Abstract
Oncolytic viruses have recently received widespread attention for their potential in innovative cancer therapy. Many telomerase promoter-regulated oncolytic adenoviral vectors retain E1A and E1B. However, the functions of E1A and E1B proteins in the oncolytic role of replication-competent adenovirus (RCAd) and RCAd enhanced transduction of replication defective adenoviruses (RDAd) have not been addressed well. In this study, we constructed viruses expressing E1A alone, E1A plus E1B-19 kDa, and E1A plus E1B-19 kDa/55 kDa. We then tested their roles in oncolysis and replication of RCAd as well as their roles in RCAd enhanced transfection rate and transgene expression of RDAd in various cancer cells in vitro and in xenografted human NCI-H460 tumors in nude mice. We demonstrated that RCAds expressing E1A alone and plus E1B-19 kDa exhibited an obvious ability in replication and oncolytic effects as well as enhanced RDAd replication and transgene expression, with the former showed more effective oncolysis, while the latter exhibited superior viral replication and transgene promotion activity. However, RCAd expressing both E1A and E1B-19 kDa/55 kDa was clearly worst in all these abilities. The effects of E1A and E1B observed through using RCAd were further validated by using plasmids expressing E1A alone, E1A plus E1B-19 kDa, and E1A plus E1B-19 kDa/55 kDa proteins. Our study provided evidence that E1A was essential for inducing replication and oncolytic effects of RCAd as well as RCAd enhanced RDAd transduction, and expression of E1B-19 kDa other than E1B-55 kDa could promote these effects. E1B-55 kDa is not necessary for the oncolytic effects of adenoviruses and somehow inhibits RCAd-mediated RDAd replication and transgene expression.
Introduction
Oncolytic viruses have recently received widespread attention for their potential in innovative cancer therapy. Through mutation and selection, some mutants, such as dl309, obtain the ability to replicate in tumor cells, spread viral particles in tumor tissues, and eventually induce tumor cell death. The early viral 1A (E1A) protein has been widely recognized to be essential for adenoviral replication and production of progeny virions in human cells. Tumor selective oncolytic adenoviruses have been developed by using tumor specific promoter elements, such as the human telomerase protein/enzyme (hTERT) gene promoter, to control gene expression essential for adenoviral replication.Citation1-Citation3 Since E1A and E1B have been identified to be important for adenoviral replication, many telomerase promoter-regulated adenoviral vectors retain both E1a and E1b genes.Citation4 This reduces the capacity of oncolytic adenoviruses to carry therapeutic genes. However, E1b encodes two proteins, E1B-55 kDa and E1B-19 kDa, which are the results of alternative splicing and can be separated at the gene level. In general, few studies have investigated the distinct roles of E1A, E1B-55 kDa, and E1B-19 kDa in the replication of oncolytic adenoviruses in tumor tissues.
Besides the increase in direct oncolytic activity, replication-competent adenoviruses (RCAd) have also been engineered with increased capacity for therapeutic genes, including suicide genes, immunomodulatory genes, and prodrug activation enzyme genes.Citation3 Therefore, nonessential viral genes are deleted to increase the carrying potential of adenoviruses for therapeutic gene.Citation5 For example, deletion of the E3 region has been successfully adopted for this purpose. Recently, the adenoviral E3 region has been recognized to contribute to evasion of the host’s antiviral response. Retention of E3 region can increase the survival of adenoviral vectors in the host’s tissues and lead to higher therapeutic efficacy. However, retention of the E3 region in oncolytic adenoviruses leaves little space for therapeutic genes.Citation6 To overcome this obstacle, a novel strategy using a combination of replication-competent adenoviruses (RCAd) and replication defective adenoviruses (RDAd) carrying therapeutic genes to treat cancer has been developed and successfully tested in preclinical animal models.Citation7
This novel combination strategy uses a low dose of RCAd and a largely decreased loading dose of RDAd compared with the routine use of RDAd alone. However, the combination significantly increased the therapeutic efficiency due to selective production of large amounts of RCAd and RDAd, wider spread invasion of neighboring cells, and increased transgene expression in tumor mass.Citation8-Citation10 Until now, the combination strategy only tested E1a driven replication of oncolytic adenoviruses and its role in enhancing RDAd production and transgene expression. The role of E1b in the combination strategy has not been tested. Whether E1B-55 kDa and E1B-19 kDa exert different roles in RCAd enhanced RDAd replication remains unclear. In this study, we investigated the role of RCAd with different replication elements on RDAd production and transgene expression in vitro and in vivo.
