Abstract
In breast cancer cells, heterodimerization of HER2 and HER3 plays important and dominant roles in the functionality and transformation of HER-mediated pathways, in particular the PI3K/Akt survival pathway. HER3 was considered as a major signaling hub in HER2-amplified cancers. Inhibition of HER3 expression may therefore represent a rational therapeutic approach to breast cancers where HER2/HER3-mediated signaling plays a role in tumorigenesis and progression. miRNAs exerts important roles in regulating gene expressions by binding to and repressing target mRNAs. Here we reported that miRNA-450b-3p inhibits HER3 expression by directly targeting 3′ UTR of HER3 mRNA and represses the downstream signal transductions of HER family. Overexpression of miRNA-450b-3p in SKBR3 cells inhibits cells clonogenic potential and enhances their sensitivity to trastuzumab, a monoclonal antibody that binds to the HER2 receptor, or doxorubicin through repressing proliferative signal pathways mediated by HER3/HER2/PI3K/AKT. Furthermore, we found that breast cancer patients with tumors that demonstrating upregulated HER3 (>2-fold) and downregulated miR-450b-3p (>2-fold) expressions compared with the paired adjacent non-tumorous tissues showed significantly poorer overall survival (P < 0.05). Our study identified miRNA-450b-3p as a new tumor repressor and also provided some evidences suggesting that downregulation of miR-450b-3p expression with concurrent overexpression of HER3 may serve as a prognostic biomarker for poor overall survival in breast cancer patients.
Keywords: :
Introduction
The human epidermal growth factor receptor family of proteins consisting of EGFR, HER2, HER3, and HER4 are type I transmembrane growth factor receptors which activate intracellular signaling pathways in response to extracellular signals.Citation1,Citation2 Over-activation of individual HER family members are etiologically linked with the pathogenesis of a panel of cancers including cancers of the breast, lung, head and neck, brain, and skin.Citation3-Citation6 The HER members always form homo- or hetero-dimers to drive signal transductions underlying their tumorigenic behavior. HER3, as the only member lacking catalytic kinase function, is an obligate partner for hetero-dimerization with the other family members and its preferred partner is HER2.Citation7 In particular, HER3 plays a critical role in HER2-mediated transformation and is indispensible for continued tumor cell growth and proliferation in tumors driven by HER2 overexpression.Citation8 Increased expression of HER3 synergistically increases the transforming potential of HER2.Citation9 In contrast, loss of HER3 abolished the transforming ability of HER2.Citation8
The HER2–HER3 signaling complex is highly effective in activating the PI3K/Akt signaling pathway,Citation10 even though HER2 is unable to directly bind PI3K and activate this pathway. The critical function of HER3 in HER2-mediated transformation is to activate PI3K/Akt signaling transduction.Citation11 In HER2-dependent cells, loss of HER3 results in reduced signaling through PI3K and cell proliferation, indicating that HER2 may be dependent on HER3 to drive growth and survival of breast cancer cells. Actually, the activation of PI3K and Akt by HER2 is mediated through the tyrosine phosphorylation of HER3 which has six tyrosine containing binding sites for PI3K.Citation12 The PI3K/Akt pathway regulates vital cell functions including proliferation, survival, glucose metabolism, epithelial–mesenchymal transition, genome stability, and angiogenesis. In particular, PI3K/Akt pathway activation by chemotherapy-rendered chemo-resistance in ovarian cancer cells.Citation13
The expression of HER3 was regulated at the level of transcription, translation, localization or phosphorylation, etc. For instance, HER3 transcription has been shown to be in part regulated through FOXO1 and FOXO3a transcription factors, rendering the resistance to TKI in HER2-driving breast cancer cells.Citation14 miR-205 and miR-106b were described to downregulate HER3 expression,Citation15,Citation16 and miR-125a and miR-125b have been shown to regulate expressions of both HER2 and HER3.Citation15 mTOR kinase inhibitor was able to increase HER3 phosphorylation through downregulating HER3 dephosphorylation.Citation16 Due to the essential roles of HER3 in HER2-driving breast cancer cells, present studies revealed that inhibition of HER3, at least in part, synergized with HER2 antagonist to breast cancer.
microRNAs (miRNAs) are single-stranded RNA molecules of 20–23 nucleotide length that control gene expression in many cellular processes. Numerous microRNA molecules have been found to typically reduce the stability of mRNAs,Citation17 including those of genes that mediate processes in tumorigenesis, such as inflammation, cell cycle regulation, stress response, differentiation, apoptosis, and invasion. miRNA targeting is mostly achieved through specific base-pairing interactions between the 5′ end (“seed” region) of the miRNA and sites within coding and untranslated regions (UTRs) of mRNAs; target sites in the 3′ UTR lead to more effective mRNA destabilization.Citation18
Given the importance of HER3 in proliferation and survival of HER2 driving breast cancer cells, we hypothesized that HER3 could be a target of specific miRNAs and be regulated by the putative miRNAs. Indeed we found the miR-450b-3p interacts with 3′ UTR of HER3 mRNA to negatively regulate HER3. It also has the capability to inhibit breast cancer cell proliferation and tumorigenesis potential and improve the sensitivity to trastuzumab and doxorubicin. Further more, in clinical practice the combination of downregulation of miR-450b-3p expression with overexpression of HER3 may serve as a prognostic biomarker predicting the patients’ overall survival.
