closure in PANC-1 and ASPC-1 cells. Meanwhile, downregulation of Ran in HUVECs did not affect cell migration (, P < 0.05). In the transwell assay, we observed similar phenomena. Downregulation of Ran could significantly suppress migration and invasion of PANC-1 and ASPC-1 cells (, P < 0.05). In addition, a tail vein injection metastasis assay was performed to examine the effects of Ran on the in vivo metastasis of PANC-1 cells. PANC-1/shRan and control cells were respectively injected into the tail veins of female nude mice. The number of the metastatic tumors in the liver was significantly reduced in those received PANC-1/shRan cells compared with those received control cells (, P < 0.05). Therefore, the in vitro and in vivo data above illustrated that Ran played significant roles in pancreatic cancer metastasis.
Figure 2. Ran promoted the migratory and invasive abilities of pancreatic cancer cells in vitro. (A and B) The migratory ability of the cells was evaluated with a wound-healing assay.The wound widths were measured at time 0 and 24 h after wounding, and the closure ratio was calculated in accordance with the following formula: wound closure (%) = (width 0 h) − width 24 h) / width 0 h. These results were then compared with those of the control cells. (C and D) The migratory (8 µm pore polycarbonate filters without matrigel) abilities and invasion (8 µm pore polycarbonate filters with matrigel) abilities of cells were evaluated by transwell assays. Representative image fields of invasive cells on the membrane are shown. The values represent the mean (SEM) from at least three separate experiments, *P < 0.05.
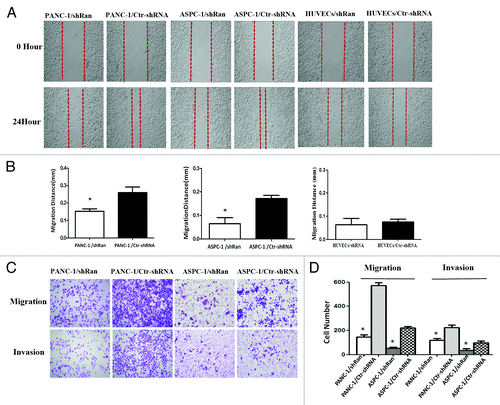
Figure 3. Ran promotes the metastatic ability of pancreatic cancer cells in vivo. Nude mice were tail vein-injected with 1 × 106 PANC-1/ shRan cells and PANC-1/Ctr-shRNA cells. All mice were sacrificed 42 d later after injection of tumor cells and examined for metastatic liver nodules. Representative images of liver tumor metastases. Metastatic loci were identified and marked by arrows; the liver tissues were sectioned serially and then stained with H&E. The number of liver metastasis data are shown in histograms. The values represent the mean (SEM) from at least three separate experiments. *P < 0.05.
The metastasis-related genes and transcription factors regulated by Ran were analyzed by RT2 Profiler™ PCR Array analysis
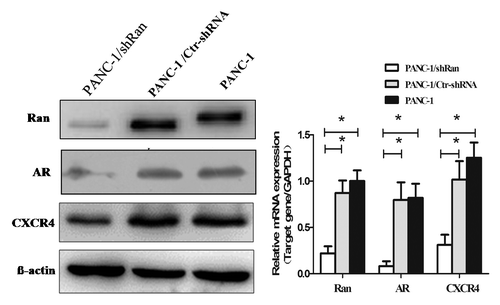
To explore the mechanism of Ran’s function, we employed a real-time PCR array to profile the expression of genes involved in the pancreatic cancer metastasis. The gene expression profiling was analyzed in the PANC-1/shRan and PANC-1/Ctr-shRNA. The tumor metastasis PCR array is designed to represent 84 genes known to be involved in metastasis. The human transcription factors PCR array profiles the expression of 84 genes. Genes with more than a 2-fold increase or decrease were considered to be regulated by Ran. Nine of the 84 genes were downregulatedin the tumor metastasis PCR array (). Seven of the 84 genes were downregulated in thetranscription factors PCR array (). To further confirm these findings, we examined the expression of these candidate molecules by western blot and real-time PCR array. It was showed that only two of these genes, androgen receptor (AR) and chemokine (C-X-C motif) receptor 4 (CXCR4) were significantly decreased by Ran silencing ().