Results
RCAds enhanced AdGFP transduction in human tumor cells with different efficiencies
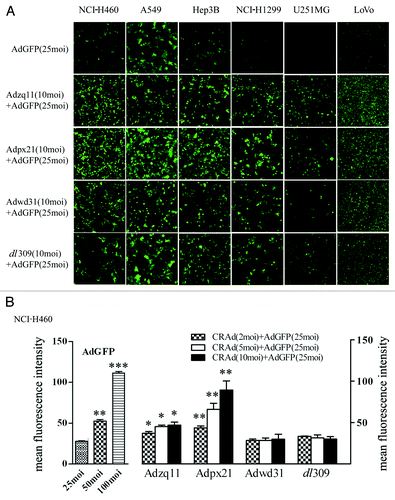
To investigate the ability of RCAds in enhancing AdGFP transduction, NCI-H460, A549, Hep3B, NCI-H1299, U251MG, and LoVo cells cultured in 24-well plates were infected with 25 MOI of AdGFP alone or co-infected with 10 MOI of Adzq11, Adpx21, Adwd31, or dl309 for 24 h. Co-infection with RCAd viruses significantly enhanced GFP expression in all tumor cells compared with AdGFP infection alone; However, Adzq11 and Adpx21 were more effective than Adwd31 and dl309 (). Next, we further tested the effect of different MOI of RCAd on GFP expression of AdGFP. About 8 × 104 NCI-H460 cells were seeded in 24-well plates and 24 h later they were infected with 25 MOI of AdGFP and 2, 5, or 10 MOI of RCAd for another 24 h. The NCI-H460 cells infected by 25, 50, and 100 MOI of AdGFP alone were used as control for the analysis of RCAd-enhanced AdGFP transduction efficiency. showed a summary about the GFP expression with or without RCAd. The GFP expression was shown as mean of fluorescence intensity and RCAd were used at different MOI (2, 5, and 10 MOI). The results demonstrated that Adzq11 or Adpx21 (2, 5, and 10 MOI) but not Adwd31 and dl309 significantly increased GFP fluorescence intensity, when combined with AdGFP (in all groups P < 0.05).
Figure 1. RCAd enhanced RDAd-carried GFP expression in cancer cells. The representative pictures of GFP expression. The different type of tumor cells were infected with 25 MOI of Ad-EGFP alone or co-infected with Ad-EGFP (25 MOI) and different RCAd (10 MOI) and the pictures were taken by epi-fluorescence microscopy (A). FACS analysis of GFP expression. The NCI-H460 cells were infected with different MOI of AdGFP (25, 50, and 100 MOI) and co-infected with Ad-EGFP (25 MOI) and different RCAd (2, 5, and 10 MOI). The mean fluorescent intensity of GFP was analyzed by FACS 72 h after infection. Data were average of 3 individual wells for each viral vector infection and at least 3 times of infection (B). *P < 0.05, **P < 0.01, ***P < 0.001.
Differential replication capacity and cytopathic effects among RCAds in human tumor cells
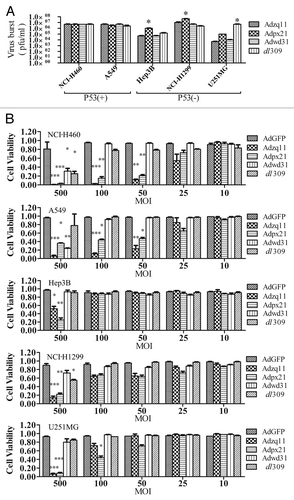
To investigate the replication ability of RCAds, NCI-H460, A549, Hep3B, NCI-H1299, and U251MG cells cultured in 24-well plates were infected with 10 MOI of Adzq11, Adpx21, Adwd31, or dl309 viruses for 4 h. After washing with 1× PBS for 3 times, cells were continually cultured in fresh medium for 72 h. The cells were then collected for viral titer measurement. All tested RCAd viruses replicated effectively in all cells. However, Adpx21 replicated more effectively than other viruses in p53-mutated tumor cells (Hep3B and NCI-H1299) (P < 0.05). The dl309 virus was more effective than other viruses in p53-mutanted U251MG cells (). To test for cytopathic effects, 4 × 103 cells were seeded in 96-well plates for 24 h and then infected with different MOIs of viruses. CCK8 assay was conducted 6 d later. The results showed that 500 MOI of Adzq11 and Adpx21 significantly decreased cell viability in all tested cells, whereas 100 and 50 MOI of Adzq11 and Adpx21 reduced cell viability in NCI-H460 and A549 cells (P < 0.05). Five hundred MOI of Adwd31 only induced cytopathic effects in NCI-H460 and A549 cells, while 500 MOI of dl309 only induced cytopathic effects in NCI-H460 and NCI-H1299 cells. Other doses of virus didn’t induce significant cytopathic effects in any of the cell lines ().
Figure 2. Virus replication and oncolytic effect of RCAd. The cancer cell lines NCI-H460, A549, Hep3B, NCI-H1299, and U251MG were infected with 10 MOI of different RCAd, wild-type adenovirus dl309 was used as a control. The cell lysates were prepared at 72 h and viral titers were measured by plaque-forming assay. The titer values are shown as mean ± SD from triplicates (A). All cancer cells mentioned above were infected with different RCAd at 10, 25, 50, 100, and 500 MOI respectively. Cell viability was measured using CCK-8 assay 7 d post-infection. Each infection were performed in 5 wells and repeated 3 times. Data are shown as the mean ± SD and compared with AdGFP alone (B). *P < 0.05, **P < 0.01, ***P < 0.001.
