Abstract
HPP1 (hyperplastic polyposis protein 1), a tumor suppressor gene, is downregulated by promoter hypermethylation in a number of tumor types including colon cancer. c-Myc is also known to play a role in the suppression of HPP1 expression via binding to a promoter region cognate E-box site. The contribution of histone deacetylation as an additional epigenetic mechanism and its potential interplay with c-Myc in the transcriptional regulation of HPP1 are unknown. We have shown that the treatment of the HPP1-non-expressing colon cancer cell lines, HCT116 and DLD-1 with HDAC inhibitors results in re-expression of HPP1. RNAi-mediated knockdown of c-Myc as well as of HDAC2 and HDAC3 in HCT116 and of HDAC1 and HDAC3 in DLD-1 also resulted in significant re-expression of HPP1. Co-immunoprecipitation (IP), chromatin IP (ChIP), and sequential ChIP experiments demonstrated binding of c-Myc to the HPP1 promoter with recruitment of and direct interaction with HDAC3. In summary, we have demonstrated that c-Myc contributes to the epigenetic regulation of HPP1 via the dominant recruitment of HDAC3. Our findings may lead to a greater biologic understanding for the application of targeted use of HDAC inhibitors for anti-cancer therapy.
Introduction
HPP1, also known as tomoregulin,Citation1TPEF,Citation2TMEFF2,Citation3 and TENB2,Citation4 is a tumor suppressor gene known to be silenced as a result of promoter methylation Citation5 in many cancers including those of the colon, rectum, stomach, esophagus and gallbladder.Citation6HPP1 is involved in modulating cell growth, maturation, and adhesion and its anti-tumorigenic effects have been demonstrated both in vitro and in vivo in prostate Citation7 and colon cancers.Citation8 The HPP1 protein is comprised of an epidermal growth factor (EGF)-like domain and two follistatin-like modules.Citation9 The EGF-like domain appears to be a ligand for c-erbB-4 and may be directly involved in cellular growth signaling.Citation1
Inactivation of HPP1 by promoter methylation is an early event in the neoplastic progression of gastrointestinal cancers. We have previously reported that aberrant HPP1 methylation occurs in 40% and 50% of ulcerative colitis-associated dysplastic lesions and carcinomas, respectively.Citation10 Furthermore, we have demonstrated that 15 of 32 (47%) gastric cancers demonstrate HPP1 hypermethylation and that it is strongly associated with concomitant hMLH1 hypermethylation.Citation5
Sequence analysis of the HPP1 promoter region has revealed a suppressive E-box that is recognized by c-Myc.Citation11 c-Myc is an oncogenic transcription factor that promotes tumorigenesis by activating or repressing its target genes which in turn, can modulate cell growth and proliferation.Citation12 There is evidence to suggest that c-Myc’s transcriptional effects may in part be mediated by an interplay with histone deacetylases. It has been reported that c-Myc suppresses the transcription of two of its target genes Id2 and Gadd153, via recruitment of histone deacetylase 3 (HDAC3).Citation13 Moreover, N-Myc had been demonstrated to act as a transrepressor by recruiting HDAC1Citation14 and HDAC2.Citation15
The contribution of histone deacetylation toward the complex transcriptional regulation of HPP1 has not been elucidated. Aberrant histone deacetylation, leading to chromatin remodeling and in turn, the functional loss of tumor-suppressor genes and/or activation of oncogenes, has been directly linked to tumorigenesis.Citation16 Histones are small basic proteins that combine with DNA to form the nucleosome core.Citation17 Histone acetylation is a posttranslational modification of the core nucleosomal histones that affects chromatin structure and gene expression. The acetylation status of histones is regulated by the opposing activities of the corresponding enzymes, histone acetylases (HATs), and HDACs.Citation18 Acetylation correlates with remodeling of nucleosomes, resulting in the relaxation of chromatin structure which facilitates the accessibility of a variety of factors to DNA causing transcriptional activation. In contrast, deacetylation of the histone tails induces transcriptional repression through chromatin condensation. Inappropriate transcriptional repression of tumor suppressor genes mediated by HDACs may be a common molecular mechanism associated with tumorigenesis.Citation19 As such, there has been growing interest in the use of HDAC inhibitors as antineoplastic agents.Citation20 In this study, we have sought to elucidate the contribution of HDAC activity and its potential interplay with c-Myc as it pertains to the transcriptional regulation of HPP1.