Table 3. mRNA expression of metastasis-related genes after downregulation of Ran
Table 4. mRNA expression of transcription factors after downregulation of Ran
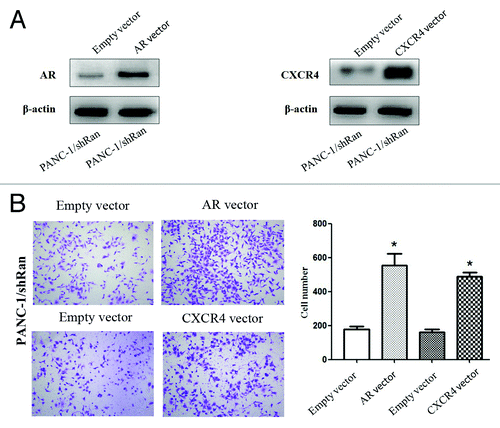
Ectopic expression of AR or CXCR4 could antagonize the anti-metastatic effects caused by Ran knockdown
The expression plasmids of AR or CXCR4 and their corresponding vectors were respectively transfected into PANC-1/shRan cells. The western blot results showed that AR or CXCR4 expression was elevated in PANC-1/shRan cells after the transfected in the former case (). In the transwell migration assay, we found elevated expression of AR or CXCR4 could significantly raise the number of cells penetrating the chamber membrane compared with the control group (, P < 0.05). This illustrates elevated expression of AR or CXCR4 were able to rescue migration of Ran knockdown cells.
Figure 5. The migratory ability of pancreatic cancer cells after transfection of AR or CXCR4 plasmid into PANC-1/shRan cells. (A) Immunoblotting analysis of AR and CXCR4 expression after transfection of plasmid into cells. β-actin was used as a loading control. (B) Cell migration was increased following transfection of AR and CXCR4 plasmid into PANC-1/shRan cells. *P < 0.05.
Discussion
One of the major hallmarks of pancreatic cancer is its systemic dissemination and extraordinary local tumor progression at early stage.Citation17,Citation18 Therefore, it was urgent to explore the mechanisms of pancreatic cancer metastasis. It was reported that, Ran promoted the metastasis of renal cell cancer and colon cancer.Citation10,Citation12 In colon carcinoma, Txl-2b could interacts with Ran via its RCC1 to regulating colon cancer cell metastasis.Citation15 Ran also involved in OPN-mediated invasion and metastatic phenotype.Citation16 However, the mechanism underlying Ran-mediated invasion and metastatic remains unclear.
In the present study, we found that the overexpression of Ran was significantly correlated with the status of metastasis of pancreatic cancer, and Ran was more highly expressed in lymph lode metastases than in primary tissues. These data suggested that Ran might serve as an oncogene in pancreatic cancer and played a positive role in cancer metastasis. We used wound-healing, transwell, and tail vein injection assays to observe the migration and invasion of pancreatic cancer cell lines with knockdown of Ran. In this study, we demonstrate that Ran could promote the invasion and metastasis of pancreatic cancer cells. To identify the downstream effectors of Ran contributing to its function, we employed a real-time PCR array to profile the expression of metastasis related genes and transcription factors. Nine of the 84 genes with more than 2-fold alteration were downregulated when Ran was knocked down and 7 of the 84 transcription factors altered >2-fold weredownregulated after the knockdown of Ran. However, only AR and CXCR4 were further confirmed by western blotting and qRT-PCR. We also found elevated expression of AR and CXCR4 were able to rescue migration of Ran knockdown cells.