Results
The expression of miR-450b-3p inversely correlates to HER3 in breast cancer cells
Due to the important roles of HER3 in breast cancer progression, we first try to identify specific miRNAs that could efficiently target the HER3 expression. We employed both the database search and manual check for the matched sequences. As a result, we found that miR-450b-3p could be one of the candidates. The 17 of 19 nucleotides of miR-450b-3p are matched to the site 971 to 989 of the HER3 3′ UTR (). To validate the correlation between the miR-450b-3p and HER3, we analyzed the expressions of miR-450b-3p and HER3 in normal breast tissue as well as breast cancer cell lines and found that the expressions of miR-450b-3p and HER3 protein are inversely correlated (). It suggested that the possible regulation of HER3 exerted by miR-450b-3p.
Figure 1. The expressions of miR-450b-3p and HER3 in breast cancer cells. (A) Schematic representation of the interaction between miR-450b-3p and the binding site on the wild-type HER3 3′ UTR, and the 6 bp modified in the mutated control. (B) The abundance of miR-450b-3p of the seven types of tissue or cell lines was evaluated by real-time PCR. The 18S rRNA was used as control. After normalization, data were transformed as log10 of relative quantity (RQ). (C) The expression of HER3, HER2, and GAPDH (the loading control) were quantified by western blot in indicated cell lines. (D) MDA-MB-453 and SKBR3 were transfected with 1nmol miR-450b-3p precursor or scrambled oligonucleotides for 36 h, and then cells were harvested and analyzed by western blot. The results are representative of three independent experiments; bars, SD, * and #, significantly different compared with scrambled control (P < 0.05).
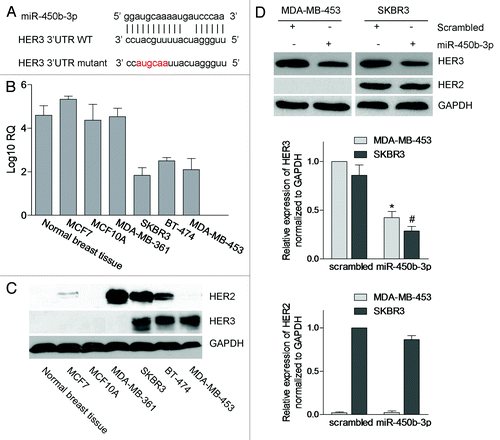
To validate the hypothesis, we transiently transfected SKBR3 and MDA-MB-453 cells with miR-450b-3p precursor and scrambled oligonucleotides, and then detected the HER3 protein expression levels. We found that the HER3 expression was significantly decreased with the transfection of miR-450b-3p, but HER2 protein levels were not affected (). These data indicated that HER3 may be a direct target of miR-450b-3p in breast cancer cells.
miR-450b-3p interacts with HER3 3′ UTR directly
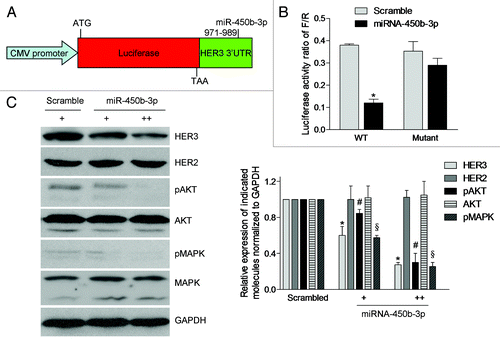
Since miRNAs modulate gene expressions through specific binding to elements of their target mRNA, a luciferase reporter assay was performed to evaluate whether miR-450b-3p regulates HER3 mRNA through this mechanism. We cloned the 1451 bp HER3 3′ UTR into the immediate downstream of the luciferase open reading frame in the pGL3-promoter vector (). As a control, we also constructed a HER3 3′ UTR mutant which lacks the putative binding site of miR-450b-3p using the site directed mutagenesis assay. The reporter vectors were co-transfected with miR-450b-3p precursor molecule or a scrambled oligonucleotide in Hela cells. The results showed that the luciferase activity of pGL3-promoter-HER3 3′ UTR (wt) was significantly decreased with miR-450b-3p expression compared with the scrambled control. But for the mutant of pGL3-promoter-HER3 3′ UTR, this inhibition of luciferase activity was impaired (). These data showed that the miR-450b-3p interacts with the 3′ UTR of HER3 directly to regulate HER3 expression.
Figure 2. miR-450b-3p directly targets 3′ UTR of HER3 and its downstream pathways. (A) Structures of the plasmid pGL3-promoter-HER3-3′ UTR. The whole HER3 3′ UTR (wild type or mutant) was fused to the immediate downstream of firefly luciferase cDNA in pGL3-promoter (Promega) to yield pGL3-promoter-HER3-3′ UTR wild type or mutant. (B) The luciferase activity of pGL3-promoter-HER3-3′ UTR wild type or mutant was measured in presence of scrambled miRNA or miR-450b-3p precursor. The renilla luciferase activity was used as the transfection control. The data was calculated from three independent experiments. Bars, SD. *, significantly different compared with scrambled control (P < 0.05). (C) The SKBR3 cells were transfected with pSUPER or increasing amount of pSUPER-miR-450b-3p for 36 h, and indicated molecules were analyzed by western blot. The results are representative of three independent experiments. Bars, SD. *, # and §, significantly different compared with scrambled control (P < 0.05).