Results
Treatment with HDAC inhibitors results in the re-expression of HPP1
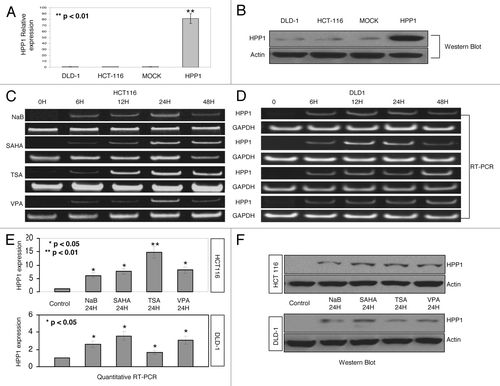
We have previously demonstrated that HPP1 is highly methylated in colon cancer cell lines and tissues and re-expressed upon exposure to the demethylating agent 5-aza-dC.Citation10 Another epigenetic mechanism by which gene expression can be repressed involves deacetylation of chromosomal histones. The baseline expression of HPP1 in DLD-1, HCT116, and Mock (HCT116 cells transfected with empty vector pcDNA 3.0) cells as compared with HPP1 overexpressing cells is shown in . Subsequently, HCT116 and DLD-1 cell lines were treated with multiple HDAC inhibitors including NaB (5 mM), TSA (200 nM), SAHA (5 μM), and VPA (2 mM). Treatment with all four inhibitors induced the re-expression of HPP1 with a consistent peak at approximately 24 h by RT-PCR () and qRT-PCR () with confirmation by western blot analysis ().
Figure 1. Expression of HPP1 in colon cancer cell lines. DLD-1, HCT116, MOCK (HCT116 transfected with empty vector control), and HPP1 (HCT116 transfected with full-length HPP1) by quantitative RT-PCR (A) and western blot (B): HPP1 is re-expressed in HCT116 (C) and DLD-1 (D) cells treated with HDAC inhibitors (SB 5 mM, SAHA 5 μM, TSA 200 nM, and VPA 2 mM) when compared with vehicle alone (control) at different time points. The re-expression of HPP1 at 24 h increased significantly by qRT-PCR in response to HDAC inhibitors (E). A similar trend in protein expression was also observed by western blot analyses (F).
HDAC inhibitors induce accumulation of acetylated H4 and downregulation of c-Myc
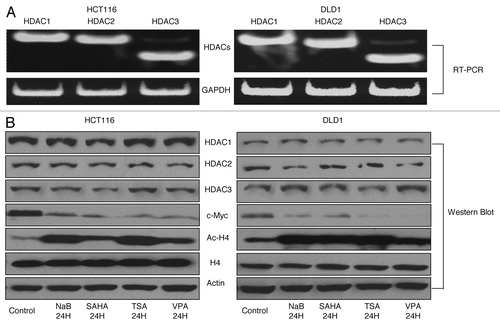
Due to the non-specific nature of HDAC inhibitors, we subsequently examined the roles of HDAC 1, 2, and 3 on HPP1 expression. These particular HDACs were selected for further study as they have been widely described as the key regulators of transcriptional suppression.Citation13,Citation14 All three HDACs were highly expressed in HCT116 and DLD-1 (). As expected, treatment with the HDAC inhibitors did not alter the detectable expression of individual HDACs in the cell lines; however the expression of Ac-H4, an activating chromatin mark, increased significantly (). Concomitantly, the expression of c-Myc, a noted suppressor of HPP1 expression, was uniformly downregulated in both cell lines in response to HDAC inhibitors. ()
Figure 2. HDAC inhibitors induced accumulation of histone acetylation in association with HPP1 re-expression and attenuation of c-Myc. HDAC 1, 2, and 3 were highly expressed by conventional RT-PCR in both HCT116 and DLD-1 cells (A). HDAC inhibitors suppressed the expression of c-Myc significantly and increased the accumulation of Ac-H4 but with no effect on the expression of HDAC1, 2, and 3 by western blot analyses (B).
HPP1 is re-expressed following siRNA-mediated knockdown of HDACs
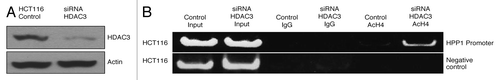
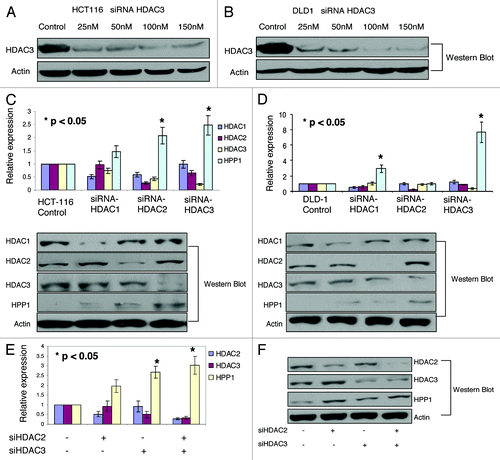
HDAC-specific siRNA knockdowns of HDACs 1–3 were performed in both HCT116 and DLD-1 cell lines. Dose-related effects of siRNA knockdown of HDAC3 are shown in . In HCT116, isolated knockdowns of HDAC2 and -3 resulted in a significant (P < 0.05) reversal of HPP1 suppression (), while in DLD-1 and HDAC1 and -3 appeared to be the critical elements (). This suggests that the contribution of individual HDACs toward the transcriptional regulation of genes such as HPP1, may be cell line-dependent. However, in both cell lines, knockdown of HDAC3 was demonstrated to have the most dominant impact on HPP1 expression. As seen in HCT116, combined knockdown of HDAC2 and -3 resulted in only a slight additive effect on HPP1 re-expression (). A similar effect was observed following combined knockdown of HDAC1 and -2 in DLD-1 (data not shown)
Figure 3. The protein expression of HPP1 in HCT116 (A) and DLD-1 (B) following the dose related knockdowns of HDAC3. By both qRT-PCR and western blot, there was upregulation of HPP1 in HCT116 (C) following knockdown of HDAC2 and HDAC3 with similar findings following abrogation of HDAC1 and HDAC3 in the DLD-1 cell line (D). Combined knockdown of both HDAC2 and HDAC3 demonstrated a slight additive effect on HPP1 re-expression in HCT116 (E and F).