Androgen receptor (AR) is a ligand-dependent transcription factor that belongs to the nuclear receptor superfamily.Citation19 In prostate cancer AR could activate the matrix metalloproteinase 2 (MMP-2) and MMP-9 or regulate the CCL2/CCR2-STAT3 axis to promote invasion and metastasis of tumors.Citation20,Citation21 Recent studies showed that IL-6 also enhanced pancreatic cancer cell migration in the presence of AR.Citation22 On the basis of what we have observed, we speculated that Ran could promote pancreatic cancer cell metastasis by regulating the downstream gene AR. CXCR4 was highly expressed in a variety of tumors and it played an important role in tumor proliferation, infiltration, vascularization, and metastasis.Citation23 Recently, some researches showed that the SDF-1/CXCR4 axis and CXCL12/CXCR4 biology axis played important roles in the invasion and metastasis of pancreatic cancer cells.Citation24,Citation25 The CXCL12/CXCR4-PI3-MAPK-NFκB signaling pathways were essential for the tumor lymphatic metastasis,Citation26 and NFκB was unable to enter the nucleus, probably because of the lack of RanGTP production.Citation27 As described above, we demonstrated that Ran enhanced pancreatic cancer cell invasion and metastasis and these effects were at least partly mediated by AR and CXCR4. The detailed mechanisms responsible for these pathways need to be further studied.
Materials and Methods
Ethics statement
For tissue specimens, all patients were informed consent to use excess pathological specimens for research purposes. The protocols used in this study were approved by the hospital’s Protection of Human Subjects Committee. The use of human tissues was approved by the institutional review board of Xijing Hospital and was conformed to the Helsinki Declaration, and to local legislation. Patients offering samples for the study signed informed consent forms. For animal research, all procedures for animal experimentation were performed in accordance with the Institutional Animal Care and Use Committee Guidelines of the Experiment Animal Center of the Fourth Military Medical University. The approval ID for using the animals was No. 12477 by Experiment Animal Center of the Fourth Military Medical University.
Cell culture and tissue collection
The human pancreatic cancer cell lines PANC-1 and ASPC-1; human umbilical vein endothelial cells (HUVECs); stably infected Ran-shRNA cells PANC-1/shRan, ASPC-1/shRan, HUVECs/shRan cells; infected scrambled non-target shRNA cells PANC-1/Ctr-shRNA, ASPC-1/Ctr-shRNA, and HUVECs/Ctr-shRNA were conserved in our institute. All of the cells were cultured in DMEM high glucose medium (Hyclone) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 mg/mL streptomycin (Hyclone).The pancreatic cancer tissues and lymph lode metastases specimens were collected from 62 patients who underwent surgery in our hospital between 2009 and 2011. In total, 38 male, 24 female; aged from 30 to 79; 16 cases were well-differentiated, 34 moderately differentiated, and 12 poorly differentiated. All cases of pancreatic cancer and adjacent non-tumor tissues were diagnosed clinically and pathologically.
Immunohistochemistry
Immunohistochemistry analysis was performed as described previously.Citation9 Primary antibodies used were polyclonal antibodies against Ran (1:500; Santa Cruz Biotechnology). The ratio of positive cells per specimen was evaluated quantitatively and scored as follows: 0, staining of ≤1%; 1, staining of 2–25%; 2, staining of 26–50%; 3, staining of 51–75%; and 4, staining of >75% of examined cells. Staining intensity was divided into four groups: 0, no signal; 1, weak staining; 2, moderate staining; and 3, strong staining. A total score of 0–12 was determined as follows: total score = ratio of positively staining cells (score) × intensity of immunoreactivity (score). Scores were graded as negative (−; score: 0–1), weak (+; score: 2–4), moderate (++; score: 5–8) or strong (+++; score: 9–12).
Western blot analysis
Western blot analysis was performed as described previously.Citation9 Primary antibodies used were as follows: polyclonal antibodies against Ran (dilution 1:500, Santa Cruz Biotechnology), monoclonal antibodies against β-actin (dilution 1:2000, Sigma), androgen receptor (AR) (diluted 1:500; Cell Signaling Technology), and CXCR4 (diluted 1:500; Abcam).