RCAds enhanced RDAd transduction efficiency in NCIH460 tumors and inhibited tumor growth in vivo
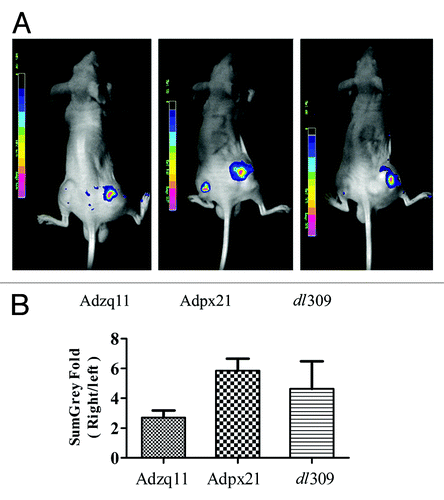
Based on results described above, Adzq11 and Adpx21 remarkably enhanced RDAd transduction efficiency and had stronger replicative and cytopathic capacity than Adwd31 in tested tumor cells. We next tested whether Adzq11 and Adpx21 were able to enhance RDAd transduction efficiency in xenograft human tumors. As shown in , combined injection of Adzq11, Adpx21, and dl309 with AdGFP resulted in much stronger GFP expression in NCI-H460 tumors. In comparison with AdGFP injection alone, GFP signal in combined injection of Adzq11, Adpx21, and dl309 with AdGFP showed 2.70 ± 0.48, 5.85 ± 0.79, and 4.64 ± 1.84-fold increment ().
Figure 3. RCAd enhanced AdGFP transduction in tumor in vivo. A total of 5 × 106 NCI-H460 cells were injected subcutaneously into both hind limbs of Balb/C nude mice. When the tumor size reached 10 mm in diameter, animals were divided into 3 groups (n = 3). A total of 5 × 107 RCAd plus 1 × 108 AdGFP were injected into the tumor on the right hind limb, while about 1 × 108 AdGFP alone were injected into the tumor on the left hind limb as control. Forty-eight hours later, in vivo imaging was performed by NightOwl LB 981 Molecular Imaging System (Berthold Technologies) (A). Comparison of fluorescence intensity in NCI-H460 tumors, data are shown as ratio of RCAd plus Ad-GFP (right leg) over AdGFP alone (left leg) (B).
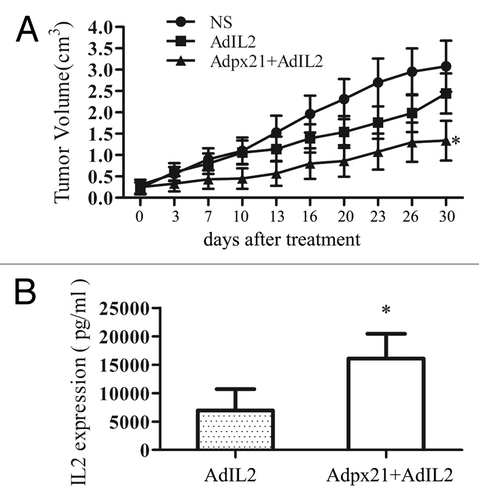
Next, we further tested whether the combined strategy could improve the therapeutic efficacy for tumors in vivo. IL-2 and interferon-α2b are two cytokines approved by the FDA for treatment of cancer. IL-2 has demonstrated activity against renal cell carcinoma, melanoma, lymphoma, and leukemia. However, systemic administration of AdIL2 can cause side effects. Therefore, we established human NCI-H460 xenografted tumors in nude mice and then intratumorally injected either saline, AdhIL2 alone or AdIL2 plus Adpx21 every other day for 3 times total. The results showed that combined injection of AdIL2 and Adpx21 significantly inhibited tumor growth compared with AdIL2 alone (n = 8, P < 0.05) (). In a separate study, we collected tumor tissues 72 h after virus injection and measured hIL2 by ELISA. The tumor tissues received Adpx21+AdIL2 injection showed significantly higher human IL2 level than that received AdhIL2 alone injection (P < 0.05) ().
Figure 4. RCAd enhanced AdIL2 transduction inhibited tumor growth. Human xenograft tumors were established by subcutaneous injection of approximately 1 × 107 NCI-H460 tumor cells into the right hind limbs of mice. When tumors reached 5 mm in diameter, mice were randomly divided into 3 groups, with 8 mice in each group. The injection of AdIL2 (5 × 107 pfu) alone or AdIL2 (5 × 107 pfu) plus Adpx21 (1 × 107 pfu) significantly inhibited tumor growth in comparison with saline injected tumors (A). In another set, 72 h after virus injection mice were sacrificed and the tumor tissues were excised and homogenized in PBS (n = 4). IL2 in the supernatant were measured by ELISA. The tumors received Adpx21 plus AdIL2 had increased IL2 expression than that received AdIL2 alone (B). Data are shown as the mean ± SD. *P < 0.05.
Distinct roles of E1a and E1b genes expression in human tumor cells
To further confirm the role of E1a and E1b, tumor cells were transfected with various plasmids carrying E1A and/or E1B 19 kDa or 55 kDa protein-encoding sequence and then infected with AdGFP 24 h later. The relative copy numbers of AdGFP and its expression were confirmed by real-time PCR. E1A or E1A plus E1B 19kDa or E1A plus the entire E1B protein encoding sequence (i.e., both 19 kDa and 55 kDa) significantly enhanced GFP expression () and increased AdGFP copy numbers () in infected tumor cells (all P < 0.05). The GFP expression also correlated with E1A expression in tumor cells ().
Figure 5. Differential effect of different element of E1 genes on AdGFP replication. SMMC7721, NCI-H460, and A549 cells grown on 6-well plates were transfected with 2 µg of plasmid DNAs using lipofectamine 2000, empty vector pZap was used as control. Twenty-four hours after transfection, cells were infected with 10 MOI of AdGFP. Four hours later AdGFP were removed and cells were washed 3 times using PBS and cultured with normal medium. The representative pictures of GFP expression were taken by epi-fluorescence microscopy 24 h later after infection (A). A summary of FACS analysis for GFP expression in cells mentioned above (B). GFP DNA detected by quantitative PCR (C). GFP mRNA detected by quantitative RT-PCR (D). E1a mRNA detected by quantitative RT-PCR (E).