Simultaneous knockdown of HDAC3 and c-Myc yields an additive effect on HPP1 re-expression
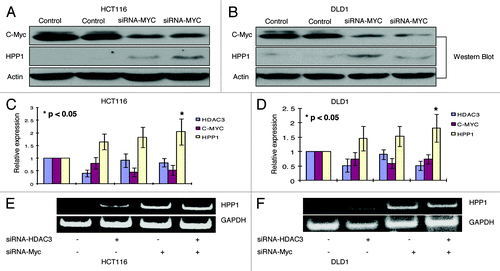
HPP1 harbors a known cognate repressive c-Myc binding site on its promoter region.Citation11 Accordingly, knockdown of c-Myc by siRNA resulted in a significant upregulation of HPP1 in both HCT116 and DLD-1 (). As noted, we have shown that HDAC3 was the most influential of the analyzed HDACs in regulating HPP1 expression in both HCT116 and DLD-1 cell lines. Simultaneous knockdown of c-Myc with HDAC3 resulted in a slight additive effect on HPP1 expression (). Given c-Myc’s recently described ability to recruit HDACs, we sought to further elucidate the interplay between c-Myc and HDACs in the transcriptional regulation of HPP1.
Figure 4.HPP1 is re-expressed in HCT116 and DLD-1 following knockdown of HDAC3 and/or c-Myc. With c-Myc knockdown, a significantly higher expression of HPP1 by Western Blot in both HCT116 (A) and DLD-1 (B) was observed as compared with controls. There is a slight additive effect on HPP1 re-expression by concomitant knockdown of HDAC3 and c-Myc as demonstrated by quantitative real-time (C and D) and conventional RT-PCR (E and F) in both cell lines.
c-Myc and HDACs localize to the HPP1 promoter
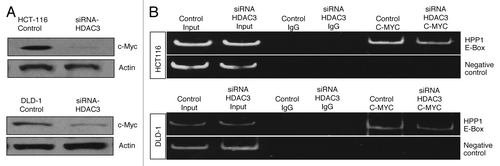
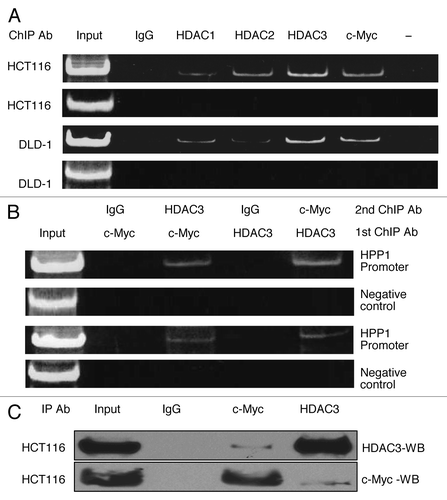
ChIP experiments were performed to determine whether c-Myc and HDAC1, -2, and -3 localize to the promoter region of HPP1. Significant enrichment of c-Myc (c-Myc IP) and HDAC3 (HDAC3 IP) chromatin immunoprecipitates was noted in both HCT116 and DLD-1 cell lines (). Interestingly, we also demonstrated separate enrichment of HDAC2 (HDAC2 IP) in HCT116 and HDAC1 (HDAC1 IP) in DLD1. These findings are in concordance with individual HDAC siRNA knockdown experiments for both of these cell lines as noted above. Conversely, there was only marginal enrichment for HDAC1 (HDAC1 IP) in HCT116 and HDAC2 (HDAC2 IP) in the DLD-1 cell line (). These results further suggest that c-Myc binds to the HPP1 promoter along with a dominant localization of HDAC3 (over HDAC1 and -2) in both cell lines. As such, further investigation of the promoter-based interaction between c-Myc and HDAC3 was performed.
Figure 5. c-Myc-mediated transcriptional repression of HPP1 in colon cancer is dependent on the dominant recruitment of HDAC3. (A) ChIP assays were performed for the HPP1 promoter in HCT116 and DLD-1 cells using antibodies to HDAC1 (lane 3), HDAC2 (lane 4), HDAC3 (lane 5), and c-Myc (lane 6). Normal rabbit IgG served as a negative control (lane 2), input chromatin (samples without IP) served as a positive control (lane 1). An unrelated promoter of c-Fos was also analyzed to confirm the specificity of the experiment. c-Myc, HDAC1, HDAC2, and HDAC3 were confirmed to bind the HPP1 promoter. HDAC3 appears to play a more dominant role in the transcriptional regulation of HPP1. (B) ChIP-re-ChIP experiments further demonstrated the co-occupancy of c-Myc and HDAC3 on the key region of the HPP1 promoter. Antibodies used for the first IP and second IP are indicated above the lanes. (C) Co-IP of c-Myc with HDAC3 from HCT116 extracts demonstrates their physical interaction. Lanes 1 and 2 are input control without IP and negative control respectively. Lanes 3 and 4 are co-immunoprecipitates from c-Myc and HDAC3 respectively.