Real-time PCR
Real-time PCR analysis was performed as described previously.Citation9 The primers used were shown in . The PCR conditions were as follows: 95 °C for 10 min followed by 40 cycles at 95 °C for 15 s and 1 min annealing. Optimal annealing temperature was determined for each primer pair. All reactions were performed in triplicate.
Table 5. Quantitative PCR primers
Wound healing assay
Cells were plated in 6-well plates at a density of 5 × 105 cells per mL and cultured to form confluent cell monolayers. Then cells were scraped with a 10 µL pipette tip and washed with PBS to remove nonadherent cells, followed by incubation in medium containing 1% FBS at 37 °C for 24 h. The wound areas were calculated and photographed under a phase contrast microscopy at 200× magnifications (Olympus, JP). The migration distance was quantified by subtracting the width of the wounds at each time point from the width of the initial wounds. All experiments were repeated three times.
Transwell assay
The migratory abilities and invasion abilities of cells were evaluated by transwell assays. A total of 4 × 104 cells in 200 µL serum free medium was added into the transwell inserts with 8 µm pore polycarbonate filters that were coated with (invasion) or without matrigel (migration) (BD Biosciences). After 24 h (migration) or 72 h (invasion) incubation, non-migrated cells on the top of the membrane were removed with a cotton swab. Cells migrated to the bottom sides of the membrane were fixed and stained. The number of migrated cells on the membrane was then counted in 3 randomly selected fields and taken the picture under a light microscope (Olympus) at 200× magnification. All the experiments were performed at least three times.
Tail vein injection
Four-week-old female nude mice obtained from the Shanghai Institute for Biological Sciences. The PANC-1/shRan cells and PANC-1/Ctr-shRNA cells were injected into tail vein with 106 cells in 0.2 mL of PBS medium. All mice were sacrificed 42 d later after injection of tumor cells and examined for metastatic liver nodules. The liver tissues from different models were subjected to histopathological analysis.
RT2 Profiler™ PCR array
To profile the gene expression associated with tumor metastasis and transcription factors, we employed the Tumor Metastasis RT2 Profiler™ PCR Array and the Human Transcription Factors RT2 Profiler™ PCR Array (SuperArray). RNA isolation, DNase treatment, and RNA clean-up were performed according to the manufacturer’s protocol (Qiagen). The isolatedRNA was reverse transcribed into cDNA using the RT2 FirstStrand Kit (Invitrogen). PCR was performed using the RT2SYBRGreen qPCR Master Mix (Invitrogen) on an ABI PRISM7900 instrument (Applied Biosystems). Data normalization was based on correcting all Ct values for the average Ct values of several constantly expressed housekeeping genes presenton the array. The analysis was completed by ShanghaiKangChen Bio-tech Company.
Construction of expression plasmids and transient transfection
The full-length pcDNA3.1 (Invitrogen) AR or CXCR4 vector was made by cloning of the full-length PCR product of AR or CXCR4 with PFU DNA polymerase (Invitrogen). All the plasmid sequences were confirmed by DNA sequencing. For transient transfection experiments, cells were plated in a 24-well plate at a density of 2 × 105 24 h before transfection. LipofectamineTM2000 (Invitrogen) was used to perform transfection with 2.0 mg pcDNA3.1 AR or CXCR4 vector or 2.0 mg pcDNA3.1 empty vector (as control) according to the manufacturer’s protocol. After 24 h, transfected cells were used in transwell migration assays.
Statistical analysis
All data were analyzed using SPSS version 17.0 software (SPSS) including the Mann–Whitney U test and Student t test. The minimal level of significance was defined as P < 0.05.