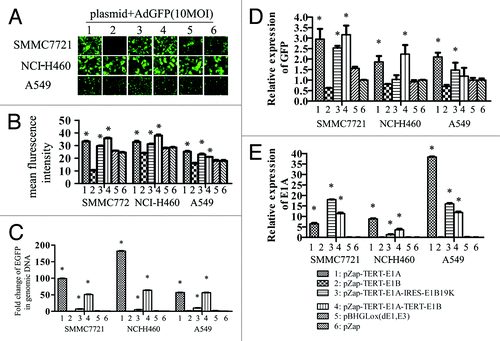
In contrast, E1B itself significantly suppressed AdGFP expression () and AdGFP copy number (). In pZap-TERT-E1B transfected cancer cells, GFP expression suppressed significantly about 0.62 ± 0.01, 0.81 ± 0.02, 0.70 ± 0.06-fold (P < 0.05) (). Similarly, a dramatically decreased AdGFP copy number was observed about 0.36 ± 0.01, 0.34 ± 0.01-fold in SMMC7721 and NCI-H460 cell respectively (P < 0.05) (). These results indicated that E1A can induce RDAd replication and expression; however, E1B can inhibit RDAd replication and expression.
Discussion
The functions of E1A and E1B proteins in adenovirus type 2 and 5 replication have been extensively studied. However, few studies compare the roles of E1A, E1B-19 kDa, and E1B-55 kDa on RCAd oncolysis and RCAd-enhanced RDAd transduction. In this study, we demonstrated that E1A alone is sufficient in mediating the oncolytic role and replication of RCAd as well as the replication and transduction of RDAd carried genes. E1B-19 kDa protein is able to enhance E1a gene expression. In contrast, E1B-55 kDa protein exerts an inhibitory effect on E1a expression and function. This study provided practical instruction for constructing an oncolytic adenovirus.
In this study, Adzq11, an adenovirus containing only TERT promoter-controlled E1a exhibited a high replication rate in cancer cells with wild-type p53 and null p53 expression. The high replication rate of Adzq11 resulted in higher oncolytic effect in all tested cancer cells compared with other adenoviruses containing TERT promoter driving both E1a and E1b expression (). Moreover, Adzq11 significantly enhanced GFP expression when it was co-transfected with a replication-defective adenovirus, AdGFP (). The enhanced GFP expression was also observed when Adzq11 was replaced by a plasmid expressing E1A protein. In addition, expression of the E1a gene significantly increased the replication of AdGEP virus (). Importantly, the viruses and plasmids that expressed only the E1a gene were more effective than the viruses and plasmids that expressed both E1a and E1b (19 kDa+55 kDa) genes. These observations suggested that E1a alone is able to effectively replicate and contribute to the oncolytic role of RCAd, RCAd-enhanced RDAd replication, and RDAd-carried transgene expression. In infected human cells, E1a is the first viral gene expressed after infection, and E1A protein is widely recognized to be sufficient in activating the cell cycle, including expression of genes required for DNA synthesisCitation11 and viral gene transcription.Citation12 Therefore, our findings on E1A are not surprising. In particular, the roles of E1A protein are not affected by p53 expression. This is crucial for establishing effective cancer therapy in all patients because most cancer patients lack functional p53 expression, while some express wild-type p53.
Many telomerase promoter-regulated adenoviral vectors retain E1B-55 kDa and/or E1B-19 kDa. However, the actual role of these two proteins in virus replication and cancer cell oncolysis has not been fully elucidated. A previous study demonstrated that E1B can enhance E1A expression.Citation13 In this study, we demonstrated that adenoviruses expressing both E1B-55 kDa and E1B-19 kDa proteins (Adwd31 and dl309) showed no oncolytic effects in all tested cancer cells, whereas Adpx21 only expressing E1B-19kDa protein showed obvious oncolytic effects in these cells (). In comparison with Adwd31 and dl309, Adpx21 dramatically increased RDAd (AdGFP) expression both in vitro and in vivo ( and ). It replicated more efficiently in Hep3B and NCI-H1299 cells, about 10–20-folds more than Adwd31 and dl309 (), and even more efficiently than Adzq11 (only having E1A) (P < 0.05). Therefore, the E1B-19 kDa protein might enhance E1A expression and its functions. E1A acts as a cue to initiate virus replication by driving the host cell into the S-phase of the cell cycle for viral DNA replication.Citation14,Citation15 An adverse effect of E1A is that it stabilizes p53, which leads to apoptosis and is unfavorable for viral replication.Citation16 E1B-19 kDa, a functional Bcl-2 homolog, directly binds Bax, Bak-inhibiting oligomerization, and mitochondrial pore-formation intrinsically inhibiting E1A-induced apoptosis through the p53-dependent and p53-independent mechanisms.Citation17-Citation20 The anti-apoptotic E1B-19 kDa protein promotes viral replication, allowing it to propagate throughout the tumor. Matsushita et al. also found that adeno-associated virus production would be reduced by at least 100-fold when adenovirus-bearing mutated E1B-19 kDa was used as a helper virus.Citation21 Polster et al. found that the number of progeny virions produced by the E1B-19 kDa mutated oncolytic viruses was 10-fold lower than that produced by viruses with intact E1B-19 kDa.Citation22 All of these observations are consistent with our results mentioned above. In contrast, Adwd31 and dl309, which expressed both E1B-55 kDa and E1B-19 kDa proteins, showed less replicating capability and impaired AdGFP expression enhancing effect than Adpx21 (expressing E1B-19 kDa protein not E1B-55 kDa). In addition, plasmids expressing E1B-55 kDa and E1B-19 kDa proteins (pZap-TERT-E1B) also showed similar effects, i.e., impaired AdGFP viral replication and GFP protein expression than other plasmids (P < 0.05) (). This suggests that E1B-55 kDa might inhibit the expression and functions of E1A.