The HER3 associated PI3K/AKT pathway is suppressed by miR-450b-3p
The HER2-HER3 heterodimer is crucial in breast cancer tumorigenesis, progression and drug resistance. Importantly, the HER3-mediated PI3K/AKT pathway activation is the primary oncogenic signaling and we tested whether the miR-450b-3p has the ability to regulate this survival pathway mediated by HER3. The mammalian expression plasmid pSUPER-miR-450b-3p, bearing miR-450b-3p coding fragment, was constructed and transfected into SKBR3 cells to overexpress miR-450b-3p. As shown in , with the increasing amount of pSUPER-miR-450b-3p, the HER3 expression was significantly decreased in a dose-dependent manner, but the HER2 expression was not affected. The levels of phosphorylated AKT and phosphorylated MAPK were decreased compared with the control vector. These data verified that miR-450b-3p was able to specifically suppress PI3K/AKT pathway.
miR-450b-3p inhibits proliferation of cancer cells
It’s well known that PI3K/AKT pathway is involved in cellular functions such as cell growth, proliferation, differentiation, survival, etc., so we assessed whether miR-450b-3p had effects on breast cancer cell growth/survival. We transfected pSUPER-miR-450b-3p or pSUPER empty vector into SKBR3 cells and selected several single stable clones with the treatment of puromycin for 14 d. Five puromycin resistant clones were sub-cultured and expanded, and the expression of miR-450b-3p was detected by qPCR. As shown in , compared with other clones, clones 2 and 3 had better expressions of miR-450b-3p and western blot showed that HER3 expressions were dramatically decreased in these two clones (). Furthermore, these two clones were used to perform clonogenic assay in vitro. After 15 d of culture, we stained the culture dishes with crystal violet and evaluated the number of colonies; as shown in , the colonies colonigenic capability of miR-450b-3p stable cells was significantly decreased. The status of PI3K/AKT pathway was also analyzed by western blot, and we found that both phosphorylated AKT and phosphorylated MAPK were obviously repressed. Furthermore, we repeated the experiment using MDA-MB-361 (without HER3 expression) and MDA-MB-453 (with HER3 expression), and found that miR-450b-3p stable transfected MDA-MB-453 proliferation activity was decreased, compared with MDA-MB-361. We also found that HER3 expression was downregulated and AKT pathway was repressed in MDA-MB-453 stable cells (Fig. S1).
Figure 3. The effects of miR-450b-3p on the proliferation of cancer cells. (A) miR-450b-3p expression of the SKBR3 cells stably transfected with pSUPER-miR-450b-3p was evaluated by real-time PCR. (B) The HER3/AKT associated pathway was assessed by western blot in stable transfected cells and control cells. (C) Clonogenic assay was used to evaluate the proliferative activity of the cells. Two hundred indicated cells were seeded in medium including 20% FBS and grew for 15 d, and then stained by crystal violet and visualized. The photographs are representative of three independent experiments; bars, SD. * and #, significantly different compared with untransfected control (P < 0.05). (D) Subcutaneous xenografts formation was used to evaluate the tumorigenesis potential of SKBR3 cells transfected with indicated plasmid. The results are representative typical photographs of SKBR3 xenograft models. (E) Mice bearing SKBR3 xenografts were measured 28 d after inoculation once every 2 d. After 14 d, the mice were sacrificed and the tumors were removed and analyzed. * and #, significantly different compared with untransfected control (P < 0.05)
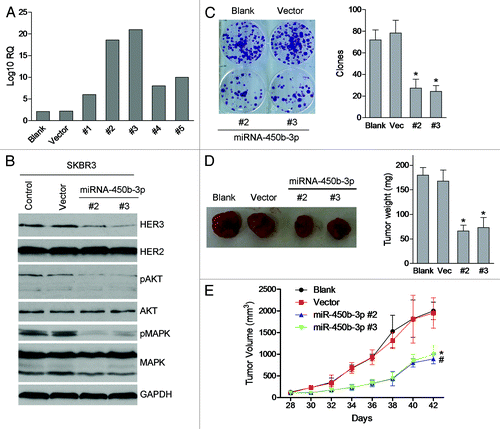
Then we established the tumor xenografts model to study the growth of miR-450b-3p expressing cells in vivo. As is shown in , the growth speed of miR-450b-3p stable SKBR3 cells was slower than the control groups. The tumor size of miR-450b-3p overexpressing stable cells was smaller than the untransfected group and the vector group. Furthermore we tested the HER3 expression and the AKT pathway in the xenografts and found the similar results that HER3 expression and AKT pathway were repressed in miR-450b-3p stable SKBR3 cells (Fig. S2).
Overall, these data suggested that overexpression of miR-450b-3p inhibits SKBR3 cell growth in vitro and in vivo through inhibiting HER3 expression and subsequently suppressing HER3/PI3K/AKT pathway.
miR-450b-3p promotes responsiveness to doxorubicin and trastusumab
HER3 forms heterodimer with HER2 and has been implicated as a key coreceptor to drive HER2-amplified breast cancer,Citation19 inhibition of HER3 by siRNA or inhibition of ligand-induced HER2/HER3 heterodimerization by pertuzumab contributed to repress cell proliferation. We suggested that inhibition of HER3 may have synergistic actions with HER2 signal repression to induce cell death. Our results showed that miR-450b-3p overexpressing cell lines are more sensitive to trastusumab compared with control cells (). These data indicated the blockage of HER2/HER3 signaling dually may augment therapeutic benefit by blocking HER2 with trastusumab.