Disclosure of Potential Conflicts of Interest
The authors have no conflict of interest or financial interest to disclose.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81201929, 30971337). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Pandol S, Gukovskaya A, Edderkaoui M, Dawson D, Eibl G, Lugea A. Epidemiology, risk factors, and the promotion of pancreatic cancer: role of the stellate cell. J GastroenterolHepatol 2012; 27:Suppl 2 127 - 34; http://dx.doi.org/10.1111/j.1440-1746.2011.07013.x; PMID: 22320930
- Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet 2011; 378:607 - 20; http://dx.doi.org/10.1016/S0140-6736(10)62307-0; PMID: 21620466
- Stathis A, Moore MJ. Advanced pancreatic carcinoma: current treatment and future challenges. Nat Rev ClinOncol 2010; 7:163 - 72; http://dx.doi.org/10.1038/nrclinonc.2009.236; PMID: 20101258
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin 2012; 62:10 - 29; http://dx.doi.org/10.3322/caac.20138; PMID: 22237781
- Dasso M. Running on Ran: nuclear transport and the mitotic spindle. Cell 2001; 104:321 - 4; http://dx.doi.org/10.1016/S0092-8674(01)00218-5; PMID: 11239388
- Güttler T, Görlich D. Ran-dependent nuclear export mediators: a structural perspective. EMBO J 2011; 30:3457 - 74; http://dx.doi.org/10.1038/emboj.2011.287; PMID: 21878989
- Dasso M. The Ran GTPase: theme and variations. CurrBiol 2002; 12:R502 - 8; http://dx.doi.org/10.1016/S0960-9822(02)00970-3; PMID: 12176353
- Sazer S, Dasso M. The ran decathlon: multiple roles of Ran. J Cell Sci 2000; 113:1111 - 8; PMID: 10704362
- Deng L, Lu Y, Zhao X, Sun Y, Shi Y, Fan H, Liu C, Zhou J, Nie Y, Wu K, et al. Ran GTPase protein promotes human pancreatic cancer proliferation by deregulating the expression of Survivin and cell cycle proteins. BiochemBiophys Res Commun 2013; 440:322 - 9; http://dx.doi.org/10.1016/j.bbrc.2013.09.079; PMID: 24076388
- Abe H, Kamai T, Shirataki H, Oyama T, Arai K, Yoshida K. High expression of Ran GTPase is associated with local invasion and metastasis of human clear cell renal cell carcinoma. Int J Cancer 2008; 122:2391 - 7; http://dx.doi.org/10.1002/ijc.23400; PMID: 18241036
- Barrès V, Ouellet V, Lafontaine J, Tonin PN, Provencher DM, Mes-Masson AM. An essential role for Ran GTPase in epithelial ovarian cancer cell survival. Mol Cancer 2010; 9:272; http://dx.doi.org/10.1186/1476-4598-9-272; PMID: 20942967
- Fan H, Lu Y, Qin H, Zhou Y, Gu Y, Zhou J, Wang X, Fan D. High Ran level is correlated with poor prognosis in patients with colorectal cancer. Int J ClinOncol 2013; 18:856 - 63; http://dx.doi.org/10.1007/s10147-012-0465-x; PMID: 22956174
- Clarke PR, Zhang C. Spatial and temporal coordination of mitosis by Ran GTPase. Nat Rev Mol Cell Biol 2008; 9:464 - 77; http://dx.doi.org/10.1038/nrm2410; PMID: 18478030
- Kau TR, Way JC, Silver PA. Nuclear transport and cancer: from mechanism to intervention. Nat Rev Cancer 2004; 4:106 - 17; http://dx.doi.org/10.1038/nrc1274; PMID: 14732865
- Lu Y, Zhao X, Li K, Luo G, Nie Y, Shi Y, Zhou Y, Ren G, Feng B, Liu Z, et al. Thioredoxin-like protein 2 is overexpressed in colon cancer and promotes cancer cell metastasis by interaction with ran. Antioxid Redox Signal 2013; 19:899 - 911; http://dx.doi.org/10.1089/ars.2012.4736; PMID: 23311631