Currently, oncolytic adenoviruses retain the E3 region to increase the survival of adenoviral vectors in host tissue and subsequently achieve higher therapeutic efficacy. However, little space is left for the insertion of therapeutic genes. To overcome this obstacle, a combination of RCAd with RDAd carrying therapeutic genes has been adopted to treat cancers. This novel strategy has been successfully tested in xenograft tumors.Citation8-Citation10 In this study, we tested the role of RCAd in enhancing RDAd-carried genes expression in cancer cells. Co-injection of RCAd and RDAd significantly enhanced GFP expression in xenograft NCI-H460 tumors (). We further tested the therapeutic effects of IL2 in xenograft NCI-H460 tumors. Co-injection of Adpx21 and AdIL2 significantly increased IL2 expression, which significantly inhibited tumor growth compared with Adpx21 and AdIL2 alone (). It is apparent that this strategy significantly increased transgene expression in vivo.
In conclusion, E1A appears to be sufficient in mediating the oncolytic role and replication of RCAd, RCAd-enhanced replication of RDAd, and transduction of RDAd-carried genes. Finally, E1B-55 kDa alone plays an inhibitory effect on adenovirus transduction.
Experimental Procedures
Adenoviruses and plasmids
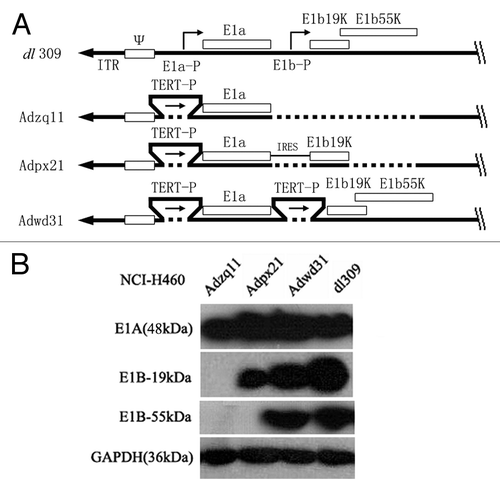
All adenoviruses used in this study were serotype 5 adenoviruses. AdGFP is an RDAd that does not contain the E1 or E3 gene but does contain a CMV driven enhanced green fluorescence (EGFP) gene expression cassette in the E3 region. Four different RCAd were used in this study. The dl309 is a naturally occurring mutant RCAd lacking several E3 region genes. The other three RCAd were constructed in our laboratory using the Adenoviral Promoterless Gateway Expression Kit (Invitrogen). Adzq11 only contained the E1a gene. Adpx21 contained the coding sequence of E1 gene expressing E1A and E1B 19 kDa protein. Adwd31 contained the coding sequence of E1 gene expressing E1A and E1B 19 kDa and 55 kDa proteins. E1a and E1b were driven by the human telomere reverse transcriptase (TRET) gene promoterCitation2 and linked by an internal ribosome entry site (IRES) in Adpx21. These adenoviral vectors were E3 deleted. The schematic diagram of adenoviral vectors used in this study is shown in . The major difference between dl309 and the other three RCAd was that dl309 expressed E1a and E1b in both normal and tumor cells. Whereas, Adzq11 expressed E1A, Adpx21 expressed E1A and E1B-19 kDa, and Adwd31 expressed E1A andintact E1B in only tumor cells that were confirmed by western blot (). Preparation and packaging of adenoviruses were performed as previously described.Citation7
Figure 6. A schematic diagram of replication competent adenovirus constructed in this study. E1a-P, E1a promoter; TERT-P, the telomerase (TERT) promoter; E1a and E1b, early E1 genes of the adenovirus; IRES, internal ribosome entry site; E1b19k, gene expresses E1b-19 kDa protein; E1b55k, gene expressing E1b-55 kDa protein (A). Confirmation of E1A, E1B-19 kDa, and E1B-55 kDa protein expression by western blot. NCI-H460 cancer cells were infected with different adenoviruses at a 10 MOI (pfu/cell) for 48 h and then the cell lysates were analyzed by western blot. GAPDH (36 kDa) was used as control (B).
To exclude the influence of viral genes in the backbone of the adenoviruses, we used various plasmids to express E1A, or E1A-E1B proteins. pZap-TERT-E1A and pZap-TERT-E1B plasmid expressed the entire length of E1A and 19 kDa/55 kDa of E1B protein under the control of the TERT gene promoter, respectively. pZap-TERT-E1A-TERT-E1B expressed the entire length of E1A and 19 kDa/55 kDa of E1B protein under the control of the TERT gene promoter separately. pZap-TERT-E1A-IRES-E1B19k expressed the entire length of the E1A protein and the 19 kDa of E1B protein under the control of one TERT gene promoter. The pBHGLox was a plasmid which contained the backbone of adenovirus but lacking E1 and E3 genes. pZap was a vector which was used to construct pZap-TERT-E1A etc.