Figure 4. miR-450b-3p overexpression sensitizes the responsiveness to trastusumab and doxorubicin. Different clones of SKBR3 cells with stably transfected miR-450b-3p or control cells were treated with indicated concentration of trastusumab for 24 h (A) or with 0.5 μM of doxorubicin for 24 h (B) respectively. And then the cells were harvested and evaluated for the percentage of apoptosis by Annexin/propidium iodied assay.
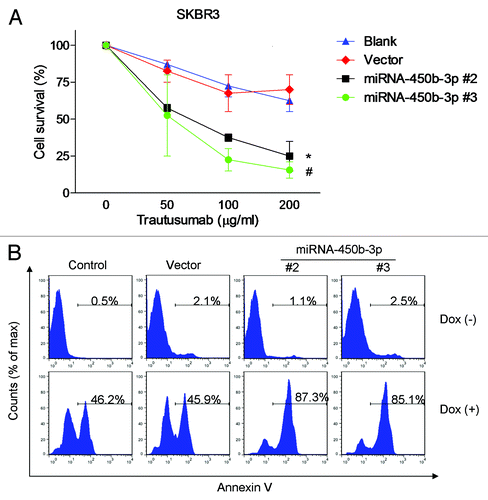
It was documented that inhibition of doxorubicin-induced HER3-PI3K-AKT signaling enhances apoptosis of ovarian cancer cells.Citation13 We hypothesized that blockage of HER3-PI3K-AKT pathway may synergistically promote cell apoptosis induced by doxorubicin. As is shown in , the control and stable cells were treated with 0.5 μM doxorubicin for 24 h, and the apoptosis percentage of control cells and vector transfected cells were 46.2% and 45.9% respectively; meanwhile the cell death of the two miR-450b-3p overexpressing cell lines are 87.3% and 85.1% respectively. The results showed that the miR-450b-3p stable cell lines were more sensitive to doxorubicin, compared with control cells.
The clinical significance of miR-450b-3p and HER3 expression
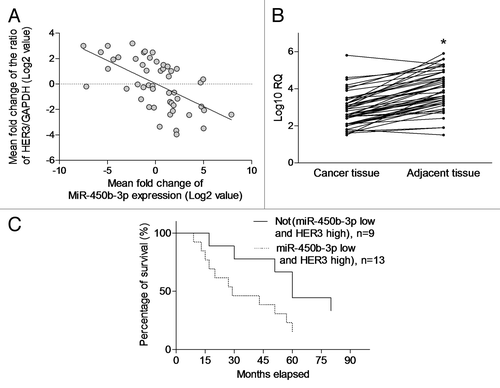
To evaluate the clinical relevance of our experimental observation, we examined the expression of miR-450b-3p and HER3 in the tumor and paired adjacent non-tumorous tissues from 50 breast cancer patients. A significant inverse correlation (P < 0.001, r = −0.3613, R2 = 0.3479) was observed between the expression of miR-450b-3p and HER3 in these breast cancer samples, suggesting that HER3 is a clinically relevant miR-450b-3p target in breast cancer (). Furthermore we found that miR-450b-3p expression was higher in paired adjacent non-cancerous tissues compared with tumor tissues (). Although neither miR-450b-3p downregulation nor HER3 upregulation alone showed significant association with patients’ overall survival (data not shown), breast cancer patients with tumors which exhibits upregulated HER3 (>2-fold) and downregulated miR-450b-3p expression (>2-fold) compared with the paired adjacent non-tumorous tissues were found to be significantly associated with poorer overall survival (P < 0.05) (). Collectively, these data suggested that downregulation of miR-450b-3p expression with concurrent overexpression of HER3 may serve as a prognostic biomarker predicting the patients’ overall survival.
Figure 5. The clinical significance of miR-450b-3p and HER3 expression. (A) The miR-450b-3p and HER3 expression in the tumor from 50 breast cancer patients was detected using q-PCR. HER3 expression was normalized to GAPDH. miR-450b-3p expression was normalized to 18S RNA. Data were transformed as log2 of relative quantity (RQ). (B) The miR-450b-3p in the tumor and paired adjacent non-tumorous tissues from 50 breast cancer patients was detected using q-PCR. The results were normalized to 18S RNA. Data were transformed as log10 of relative quantity. *, significantly different compared with adjacent non-tumorous tissues. (C) Kaplan–Meier curve was used to evaluate survival rate difference in the patients with or without more than 2-fold upregulated HER3 and 2-fold downregulated miR-450b-3p expression.
Discussion
Recently the modulation of HER family members by miRNAs becomes a field of great interest. It was reported that enforced expression of miR-125a or miR-125b coordinately suppressed HER2 and HER3 mediated signaling transduction in breast cancer cells.Citation15 miR-331-3p regulated HER2 expression and androgen receptor signaling in prostate cancer.Citation20 Many studies have focused on HER2 modulation, however, the modulation of HER3 by miRNA remains largely unknown. To our knowledge, only miR-205 has been reported to be able to destabilize the mRNA of HER3 and repress the HER3 mediated PI3K/AKT pathway in breast cancer cells.Citation21 Recently miRNA106b has been reported to inhibit HER3 expression and synergize with TKI to SKBR3 cells.Citation16 Cancers with driving HER3 amplifications or mutations have not been found, and studies of its expression in tumors have been only weakly provocative. We suspected that the overexpression of HER3 in breast cancer cells was dominantly controlled on transcriptional or posttranscriptional level by other molecules, especially by miRNA. We combined the database search and manual check and found that miR-450b-3p is one of the potential candidates.