Sequential ChIP and Co-IP demonstrate co-occupancy and interaction of c-Myc and HDAC3 at the HPP1 promoter
ChIP-re-ChIP experiments were further performed to demonstrate the co-occupancy of c-Myc and HDAC3 on the HPP1 promoter. ChIP assay lysates were prepared from HCT116 and DLD-1 and immunoprecipitated with c-Myc or HDAC3 antibodies to bring down all promoter elements bound by c-Myc or HDAC3. The chromatin complexes were eluted and re-immunoprecipitated with the converse antibody (HDAC3 or c-Myc). PCR assays were performed on the second immunoprecipitated DNA to amplify promoter fragments. There was positive promoter amplification from both cell lines suggesting co-localization of c-Myc and HPP1 (). As a negative control, IP was performed using c-Myc or HDAC3 antibody followed by a second IP using anti-rabbit secondary IgG antibody. An unrelated promoter c-Fos was also analyzed to confirm the specificity of the experiment. The direct physical interaction between c-Myc and HDAC3 was further confirmed using co-IP ().
HDAC3 contributes to H4 acetylation at the HPP1 promoter
Histone H4 acetylation is a means by which Myc activates transcription.Citation21 Myc-dependent repression has been shown to correlate with reduced histone H4 acetylation.Citation13 However, the contribution of HDAC3 to H4 acetylation remains largely unknown. We used ChIP to probe for changes in H4 acetylation status at the HPP1 promoter site at baseline and following knockdown of HDAC3. Knockdown of HDAC3 results in a significant increase in H4 acetylation at the HPP1 promoter providing additional evidence supporting the role of HDAC3 in regulating HPP1 expression ().
c-Myc recruits HDAC3 to the HPP1 promoter
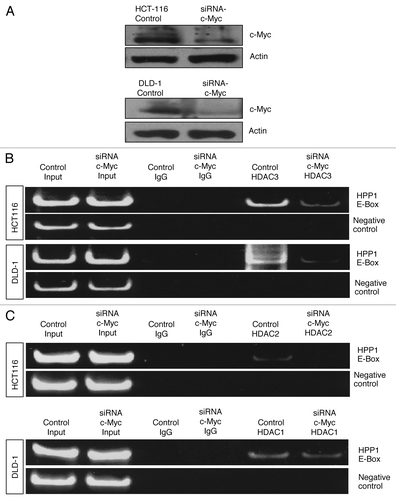
To further confirm that c-Myc primarily binds to the HPP1 promoter with subsequent recruitment of HDACs, ChIP experiments were performed in association with knockdowns of c-Myc () and HDAC3 (). With abrogation of c-Myc, significant reduction of HDAC3 binding to the HPP1 promoter was observed for both cell lines (). c-Myc knockdown was also associated with reduced enrichment of HDAC2 and -1 chromatin immunoprecipitates in HCT116 and DLD-1 respectively (). Conversely, HDAC3 knockdown () had little effect on the ChIP of c-Myc () thus suggesting that c-Myc is the critical binding element and is necessary for the recruitment of HDACs to the HPP1 promoter.
Figure 7. Impact of c-Myc knockdown on HDAC3 binding to the promoter region of HPP1: As a consequence of c-Myc attenuation in HCT116 and DLD-1 (A), significantly less enrichment of HDAC3 (HDAC3 IP) chromatin immunoprecipitates was noted in both HCT116 and DLD-1 cell lines (B) when compared with control. There was a loss of detectable enrichment of HDAC2 in HCT116 and minimal reduction in HDAC1 enrichment in DLD1 (C).
Discussion
HPP1 is a tumor suppressor gene that is inactivated in over 80% of colorectal cancers and 2/3 of polyps.Citation9 There is growing interest in the role of HPP1 as a serum and stool-based biomarker for colorectal cancerCitation22,Citation23 and we have previously described that its tumor suppressive behavior is primarily mediated by STAT1 pathway activation.Citation24 The role of methylation in the transcriptional inactivation of HPP1 is well described.Citation5,Citation11,Citation25 Furthermore, the HPP1 promoter harbors a cognate repressive E-box c-Myc binding site.Citation11 The contributions of HDACs to the complex transcriptional regulation of HPP1 and its interplay with c-Myc have not been elucidated.
We have demonstrated, as is the case with a number of epigenetically silenced genes,Citation26,Citation27 that the treatment of colon cancer cell lines with HDAC inhibitors resulted in the re-expression of HPP1. From our targeted screening of HDAC candidates, we have demonstrated that HDAC3 makes the most prominent contribution toward the regulation of HPP1 repression. HDAC3 is a core component of the nuclear receptor co-repressor complexes N-Cor and SMRTCitation28,Citation29 and its inhibition induced re-expression of HPP1 in both HCT116 and DLD-1 cell lines.