- Kurisetty VV, Johnston PG, Johnston N, Erwin P, Crowe P, Fernig DG, Campbell FC, Anderson IP, Rudland PS, El-Tanani MK. RAN GTPase is an effector of the invasive/metastatic phenotype induced by osteopontin. Oncogene 2008; 27:7139 - 49; http://dx.doi.org/10.1038/onc.2008.325; PMID: 18794800
- Neoptolemos JP, Dunn JA, Stocken DD, Almond J, Link K, Beger H, Bassi C, Falconi M, Pederzoli P, Dervenis C, et al, European Study Group for Pancreatic Cancer. Adjuvant chemoradiotherapy and chemotherapy in resectable pancreatic cancer: a randomised controlled trial. Lancet 2001; 358:1576 - 85; http://dx.doi.org/10.1016/S0140-6736(01)06651-X; PMID: 11716884
- Keleg S, Büchler P, Ludwig R, Büchler MW, Friess H. Invasion and metastasis in pancreatic cancer. Mol Cancer 2003; 2:14; http://dx.doi.org/10.1186/1476-4598-2-14; PMID: 12605717
- Mellado B, Codony J, Ribal MJ, Visa L, Gascón P. Molecular biology of androgen-independent prostate cancer: the role of the androgen receptor pathway. ClinTranslOncol 2009; 11:5 - 10; http://dx.doi.org/10.1007/s12094-009-0304-3; PMID: 19155198
- Hara T, Miyazaki H, Lee A, Tran CP, Reiter RE. Androgen receptor and invasion in prostate cancer. Cancer Res 2008; 68:1128 - 35; http://dx.doi.org/10.1158/0008-5472.CAN-07-1929; PMID: 18281488
- Izumi K, Fang LY, Mizokami A, Namiki M, Li L, Lin WJ, Chang C. Targeting the androgen receptor with siRNA promotes prostate cancer metastasis through enhanced macrophage recruitment via CCL2/CCR2-induced STAT3 activation. EMBO Mol Med 2013; 5:1383 - 401; http://dx.doi.org/10.1002/emmm.201202367; PMID: 23982944
- Okitsu K, Kanda T, Imazeki F, Yonemitsu Y, Ray RB, Chang C, Yokosuka O. Involvement of interleukin-6 and androgen receptor signaling in pancreatic cancer. Genes Cancer 2010; 1:859 - 67; http://dx.doi.org/10.1177/1947601910383417; PMID: 21779469
- Balkwill F. The significance of cancer cell expression of the chemokine receptor CXCR4. Semin Cancer Biol 2004; 14:171 - 9; http://dx.doi.org/10.1016/j.semcancer.2003.10.003; PMID: 15246052
- Roy I, Zimmerman NP, Mackinnon AC, Tsai S, Evans DB, Dwinell MB. CXCL12 chemokine expression suppresses human pancreatic cancer growth and metastasis. PLoS One 2014; 9:e90400; http://dx.doi.org/10.1371/journal.pone.0090400; PMID: 24594697
- Li X, Ma Q, Xu Q, Liu H, Lei J, Duan W, Bhat K, Wang F, Wu E, Wang Z. SDF-1/CXCR4 signaling induces pancreatic cancer cell invasion and epithelial-mesenchymal transition in vitro through non-canonical activation of Hedgehog pathway. Cancer Lett 2012; 322:169 - 76; http://dx.doi.org/10.1016/j.canlet.2012.02.035; PMID: 22450749
- Lu DY, Tang CH, Yeh WL, Wong KL, Lin CP, Chen YH, Lai CH, Chen YF, Leung YM, Fu WM. SDF-1alpha up-regulates interleukin-6 through CXCR4, PI3K/Akt, ERK, and NF-kappaB-dependent pathway in microglia. Eur J Pharmacol 2009; 613:146 - 54; http://dx.doi.org/10.1016/j.ejphar.2009.03.001; PMID: 19285061
- Wong CH, Chan H, Ho CY, Lai SK, Chan KS, Koh CG, Li HY. Apoptotic histone modification inhibits nuclear transport by regulating RCC1. Nat Cell Biol 2009; 11:36 - 45; http://dx.doi.org/10.1038/ncb1810; PMID: 19060893