It’s well known that the miRNAs generally bind to the coding region or 3′ UTR of their target mRNAs and repress protein production by destabilizing the mRNA and translational silencing. The putative function of miR-450b-3p has not been fully understood. It was reported that miR-450b-5p was downregulated in the liver of mouse primary infected with Plasmodium chabaudi malaria.Citation22 Recently higher expression of miR-450b-5p has been reported to be associated with better response to neoadjuvant chemoradiotherapy in the locally advanced rectal cancer patients; however, the mechanisms remained unknown.Citation23 Our results provided the new clues that miR-450b-3p interacts with HER3 mRNA at 3′ UTR to repress HER3 expression and to modulate HER3 mediated PI3K/AKT pathway. To our knowledge, this is the first article to discuss the regulation of HER3 by miR-450b-3p in breast cancers.
To date, every type of breast tumor analyzed by miRNA profiling has shown significantly different miRNA profiles (for mature and/or precursor miRNAs) compared with normal cells from the same tissue.Citation24 The published large-scale miRNA profiles revealed that several miRNAs are consistently deregulated in tumors from breast cancer patients.Citation25 Furthermore the key pathway involved with the individual miRNA was indentified in breast cancer. In fact a global decrease in miRNA expression has been observed in human cancers, suggesting that most miRNAs may act as tumor suppressors to downregulate the survival/proliferation pathways.Citation26 Our result provided a new example for this assumption. We found that the profiles of miR-450b-3p are varied in different cancer cells and are strongly and reversely association of the HER3 expression.
It was reported that PI3K inhibitors can upregulate HER3 expression and activity, as a feedback, to attenuate antitumor effect of PI3K inhibitors in HER2-overepressing cells.Citation14,Citation27 We suggested that the HER2 antagonists may induce the HER3 expression and activity, following the PI3K/AKT pathway suppression, to attenuate the response to these agents. Inhibition of HER3 expression or associated pathway may synergize with HER2 antagonists to repress the PI3K/AKT pathway and the feedback loop. Our results proved that miR-450b-3p enhanced the responsiveness of the breast cancer cells to trastuzumab and indicated that combination with HER2/HER3 antagonists should have better therapeutic effects.
Resistance to chemotherapy is a serious problem for the successful treatment during many cancers treatment such as ovarian cancer and breast cancer. The widely used chemotherapeutic drug doxorubicin has dual functions of inducing apoptosis signals such as p53 pathway, as well as survival signals such as PI3K/AKT pathway. It was demonstrated that activation of the HER3/PI3K/AKT signaling cascade induced by doxorubicin plays critical roles in chemotherapy resistance.Citation13 Our results showed that the blockage of HER3-PI3K-AKT pathway with the miR-450b-3p overexpression enhanced the response to doxorubicin. Giving the complication of function of doxorubicin, whether other pathways are involved in cannot be excluded. But at least in the cells with HER3 high-expression, the inhibitory of HER3-PI3K-AKT pathway should be an effective method to improve the efficacy of doxorubicin.
Up to now, the results are controversial about the prognostic impact of HER3 overexpression in cancers. The predictive significance of HER3 expression depends on the HER3 cellular localization and depends on the cancer type where the HER2 is overexpressed or not. For example, a metaanalysis showed that expression of HER3 is associated with worse survival in solid tumors. The influence of HER3 may be greater in those tumors where HER2 is commonly overexpressed.Citation28 Membranous HER3 expression is strongly associated with poor prognosis in head and neck squamous cell carcinoma and nucleus HER3 predicts a more favorable overall survival in uveal melanoma.Citation29,Citation30 Co-overexpression of HER3 and HER2 is associated with significantly decreased survival in breast cancer, but individual overexpression is not.Citation31,Citation32 Our finding consists with the others’ results that HER3 overexpression alone is not associated with overall survival in breast cancer patients. Therefore, we further found that downregulation of miR-450b-3p expression combined with overexpression of HER3 may have prognostic significance in breast cancer. The higher expression of miR-450b-3p in adjacent non-cancerous tissues indicated that the status of miR-450b-3p may contribute to tumor occurrence or progressive.
In summary, we identify miR-450b-3p as a new oncosuppressor molecule in breast cancer, which is able to interfere with the proliferative pathway mediated by the HER receptor. Our study also provides experimental evidence that miR-450b-3p can improve the responsiveness to trastuzumab and chemotherapeutic drug treatment, indicating the possibility of using this microRNA as a new biomarker for response to specific therapies and as a tool of an innovative and promising therapy.
Materials and Methods
Chemicals
Dulbecco’s modified Eagle medium (D-MEM), penicillin, streptomycin, and trypsin-EDTA were obtained from GIBCO. Fetal bovine serum was obtained from Sijiqing Beijin. Lipofectamine 2000 was purchased from Invitrogen. Trastuzumab was purchased from Roche. The remaining reagents were obtained from Sigma, unless otherwise specified.
Cell lines and patient samples
The normal breast tissue was collected at. HeLa, MCF7, MCF10A, MDA-MB-361, BT-474, and MDA-MB-453 cells were cultured in D-MEM supplemented with 20% FBS and 2 mmol/L glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin at 37 °C in 5% CO2. SKBR3 cells were cultured in McCoy’s 5A medium supplemented with 20% FBS and 2 mmol/L glutamine, 100 U/mL penicillin, and 100 mg/mL streptomycin at 37 °C in 5% CO2. Fresh breast cancer tissue samples, together with their paired adjacent non-cancerous tissues from each patient, were collected from breast cancer curative resection surgery, snap frozen, and stored in liquid nitrogen until use for experimental purposes. For the use of these clinical materials for research purposes, prior patients’ written informed consents and approval from the Institutional Research Ethics Committee of the Affiliated Jiangyin Hospital of Southeast University Medical College were obtained.