Cell culture
Human glioma U251MG, hepatoma Hep3B and SMMC7721, non-small lung cancer NCI-H460 and NCI-H1299, lung adenocarcinoma A549, and colon adenocarcinoma LoVo cell lines were purchased from the Chinese Academy of Science. All cells were cultured in either RPMI 1640 or DMEM with 10% fetal bovine serum, 100 units/mL penicillin, and 100 µg/mL streptomycin at 37 °C, 5% CO2.
Western blot analysis
NCI-H460 tumor cells at 60–70% confluence were infected with different viruses at 10 MOI. After 48 h, cell lysates were prepared using lysis buffer containing 2% sodium dodecyl sulfate (SDS) and 0.125 M TRIS-HCl (pH 6.8) and proteinase inhibitor. The total protein of the cell lysates was measured using the Bio-Rad Protein Assay kit (Bio-Rad). About 40 μg proteins per sample were separated on 10% SDS-polyacrylamide gel and blotted onto a polyvinylidene difluoride membrane (Roche). The membrane was incubated with antibodies against E1A, E1B-19 kDa, E1B-55 kDa and GAPDH (Santa Cruz Biotechnology Inc.) overnight at 4 °C respectively. After washing, the membrane was probed with the appropriate secondary peroxidase-conjugated antibodies and subsequently visualized using a chemiluminescence method (ECL, Roche). Images of the bands were captured using an image acquisition software system (ChemiDoc™ XRS+; Bio-Rad).
Evaluation of virus infection and transduction in vitro
Tumor cells with wild-type p53 gene (NCI-H460, LoVo and A549) and mutant p53 gene (U251MG, Hep3B and NCI-H1299) were cultured in 24-well plates and infected with 25 multiplicity of infection (MOI) of AdGFP alone or 10 MOI of different RCAd. The infection and transduction efficiency were evaluated by GFP expression, which was photographed by epi-fluorescence microscopy (Zeiss, Axiovert S100) and quantified by flow cytometry (FACS, Beckman-Coulter). To further verify RCAd enhanced AdGFP expression, NCIH460 cells were infected with 25 MOI of AdGFP and 2, 5, or 10 MOI of different RCAd and analyzed by FACS.
Adenovirus titering
Adenovirus titering was performed by virus plaque assay.Citation23 Briefly, U251MG, Hep3B, NCI-H460, NCI-H1299, SMMC7721, and A549 tumor cells in the exponential growth phase were infected with 10 MOI of different RCAd for 4 h, washed 3 times using PBS, and then cultured in fresh growth medium. Three days later, cells were harvested and subjected to the plaque assay.
Cell viability assay
Cell Counting Kit-8 (CCK-8, Dojindo) was used for cell viability assay. Briefly, U251MG, Hep3B, NCI-H460, NCI-H1299, or A549 cells were seeded into 96-well plates at 4 × 103 cells per well. Twenty-four hours later, cells were infected with adenovirus at different MOI (10, 25, 50, 100, and 500 MOI, respectively) for 6 d. Ten microliters of the reagent provided with the kit was added to the cells followed by incubation for 1 h. Cell viability was assessed based on colorimetric properties using the microplate reader at 450 nm. Cell viability was calculated according to OD values.
Animals
All animal experiments were conducted in accordance with an approved protocol from the Shanghai Jiaotong University School of Medicine. Balb/C nude mice (BALB/c, nu/nu) weighing 18–20 g were provided by the animal center of the Shanghai Institute for Biological Science, Chinese Academy of Sciences and housed in rooms under standard lighting conditions and temperature. Water and food were provided ad libitum.
In vivo imaging assay for RCAd enhanced adenovirus transduction
NCI-H460 xenograft tumors were introduced in Balb/C nude mice by subcutaneous injection of 5 × 106 NCI-H460 cells in 50 µL of PBS into both hind limbs. When the tumor size reached 10 mm in diameter, animals were divided into 3 groups (n = 3). About 1 × 108 AdGFP were injected into the tumor on the left hind limb as control, and 5 × 107 RCAd + 1 × 108 AdGFP were injected into the tumor on the right hind limb (group 1: Adzq11 + AdGFP; group 2: Adpx21 + AdGFP; group 3: dl309 + AdGFP). Forty-eight hours later, in vivo imaging was performed as previously described.Citation24
RCAd enhanced IL2 expression and antitumor effect of IL2 in vivo
Human xenograft tumor models were established by subcutaneous injection of approximately 1 × 107 NCI-H460 tumor cells into the right hind limbs of mice. To test RCAd-enhanced IL2 expression in vivo, animals with 10 mm in diameter NCI-H460 tumors were divided into 2 groups (n = 4) and intratumorally injected with AdIL2 (5 × 107 pfu) or Adpx21 (1 × 107 pfu) + AdIL2 (5 × 107 pfu). Seventy-two hours after virus injection, mice were sacrificed and the tumor tissues were excised and homogenized in PBS. IL2 levels in the supernatant were measured by ELISA. To test the antitumor effect of IL2 in vivo, mice were randomly divided into 3 groups (n = 8). When tumors reached 5 mm in diameter, they were injected with saline (NS), AdIL2 (5 × 107 pfu) or Adpx21 (1 × 107 pfu) + AdIL2 (5 × 107 pfu) on day 0, 3, and 7, respectively. Tumors were measured in two dimensions 2 times per week, and tumor volume (V) was calculated using the following formula: V = 0.52 × S2 × L (S, the shortest dimension; L, the longest dimension). Animals were euthanized 30 d after treatment.