Given c-Myc’s known role as a suppressive factor in the regulation of HPP1, its relationship with HDACs warranted further investigation. c-Myc has traditionally been considered a transcriptional activator, however it is now appreciated that Myc-driven transcriptional repression is also critical for its oncogenic properties.Citation30 Furthermore, there is growing evidence to suggest that histone deacetylation plays a role in the these repressive functions.Citation13,Citation31
Herein we have demonstrated a physical interaction between c-Myc and HDAC3 and that they both bind to and co-occupy the key HPP1 promoter region. It is possible that c-Myc interaction could be direct or indirect through other components of the N-CoR-HDAC3 complex. By sequential ChIP experiments, we have further demonstrated that depletion of c-Myc results in a reduction of HDAC3 association with the HPP1 promoter. However, in the converse experiment, depletion of HDAC3 did not significantly affect c-Myc binding, thus suggesting that it is c-Myc that is essential for the recruitment of HDAC3 to the HPP1 promoter. Our findings are similar to those of Kurland et al. in which recruitment of HDAC3 was found to be necessary for the c-Myc suppressive effect on the Id2 and Gadd153 genes and that the MbIII co-factor was critical for this relationship.Citation13 In similar fashion, we have also observed enhanced H4 acetylation at the HPP1 promoter region following siRNA-mediated attenuation of HDAC3, adding further evidence to support the active role of functional deacetylation in the regulation of HPP1.
Kurland et al. reported that HDAC3 was likely not the only HDAC that contributes to c-Myc’s role in transcriptional suppression.Citation13 Given that we have demonstrated that non-specific HDAC inhibiting agents resulted in a strong re-expression of HPP1 and that targeted knockdown of HDAC3 resulted in only a partial restoration of expression, we also felt it likely that the latter was not the sole HDAC relevant for the transcriptional regulation of HPP1. Accordingly, we have demonstrated that HDAC2 and HDAC1 also contribute in part, toward HPP1 transcriptional repression in the HCT116 and DLD-1 cell lines, respectively. Recent studies have revealed a role for HDAC1 in Myc-dependent repression at the HIV-1Citation31 and transglutaminaseCitation14 genes. Similarly, there is growing evidence to suggest an additional role for HDAC2 in mediating c-Myc’s transcriptional effects.Citation15,Citation32 For example, c-Myc overexpression correlated with HDAC2 upregulation, which resulted in tumor cell proliferation through inhibition of cyclin G2.Citation15 Our findings also support the notion of functional redundancy among HDACs.Citation33 In genome-wide analyses of Drosophila cells, RNAi-mediated knockdown of HDAC1 or HDAC3 produced overlapping changes in gene expression profiles, suggesting that these two enzymes have partially redundant functions.Citation32
An additional interesting observation in our study was the significant uniform downregulation of c-Myc in colon cancer cell lines upon exposure to HDAC inhibitors. This suggests that HDAC inhibitors may restore the expression of epigenetically silenced genes such as HPP1, not only via the it classical functional repression of HDAC activity but also possibly through the inhibition of c-Myc. Knockdown of HDAC3 in both HCT116 and DLD-1 only slightly downregulated the expression of c-Myc, suggesting that repression may involve additional mechanisms. Takashi et al. reported that SAHA caused both β-catenin accumulation in the nucleus associated with enhanced TCF-mediated transcriptional activities and decreased levels of c-Myc in pancreatic cancer cells (PANC-1), suggesting independence from an activated β-catenin pathway.Citation34 Notably, inactivation of HDAC6 has been shown to inhibit β-catenin nuclear localization which in turn, is important for the induction of c-Myc.Citation35 The exact mechanism by which HDAC inhibitors decrease the expression c-Myc requires further study.
Among the oncogenic mechanisms mediated by c-Myc is the transcriptional repression and downregulation of tumor suppressor genes. We have shown that c-Myc contributes to the complex epigenetic transcriptional regulation of HPP1 by dominant recruitment of HDAC3. Specific HDAC-directed therapy that reverses the silencing of similarly regulated tumor suppressor genes may represent an area of investigation for future targeted combination cancer treatments.
Experimental Procedures
Antibodies and reagents
Commercially available antibodies were obtained for HPP1 (Novus Biologicals), HDAC1, -2, -3, -4, Ac-H4, β-actin (Santa Cruz Biotech), and c-Myc (Cell Signaling Technology). Four HDAC inhibitors were utilized and included vorinostat (SAHA; Calbiochem), trichostatin (TSA), sodium butyrate (NaB; Sigma), and valproic acid (VPA; Tocris Bioscience).