Plasmid construction and transfection
To generate a plasmid expressing miR-450b-3p, we annealed the following long primers, 5′-GATCCCCGGA UGCAAAAUGA UCCCAATTCA AGAGATTGGG AUCATTTTGC AUCCTTTTTA-3′ and 5′-AGCTTAAAAA GGAUGCAAAA UGAUCCCAAT CTCTTGAATT GGGAUCATTT TGCAUCCGGG-3′ and then inserted into pSUPER-retro-puro vector at the BamHI and HindIII sites. The pGL3-HER3 3′ UTR reporter plasmid was constructed by inserting the full 1451-bp segment of the HER3 3′ UTR into a pGL3-Promoter plasmid at XbaI site. The following primers were used to amplify the HER3 3′ UTR: sense, 5′-AAATCTAGAC TCCTGCTCCCT GTGGCACTCA GG-3′ and antisense, 5′-GGGTCTAGAT GAAAAAAAAC TTTGGCATTT TTAT-3′. The sense and antisense orientations of the HER3 3′ UTR in the luciferase reporter were identified by both enzyme digestion and DNA sequencing. A pGL3-HER3 3′ UTR plasmid containing mutated miR-450b-3p binding site within the 3′ UTR was generated using Stratagene’s QuikChange II Site Directed Mutagenesis Kit according to the manufacturer’s instructions. The primers for mutant were sense, 5′-ATTGGGAGGAT TAACGTACCT GGATCTACT-3′ and antisense, 5′-AGTAGATCCA GGTACGTTAA TCCTCCCAAT-3′.
Transient transfection was performed with Lipofectamine 2000 according to the manufacturer’s instructions. SKBR3 or MDA-MB-453 cells were seeded at a density of 5 × 105 cells per well of a 6-well plate. After allowing cells to grow overnight, the cells were transfected with noted plasmids. Twenty-eight hours after transfection, cells were harvested for western blot analysis. For luciferase assay, the time before harvest was 20 h.
To establish the miR-450b-3p stable expression cell line, pSUPER-retro-puro- miR-450b-3p plasmid was transfected to SKBR3 cells according to the manuals and the empty vector was used as control. Forty-eight hours later, the transfected cells were cultured in the medium with 2 μg/mL puromycin to establish the stable cell line. Several puromycin resistant colonies were picked up and expanded for conformation by analyzing the miR-450b-3p expression.
Flow cytometry assays
The flow cytometry assay was used to quantify cell apoptosis. Briefly, cells with or without treatment were washed twice with phosphate-buffered saline and then stained with fluorescein isothiocyanate-Annexin-V and propidium iodide for 1 h, according to the manufacturer’s instructions. Stained cells were detected by using the FACScan (Becton Dickinson) and analyzed by using the FlowJo software (Tree star Inc.).
RNA extraction and real-time PCR
Total RNA was extracted from cell lines and human samples with Trizol (Invitrogen) according to the manufacturer’s instructions. The concentration of RNA was quantified using NanoDrop Specthophotometer (NanoDrop Technologies). TaqMan MicroRNA Reverse Transcription kit and TaqMan MicroRNA Assay were used to detect and quantify mature miR-450b-3p in accordance with manufacturer’s instructions (Applied Biosystems). Normalization was performed with18S rRNA.
Western blot
Cells from different experimental conditions were collected and pelleted. Western blot analysis was performed as described.Citation33 The blots were incubated with noted antibody. All the antibodies were purchased from Santa Cruz.
Clonogenic assay of cells in vitro
Clonogenic assay was used to assess the proliferation of cancer cells according the reference.Citation34 Briefly, exponentially growing cells were harvested and 200 cells were re-plated in a 6-well plate, allowing them to grow for 14 d to form colonies, which were then stained with crystal violet (0.5 g/L) and visualized under the microscope.
Xenograft tumor studies
The Institutional Animal Care and Use Committee of the Affiliated Jiangyin Hospital of Southeast University Medical College approved the protocol for animal experiments. All mice were handled in strict accordance with good animal practice as defined by the relevant national and/or local animal welfare bodies. Five mice each group were used for study. Six-week-old Nu/Nu nude mice were housed under pathogen-free conditions in microisolator cages. Xenografts were raised by injecting 5 × 106 of SKBR3 cells stably transfected with indicated plasmid in a balanced salt solution into subcutaneous tissue over the flank region of nude mice. Tumor volume was assessed by caliper measurements once every two days from 28th day and calculated with the formula: V = (L × W2) / 2 (L, length; W, width) as previously described.Citation35 Forty-two days after inoculation, mice were sacrificed by inhaled CO2. Harvested tumors were used for further analysis.
Statistical analysis
Significant differences between two groups were analyzed using the two-sided unpaired Student t test and P value < 0.05 was considered statistically significant. Statistical analysis was performed with Graphpad Prism 5 software.
Additional material
Download Zip (373.8 KB)Disclosure of Potential Conflicts of Interest
The authors confirm that there are no conflicts of interest.