Plasmid transfection, AdGFP copy number analyses, and quantitative PCR assessment for E1a and EGFP mRNA level
To exclude the possible influence of genes located in the backbone of the adenoviruses in the virus based experiments, 2 µg of plasmid DNAs (pZap-TERT-E1A; pZap-TERT-E1B; pZap-TERT-E1A-IRES-E1B19K, and pZap-TERT-E1A-TERT-E1B) were transfected into SMMC7721, NCI-H460, and A549 cells using lipofectamine 2000. The empty vector pZap was used as a control. Twenty-four hours after transfection, cells were infected with 10 MOI of AdGFP. Four hours later the viruses were removed and the cells were washed with PBS for 3 times and then cultured with normal growth medium. Twenty-four hours later, GFP expression was observed under an epi-fluorescence microscope, and cells were collected for further study. The detection of adenoviral genomic DNA was performed using real-time PCR. The genomic DNA of AdGFP was defined by the GFP gene. The GFP gene was amplified by forward primer 5′-AGAAGAACGG CATCAAGGTG-3′ and reverse primer 5′-GAACTCCAGCA GGACCATGT-3′. The DNA was extracted using DNA extraction kit (Qiagen). The quantitative PCR reactions were performed by use of the SYBR Premix Ex TaqTM kit (TaKaRa). The human genomic GAPDH gene was amplified at same time as an internal control using forward primer 5′-AGGGCCCTGA CAACTCTTTT-3′ and reverse primer 5′-AGGGGTCTAC ATGGCAACTG-3′. The copy numbers of GFP gene was determined relative to GAPDH CT value.
Total RNA was isolated using Trizol reagent (Invitrogen) following the manufacturer’s instructions. To remove the possible contamination of adenovirus genomic DNA, DNase digestion was performed before reverse transcription. Reverse transcription and cDNA synthesis were accomplished by using Access RT-PCR System (Promega). The cDNA was then used as templates in quantitative PCR reactions as described above. Relative quantification of GFP and E1a was determined by comparative CT method (RQ = 2−ΔΔCT) and normalized to the GAPDH level. The average value from group pZap was used as baseline. The fold changes over pZap were calculated. The E1a gene was amplified by forward primer: 5′-TCCGGTCCTT CTAACACACC-3′ and reverse primer: 5′-GGCGTTTACA GCTCAAGTCC-3′. The GFP gene was amplified by forward primer and reverse primer above. Human GAPDH gene was amplified at the same time as an internal control using forward primer: 5′- ATGGAAATCC CATCACCATC TT -3′ and reverse primer: 5′- CGCCCACTTG ATTTTGG -3′.
Statistical analysis
All in vitro experiments were completed three times under separate conditions, and the in vitro and in vivo experimental data are presented as the mean plus or minus standard deviation (mean ± SD). All analyses were performed using SPSS v13.0 (SPSS Inc.). Differences were assessed using one-way ANOVA. A P < 0.05 was considered statistically significant.
Disclosure of Potential Conflicts of Interest
All authors declared no conflicts of interest.
Acknowledgments
This project was supported by grants from National Natural Science Foundation of China (30325043, 30672440, and 30500553), the National Basic Research Program of China (2010CB529902), Science and Technology Committee of Shanghai (064119539), and Leading Medical Talent of Shanghai (040308).
References
- Fukuda K, Abei M, Ugai H, Kawashima R, Seo E, Wakayama M, Murata T, Endo S, Hamada H, Hyodo I, et al. E1A, E1B double-restricted replicative adenovirus at low dose greatly augments tumor-specific suicide gene therapy for gallbladder cancer. Cancer Gene Ther 2009; 16:126 - 36; http://dx.doi.org/10.1038/cgt.2008.67; PMID: 18818710
- Huang Q, Zhang X, Wang H, Yan B, Kirkpatrick J, Dewhrist MW, Li CY. A novel conditionally replicative adenovirus vector targeting telomerase-positive tumor cells. Clin Cancer Res 2004; 10:1439 - 45; http://dx.doi.org/10.1158/1078-0432.CCR-03-0122; PMID: 14977847
- Jounaidi Y, Doloff JC, Waxman DJ. Conditionally replicating adenoviruses for cancer treatment. Curr Cancer Drug Targets 2007; 7:285 - 301; http://dx.doi.org/10.2174/156800907780618301; PMID: 17504125
- Wirth T, Kühnel F, Kubicka S. Telomerase-dependent gene therapy. Curr Mol Med 2005; 5:243 - 51; http://dx.doi.org/10.2174/1566524053586536; PMID: 15974879
- Doloff JC, Waxman DJ, Jounaidi Y. Human telomerase reverse transcriptase promoter-driven oncolytic adenovirus with E1B-19 kDa and E1B-55 kDa gene deletions. Hum Gene Ther 2008; 19:1383 - 400; http://dx.doi.org/10.1089/hum.2008.056; PMID: 18771358