Cell culture, treatments, and transfection
The HCT116 and DLD-1 colon cancer cell lines were used to study the epigenetic regulation of HPP1 as they show negligible expression. Cells were cultured in RPMI 1640 (Invitrogen) and supplemented with heat-inactivated 10% fetal bovine serum (Invitrogen) at 37 °C in a humidified incubator containing 5% CO2. Both cell lines were treated with HDAC inhibitors, NaB (5 mM), SAHA (5 µM), TSA (200 nM), and VPA (2 mM) at different time points (6–48 h). The control cultures were treated with dimethyl sulfoxide (DMSO) or ethanol depending on the solubility of the specific HDAC inhibitor. For siRNA transfection 2 × 105 cells were seeded into 6-well plates, incubated overnight and then transfected with control or targeted (HDAC 1–3, Thermo Scientific; c-Myc; Dharmacon) siRNAs using Lipofectamine 2000 (Invitrogen) at a final concentration of 100 nM according to the manufacturer’s instructions.
RNA extraction and RT-PCR
Total RNA was extracted using RNeasy Mini Kits and QIAshredder columns (Qiagen). According to manufacturer’s protocol, 1 µg of total RNA was converted to DNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The Taqman Gene Expression Master Mix and primer/probes for HDAC1 (Assay ID: Hs02621185_s1), HDAC2 (Assay ID: Hs00231032_m1), HDAC3 (Assay ID: Hs00187320_m1), c-Myc (Assay ID: Hs00905030_m1), HPP1 (Assay ID: Hs00249367_m1), and GAPDH (Assay ID: Hs02758991_g1) were purchased from Applied Biosystems. PCR products were analyzed using the ABI Prism 7500 Sequence Detection System (Applied Biosystems) according to the manufacturer’s instructions.
Western blot
Western blots were performed as previously described.Citation8 The concentrations of primary antibodies were as follows: HDAC1 (1:2000), HDAC2 (1:2000), HDAC3 (1:2000), c-Myc (1:3000), HPP1 (1:3000), and β-actin (1:2000).
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed as previously described.Citation36 Co-precipitated DNA was quantified using PCR. The HPP1 promoter sequence (see below) was obtained from GenBank (NCBI reference sequence NT_005403) and specific primers (underlined) were designed to include the c-Myc E-box binding region (bolded). HPP1 promoter sequence; TTTGGAAGCA GCAGGTCCTC AGCCCGCCCG GGGTCACGTG GGAAGAGGCA GTCGGGCTCT GATTGGTGGA GCAGGATGCA GGTCCCGGGA GGGAGGGGTC GACGAGGAGG TGCAAGGATG CAAGGAGGAG GCGGCCGCGG AAGCCACAGA TGGGCTCGCT CGCCAGGCGC TGGCCCGAGT GGGGCTAGGC GGGGATGGCT CAAATGAGAA. The 152–bp amplicon products were electrophoresed in 8% native polyacrylamide gels and signals were quantitated on a PhosphorImager using IMAGEQUANT software (GE Healthcare Biosciences).
Sequential ChIP
ChIP-re-ChIP assays were performed as described by Reid et al.Citation37 Briefly, the primary immunoprecipitation (IP) was performed using anti-c-Myc or anti-HDAC3 antibodies. The immunoprecipitated complexes were eluted with re-ChIP buffer. The elutions from the c-Myc or HDAC3 IPs were then re-immunoprecipitated with the converse HDAC3 or c-Myc antibody. The resulting re-ChIP immunoprecipitates were examined for the presence of the promoter sequences as described above for the one step ChIP.
Co-immunoprecipitation and western blot analysis
The Pierce Co-Immunoprecipitation (Co-IP) Kit (26149) was used to analyze the interaction between c-Myc and HDAC3. Equal cell numbers (20 × 106/sample) were lysed in 10× cell volumes of Co-IP buffer and pre-cleared with amine-reactive resin. Samples were then incubated with a 1:50 dilution of anti-c-Myc (C33) and anti-HDAC3 antibodies. Precipitated proteins were washed three times in 10× cell volumes of Co-IP buffer, released by boiling in SDS sample buffer and separated by SDS-PAGE. Gels were transferred and western blot analysis was performed as noted in section 4.4.
| Abbreviations: | ||
| HPP1 | = | hyperplastic polyposis protein 1 |
| HDAC | = | histone deacetylase |
| IP | = | immunoprecipitation |
| ChIP | = | chromatin immunoprecipitation |
| SB | = | sodium butyrate |
| SAHA | = | vorinostat |
| TSA | = | trichostatin A |
| VPA | = | valproic acid |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed by all authors in this manuscript.
Acknowledgments
This work was supported by NIH grant R01 CA131398 to D.S.