Acknowledgments
This study was supported by National Natural Science Foundation of China (81060048).
Author Contributions
Z.Z. and T.L. conceived and designed the experiments. Z.Z., S.S., and W.M. performed the experiments. R.L. and Q.W. contributed reagents/materials/analysis tools. Z.Z., R.L., and T.L. analyzed the data. Z.Z., R.L., and T.L. wrote the paper.
Reference
- Ullrich A, Coussens L, Hayflick JS, Dull TJ, Gray A, Tam AW, Lee J, Yarden Y, Libermann TA, Schlessinger J, et al. Human epidermal growth factor receptor cDNA sequence and aberrant expression of the amplified gene in A431 epidermoid carcinoma cells. Nature 1984; 309:418 - 25; http://dx.doi.org/10.1038/309418a0; PMID: 6328312
- Ullrich A, Schlessinger J. Signal transduction by receptors with tyrosine kinase activity. Cell 1990; 61:203 - 12; http://dx.doi.org/10.1016/0092-8674(90)90801-K; PMID: 2158859
- Shiraishi M, Noguchi M, Shimosato Y, Sekiya T. Amplification of protooncogenes in surgical specimens of human lung carcinomas. Cancer Res 1989; 49:6474 - 9; PMID: 2573414
- Gerosa MA, Talarico D, Fognani C, Raimondi E, Colombatti M, Tridente G, De Carli L, Della Valle G. Overexpression of N-ras oncogene and epidermal growth factor receptor gene in human glioblastomas. J Natl Cancer Inst 1989; 81:63 - 7; http://dx.doi.org/10.1093/jnci/81.1.63; PMID: 2908920
- Cowley GP, Smith JA, Gusterson BA. Increased EGF receptors on human squamous carcinoma cell lines. Br J Cancer 1986; 53:223 - 9; http://dx.doi.org/10.1038/bjc.1986.39; PMID: 2420349
- Rao VH, Kandel A, Lynch D, Pena Z, Marwaha N, Deng C, Watson P, Hansen LA. A positive feedback loop between HER2 and ADAM12 in human head and neck cancer cells increases migration and invasion. Oncogene 2012; 31:2888 - 98; http://dx.doi.org/10.1038/onc.2011.460; PMID: 21986939
- Berger MB, Mendrola JM, Lemmon MA. ErbB3/HER3 does not homodimerize upon neuregulin binding at the cell surface. FEBS Lett 2004; 569:332 - 6; http://dx.doi.org/10.1016/j.febslet.2004.06.014; PMID: 15225657
- Vaught DB, Stanford JC, Young C, Hicks DJ, Wheeler F, Rinehart C, Sánchez V, Koland J, Muller WJ, Arteaga CL, et al. HER3 is required for HER2-induced preneoplastic changes to the breast epithelium and tumor formation. Cancer Res 2012; 72:2672 - 82; http://dx.doi.org/10.1158/0008-5472.CAN-11-3594; PMID: 22461506
- Alimandi M, Romano A, Curia MC, Muraro R, Fedi P, Aaronson SA, Di Fiore PP, Kraus MH. Cooperative signaling of ErbB3 and ErbB2 in neoplastic transformation and human mammary carcinomas. Oncogene 1995; 10:1813 - 21; PMID: 7538656
- Soltoff SP, Carraway KL 3rd, Prigent SA, Gullick WG, Cantley LC. ErbB3 is involved in activation of phosphatidylinositol 3-kinase by epidermal growth factor. Mol Cell Biol 1994; 14:3550 - 8; PMID: 7515147
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007; 316:1039 - 43; http://dx.doi.org/10.1126/science.1141478; PMID: 17463250
- Prigent SA, Gullick WJ. Identification of c-erbB-3 binding sites for phosphatidylinositol 3′-kinase and SHC using an EGF receptor/c-erbB-3 chimera. EMBO J 1994; 13:2831 - 41; PMID: 8026468
- Bezler M, Hengstler JG, Ullrich A. Inhibition of doxorubicin-induced HER3-PI3K-AKT signalling enhances apoptosis of ovarian cancer cells. Mol Oncol 2012; 6:516 - 29; http://dx.doi.org/10.1016/j.molonc.2012.07.001; PMID: 22841590
- Chakrabarty A, Sánchez V, Kuba MG, Rinehart C, Arteaga CL. Feedback upregulation of HER3 (ErbB3) expression and activity attenuates antitumor effect of PI3K inhibitors. Proc Natl Acad Sci U S A 2012; 109:2718 - 23; http://dx.doi.org/10.1073/pnas.1018001108; PMID: 21368164
- Scott GK, Goga A, Bhaumik D, Berger CE, Sullivan CS, Benz CC. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem 2007; 282:1479 - 86; http://dx.doi.org/10.1074/jbc.M609383200; PMID: 17110380
- Amin DN, Sergina N, Lim L, Goga A, Moasser MM. HER3 signalling is regulated through a multitude of redundant mechanisms in HER2-driven tumour cells. Biochem J 2012; 447:417 - 25; http://dx.doi.org/10.1042/BJ20120724; PMID: 22853430
- Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature 2008; 455:64 - 71; http://dx.doi.org/10.1038/nature07242; PMID: 18668037
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004; 5:522 - 31; http://dx.doi.org/10.1038/nrg1379; PMID: 15211354