- Costantini LC, Bakowska JC, Breakefield XO, Isacson O. Gene therapy in the CNS. Gene Ther 2000; 7:93 - 109; http://dx.doi.org/10.1038/sj.gt.3301119; PMID: 10673714
- Wang H, Wei F, Zhang J, Wang F, Li H, Chen X, Xie K, Wang Y, Li C, Huang Q. A novel immunocompetent murine tumor model for the evaluation of RCAd-enhanced RDAd transduction efficacy. Tumour Biol 2012; 33:1245 - 53; http://dx.doi.org/10.1007/s13277-012-0374-7; PMID: 22627833
- Kirn D. Clinical research results with dl1520 (Onyx-015), a replication-selective adenovirus for the treatment of cancer: what have we learned?. Gene Ther 2001; 8:89 - 98; http://dx.doi.org/10.1038/sj.gt.3301377; PMID: 11313778
- Lee CT, Park KH, Yanagisawa K, Adachi Y, Ohm JE, Nadaf S, Dikov MM, Curiel DT, Carbone DP. Combination therapy with conditionally replicating adenovirus and replication defective adenovirus. Cancer Res 2004; 64:6660 - 5; http://dx.doi.org/10.1158/0008-5472.CAN-04-1200; PMID: 15374981
- Lee CT, Lee YJ, Kwon SY, Lee J, Kim KI, Park KH, Kang JH, Yoo CG, Kim YW, Han SK, et al. In vivo imaging of adenovirus transduction and enhanced therapeutic efficacy of combination therapy with conditionally replicating adenovirus and adenovirus-p27. Cancer Res 2006; 66:372 - 7; http://dx.doi.org/10.1158/0008-5472.CAN-05-1515; PMID: 16397251
- Moran E. Interaction of adenoviral proteins with pRB and p53. FASEB J 1993; 7:880 - 5; PMID: 8344487
- Turnell AS, Mymryk JS. Roles for the coactivators CBP and p300 and the APC/C E3 ubiquitin ligase in E1A-dependent cell transformation. Br J Cancer 2006; 95:555 - 60; http://dx.doi.org/10.1038/sj.bjc.6603304; PMID: 16880778
- Yang Y, McKerlie C, Lu Z, Wang L, Buchwald M. In vivo potential effects of adenovirus type 5 E1A and E1B on lung carcinogenesis and lymphoproliferative inflammation. J Virol 2008; 82:8105 - 11; http://dx.doi.org/10.1128/JVI.00536-08; PMID: 18524829
- Lavery DJ, Chen-Kiang S. Adenovirus E1A and E1B genes are regulated posttranscriptionally in human lymphoid cells. J Virol 1990; 64:5349 - 59; PMID: 2145444
- Ries SJ, Brandts CH. Oncolytic viruses for the treatment of cancer: current strategies and clinical trials. Drug Discov Today 2004; 9:759 - 68; http://dx.doi.org/10.1016/S1359-6446(04)03221-0; PMID: 15450242
- Lowe SW, Ruley HE, Jacks T, Housman DE. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell 1993; 74:957 - 67; http://dx.doi.org/10.1016/0092-8674(93)90719-7; PMID: 8402885
- White E. Mechanisms of apoptosis regulation by viral oncogenes in infection and tumorigenesis. Cell Death Differ 2006; 13:1371 - 7; http://dx.doi.org/10.1038/sj.cdd.4401941; PMID: 16676007
- Cross JR, Postigo A, Blight K, Downward J. Viral pro-survival proteins block separate stages in Bax activation but changes in mitochondrial ultrastructure still occur. Cell Death Differ 2008; 15:997 - 1008; http://dx.doi.org/10.1038/cdd.2008.14; PMID: 18274554
- Yoon AR, Kim JH, Lee YS, Kim H, Yoo JY, Sohn JH, Park BW, Yun CO. Markedly enhanced cytolysis by E1B-19kD-deleted oncolytic adenovirus in combination with cisplatin. Hum Gene Ther 2006; 17:379 - 90; http://dx.doi.org/10.1089/hum.2006.17.379; PMID: 16610926
- Lomonosova E, Subramanian T, Chinnadurai G. Mitochondrial localization of p53 during adenovirus infection and regulation of its activity by E1B-19K. Oncogene 2005; 24:6796 - 808; http://dx.doi.org/10.1038/sj.onc.1208836; PMID: 16007153
- Matsushita T, Okada T, Inaba T, Mizukami H, Ozawa K, Colosi P. The adenovirus E1A and E1B19K genes provide a helper function for transfection-based adeno-associated virus vector production. J Gen Virol 2004; 85:2209 - 14; http://dx.doi.org/10.1099/vir.0.79940-0; PMID: 15269360
- Polster BM, Pevsner J, Hardwick JM. Viral Bcl-2 homologs and their role in virus replication and associated diseases. Biochim Biophys Acta 2004; 1644:211 - 27; http://dx.doi.org/10.1016/j.bbamcr.2003.11.001; PMID: 14996505
- Graham FL, Prevec L. Methods for construction of adenovirus vectors. Mol Biotechnol 1995; 3:207 - 20; http://dx.doi.org/10.1007/BF02789331; PMID: 7552690
- Ji X, Cheng L, Wei F, Li H, Wang M, Tian Y, Chen X, Wang Y, Wolf F, Li C, et al. Noninvasive visualization of retinoblastoma growth and metastasis via bioluminescence imaging. Invest Ophthalmol Vis Sci 2009; 50:5544 - 51; http://dx.doi.org/10.1167/iovs.08-3258; PMID: 19608529