References
- Uchida T, Wada K, Akamatsu T, Yonezawa M, Noguchi H, Mizoguchi A, Kasuga M, Sakamoto C. A novel epidermal growth factor-like molecule containing two follistatin modules stimulates tyrosine phosphorylation of erbB-4 in MKN28 gastric cancer cells. Biochem Biophys Res Commun 1999; 266:593 - 602; http://dx.doi.org/10.1006/bbrc.1999.1873; PMID: 10600548
- Liang G, Robertson KD, Talmadge C, Sumegi J, Jones PA. The gene for a novel transmembrane protein containing epidermal growth factor and follistatin domains is frequently hypermethylated in human tumor cells. Cancer Res 2000; 60:4907 - 12; PMID: 10987305
- Lin K, Taylor JR Jr., Wu TD, Gutierrez J, Elliott JM, Vernes JM, Koeppen H, Phillips HS, de Sauvage FJ, Meng YG. TMEFF2 is a PDGF-AA binding protein with methylation-associated gene silencing in multiple cancer types including glioma. PLoS One 2011; 6:e18608; http://dx.doi.org/10.1371/journal.pone.0018608; PMID: 21559523
- Glynne-Jones E, Harper ME, Seery LT, James R, Anglin I, Morgan HE, Taylor KM, Gee JM, Nicholson RI. TENB2, a proteoglycan identified in prostate cancer that is associated with disease progression and androgen independence. Int J Cancer 2001; 94:178 - 84; http://dx.doi.org/10.1002/ijc.1450; PMID: 11668495
- Shibata DM, Sato F, Mori Y, Perry K, Yin J, Wang S, Xu Y, Olaru A, Selaru F, Spring K, et al. Hypermethylation of HPP1 is associated with hMLH1 hypermethylation in gastric adenocarcinomas. Cancer Res 2002; 62:5637 - 40; PMID: 12384516
- Takahashi T, Shivapurkar N, Riquelme E, Shigematsu H, Reddy J, Suzuki M, Miyajima K, Zhou X, Bekele BN, Gazdar AF, et al. Aberrant promoter hypermethylation of multiple genes in gallbladder carcinoma and chronic cholecystitis. Clin Cancer Res 2004; 10:6126 - 33; http://dx.doi.org/10.1158/1078-0432.CCR-04-0579; PMID: 15447999
- Gery S, Sawyers CL, Agus DB, Said JW, Koeffler HP. TMEFF2 is an androgen-regulated gene exhibiting antiproliferative effects in prostate cancer cells. Oncogene 2002; 21:4739 - 46; http://dx.doi.org/10.1038/sj.onc.1205142; PMID: 12101412
- Elahi A, Zhang L, Yeatman TJ, Gery S, Sebti S, Shibata D. HPP1-mediated tumor suppression requires activation of STAT1 pathways. Int J Cancer 2008; 122:1567 - 72; http://dx.doi.org/10.1002/ijc.23202; PMID: 18059030
- Young J, Biden KG, Simms LA, Huggard P, Karamatic R, Eyre HJ, Sutherland GR, Herath N, Barker M, Anderson GJ, et al. HPP1: a transmembrane protein-encoding gene commonly methylated in colorectal polyps and cancers. Proc Natl Acad Sci U S A 2001; 98:265 - 70; http://dx.doi.org/10.1073/pnas.98.1.265; PMID: 11120884
- Sato F, Shibata D, Harpaz N, Xu Y, Yin J, Mori Y, Wang S, Olaru A, Deacu E, Selaru FM, et al. Aberrant methylation of the HPP1 gene in ulcerative colitis-associated colorectal carcinoma. Cancer Res 2002; 62:6820 - 2; PMID: 12460892
- Gery S, Koeffler HP. Repression of the TMEFF2 promoter by c-Myc. J Mol Biol 2003; 328:977 - 83; http://dx.doi.org/10.1016/S0022-2836(03)00404-2; PMID: 12729735
- Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer 2008; 8:976 - 90; http://dx.doi.org/10.1038/nrc2231; PMID: 19029958
- Kurland JF, Tansey WP. Myc-mediated transcriptional repression by recruitment of histone deacetylase. Cancer Res 2008; 68:3624 - 9; http://dx.doi.org/10.1158/0008-5472.CAN-07-6552; PMID: 18483244
- Liu T, Tee AE, Porro A, Smith SA, Dwarte T, Liu PY, Iraci N, Sekyere E, Haber M, Norris MD, et al. Activation of tissue transglutaminase transcription by histone deacetylase inhibition as a therapeutic approach for Myc oncogenesis. Proc Natl Acad Sci U S A 2007; 104:18682 - 7; http://dx.doi.org/10.1073/pnas.0705524104; PMID: 18003922
- Marshall GM, Gherardi S, Xu N, Neiron Z, Trahair T, Scarlett CJ, Chang DK, Liu PY, Jankowski K, Iraci N, et al. Transcriptional upregulation of histone deacetylase 2 promotes Myc-induced oncogenic effects. Oncogene 2010; 29:5957 - 68; http://dx.doi.org/10.1038/onc.2010.332; PMID: 20697349
- Fotheringham S, Epping MT, Stimson L, Khan O, Wood V, Pezzella F, Bernards R, La Thangue NB. Genome-wide loss-of-function screen reveals an important role for the proteasome in HDAC inhibitor-induced apoptosis. Cancer Cell 2009; 15:57 - 66; http://dx.doi.org/10.1016/j.ccr.2008.12.001; PMID: 19111881
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997; 389:251 - 60; http://dx.doi.org/10.1038/38444; PMID: 9305837
- Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Histone deacetylases and cancer: causes and therapies. Nat Rev Cancer 2001; 1:194 - 202; http://dx.doi.org/10.1038/35106079; PMID: 11902574
- Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet 2005; 37:1289 - 95; PMID: 16200064
- Khan O, La Thangue NB. Drug Insight: histone deacetylase inhibitor-based therapies for cutaneous T-cell lymphomas. Nat Clin Pract Oncol 2008; 5:714 - 26; http://dx.doi.org/10.1038/ncponc1238; PMID: 18839006
- Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev 2001; 15:2069 - 82; http://dx.doi.org/10.1101/gad.906601; PMID: 11511539
- Belshaw NJ, Elliott GO, Williams EA, Bradburn DM, Mills SJ, Mathers JC, Johnson IT. Use of DNA from human stools to detect aberrant CpG island methylation of genes implicated in colorectal cancer. Cancer Epidemiol Biomarkers Prev 2004; 13:1495 - 501; PMID: 15342451
- Sabbioni S, Miotto E, Veronese A, Sattin E, Gramantieri L, Bolondi L, Calin GA, Gafà R, Lanza G, Carli G, et al. Multigene methylation analysis of gastrointestinal tumors: TPEF emerges as a frequent tumor-specific aberrantly methylated marker that can be detected in peripheral blood. Mol Diagn 2003; 7:201 - 7; PMID: 15068392
- Elahi A, Zhang L, Yeatman TJ, Gery S, Sebti S, Shibata D. HPP1-mediated tumor suppression requires activation of STAT1 pathways. Int J Cancer 2008; 122:1567 - 72; http://dx.doi.org/10.1002/ijc.23202; PMID: 18059030
- Ivanauskas A, Hoffmann J, Jonaitis LV, Markelis R, Juozaityte E, Kupcinskas L, Lofton-Day C, Röcken C, Malfertheiner P. Distinct TPEF/HPP1 gene methylation patterns in gastric cancer indicate a field effect in gastric carcinogenesis. Dig Liver Dis 2008; 40:920 - 6; http://dx.doi.org/10.1016/j.dld.2008.05.004; PMID: 18799374
- Coombes MM, Briggs KL, Bone JR, Clayman GL, El-Naggar AK, Dent SY. Resetting the histone code at CDKN2A in HNSCC by inhibition of DNA methylation. Oncogene 2003; 22:8902 - 11; http://dx.doi.org/10.1038/sj.onc.1207050; PMID: 14654786
- Dong W, Wang L, Shen R. MYO5B is epigenetically silenced and associated with MET signaling in human gastric cancer. Dig Dis Sci 2013; 58:2038 - 45; http://dx.doi.org/10.1007/s10620-013-2600-6; PMID: 23456500
- Ishizuka T, Lazar MA. The N-CoR/histone deacetylase 3 complex is required for repression by thyroid hormone receptor. Mol Cell Biol 2003; 23:5122 - 31; http://dx.doi.org/10.1128/MCB.23.15.5122-5131.2003; PMID: 12861000
- Watson PJ, Fairall L, Santos GM, Schwabe JW. Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature 2012; 481:335 - 40; PMID: 22230954
- Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol 2005; 6:635 - 45; http://dx.doi.org/10.1038/nrm1703; PMID: 16064138
- Jiang G, Espeseth A, Hazuda DJ, Margolis DM. c-Myc and Sp1 contribute to proviral latency by recruiting histone deacetylase 1 to the human immunodeficiency virus type 1 promoter. J Virol 2007; 81:10914 - 23; http://dx.doi.org/10.1128/JVI.01208-07; PMID: 17670825
- Bhandari DR, Seo KW, Jung JW, Kim HS, Yang SR, Kang KS. The regulatory role of c-MYC on HDAC2 and PcG expression in human multipotent stem cells. J Cell Mol Med 2011; 15:1603 - 14; http://dx.doi.org/10.1111/j.1582-4934.2010.01144.x; PMID: 20716118
- Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell 2002; 9:45 - 57; http://dx.doi.org/10.1016/S1097-2765(01)00429-4; PMID: 11804585
- Kumagai T, Wakimoto N, Yin D, Gery S, Kawamata N, Takai N, Komatsu N, Chumakov A, Imai Y, Koeffler HP. Histone deacetylase inhibitor, suberoylanilide hydroxamic acid (Vorinostat, SAHA) profoundly inhibits the growth of human pancreatic cancer cells. Int J Cancer 2007; 121:656 - 65; http://dx.doi.org/10.1002/ijc.22558; PMID: 17417771
- Li Y, Zhang X, Polakiewicz RD, Yao TP, Comb MJ. HDAC6 is required for epidermal growth factor-induced beta-catenin nuclear localization. J Biol Chem 2008; 283:12686 - 90; http://dx.doi.org/10.1074/jbc.C700185200; PMID: 18356165
- Villagra A, Ulloa N, Zhang X, Yuan Z, Sotomayor E, Seto E. Histone deacetylase 3 down-regulates cholesterol synthesis through repression of lanosterol synthase gene expression. J Biol Chem 2007; 282:35457 - 70; http://dx.doi.org/10.1074/jbc.M701719200; PMID: 17925399
- Reid G, Hübner MR, Métivier R, Brand H, Denger S, Manu D, Beaudouin J, Ellenberg J, Gannon F. Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol Cell 2003; 11:695 - 707; http://dx.doi.org/10.1016/S1097-2765(03)00090-X; PMID: 12667452