- Lee-Hoeflich ST, Crocker L, Yao E, Pham T, Munroe X, Hoeflich KP, Sliwkowski MX, Stern HM. A central role for HER3 in HER2-amplified breast cancer: implications for targeted therapy. Cancer Res 2008; 68:5878 - 87; http://dx.doi.org/10.1158/0008-5472.CAN-08-0380; PMID: 18632642
- Epis MR, Giles KM, Barker A, Kendrick TS, Leedman PJ. miR-331-3p regulates ERBB-2 expression and androgen receptor signaling in prostate cancer. J Biol Chem 2009; 284:24696 - 704; http://dx.doi.org/10.1074/jbc.M109.030098; PMID: 19584056
- Iorio MV, Casalini P, Piovan C, Di Leva G, Merlo A, Triulzi T, Ménard S, Croce CM, Tagliabue E. microRNA-205 regulates HER3 in human breast cancer. Cancer Res 2009; 69:2195 - 200; http://dx.doi.org/10.1158/0008-5472.CAN-08-2920; PMID: 19276373
- Delić D, Dkhil M, Al-Quraishy S, Wunderlich F. Hepatic miRNA expression reprogrammed by Plasmodium chabaudi malaria. Parasitol Res 2011; 108:1111 - 21; http://dx.doi.org/10.1007/s00436-010-2152-z; PMID: 21085987
- Svoboda M, Sana J, Fabian P, Kocakova I, Gombosova J, Nekvindova J, Radova L, Vyzula R, Slaby O. MicroRNA expression profile associated with response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer patients. Radiat Oncol 2012; 7:195; http://dx.doi.org/10.1186/1748-717X-7-195; PMID: 23167930
- Volinia S, Galasso M, Sana ME, Wise TF, Palatini J, Huebner K, Croce CM. Breast cancer signatures for invasiveness and prognosis defined by deep sequencing of microRNA. Proc Natl Acad Sci U S A 2012; 109:3024 - 9; http://dx.doi.org/10.1073/pnas.1200010109; PMID: 22315424
- Ferracin M, Querzoli P, Calin GA, Negrini M. MicroRNAs: toward the clinic for breast cancer patients. Semin Oncol 2011; 38:764 - 75; http://dx.doi.org/10.1053/j.seminoncol.2011.08.005; PMID: 22082762
- Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, Ambros VR, Israel MA. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res 2007; 67:2456 - 68; http://dx.doi.org/10.1158/0008-5472.CAN-06-2698; PMID: 17363563
- Garrett JT, Olivares MG, Rinehart C, Granja-Ingram ND, Sánchez V, Chakrabarty A, Dave B, Cook RS, Pao W, McKinely E, et al. Transcriptional and posttranslational up-regulation of HER3 (ErbB3) compensates for inhibition of the HER2 tyrosine kinase. Proc Natl Acad Sci U S A 2011; 108:5021 - 6; http://dx.doi.org/10.1073/pnas.1016140108; PMID: 21385943
- Ocana A, Vera-Badillo F, Seruga B, Templeton A, Pandiella A, Amir E. HER3 overexpression and survival in solid tumors: a meta-analysis. J Natl Cancer Inst 2013; 105:266 - 73; http://dx.doi.org/10.1093/jnci/djs501; PMID: 23221996
- Takikita M, Xie R, Chung JY, Cho H, Ylaya K, Hong SM, Moskaluk CA, Hewitt SM. Membranous expression of Her3 is associated with a decreased survival in head and neck squamous cell carcinoma. J Transl Med 2011; 9:126; http://dx.doi.org/10.1186/1479-5876-9-126; PMID: 21801427
- Trocmé E, Mougiakakos D, Johansson CC, All-Eriksson C, Economou MA, Larsson O, Seregard S, Kiessling R, Lin Y. Nuclear HER3 is associated with favorable overall survival in uveal melanoma. Int J Cancer 2012; 130:1120 - 7; http://dx.doi.org/10.1002/ijc.26118; PMID: 21484789
- Abd El-Rehim DM, Pinder SE, Paish CE, Bell JA, Rampaul RS, Blamey RW, Robertson JF, Nicholson RI, Ellis IO. Expression and co-expression of the members of the epidermal growth factor receptor (EGFR) family in invasive breast carcinoma. Br J Cancer 2004; 91:1532 - 42; http://dx.doi.org/10.1038/sj.bjc.6602184; PMID: 15480434
- Suo Z, Risberg B, Kalsson MG, Willman K, Tierens A, Skovlund E, Nesland JM. EGFR family expression in breast carcinomas. c-erbB-2 and c-erbB-4 receptors have different effects on survival. J Pathol 2002; 196:17 - 25; http://dx.doi.org/10.1002/path.1003; PMID: 11748637
- Renart J, Reiser J, Stark GR. Transfer of proteins from gels to diazobenzyloxymethyl-paper and detection with antisera: a method for studying antibody specificity and antigen structure. Proc Natl Acad Sci U S A 1979; 76:3116 - 20; http://dx.doi.org/10.1073/pnas.76.7.3116; PMID: 91164
- Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc 2006; 1:2315 - 9; http://dx.doi.org/10.1038/nprot.2006.339; PMID: 17406473
- Liu AW, Cai J, Zhao XL, Jiang TH, He TF, Fu HQ, Zhu MH, Zhang SH. ShRNA-targeted MAP4K4 inhibits hepatocellular carcinoma growth. Clin Cancer Res 2011; 17:710 - 20; http://dx.doi.org/10.1158/1078-0432.CCR-10-0331; PMID: 21196414
