Abstract
Multiple myeloma (MM) is a malignant plasma cells proliferative disease. The intricate cross-talk of myeloma cells with bone marrow microenvironment plays an important role in facilitating growth and survival of myeloma cells. Bone marrow mesenchymal stem cells (BMMSCs) are important cells in MM microenvironment. In solid tumors, BMMSCs can be educated by tumor cells to become cancer-associated fibroblasts (CAFs) with high expression of fibroblast activation protein (FAP). FAP was reported to be involved in drug resistance, tumorigenesis, neoplastic progression, angiogenesis, invasion, and metastasis of tumor cells. However, the expression and the role of FAP in MM bone marrow microenvironment are still less known. The present study is aimed to investigate the expression of FAP, the role of FAP, and its relevant signaling pathway in regulating apoptosis induced by bortezomib in MM cells. In this study, our data illustrated that the expression levels of FAP were not different between the cultured BMMSCs isolated from MM patients and normal donors. The expression levels of FAP can be increased by tumor cells conditioned medium (TCCM) stimulation or coculture with RPMI8226 cells. FAP has important role in BMMSCs mediated protecting MM cell lines from apoptosis induced by bortezomib. Further study showed that this process may likely through β-catenin signaling pathway in vitro. The activation of β-catenin in MM cell lines was dependent on direct contact with BMMSCs other than separated by transwell or additional condition medium from BMMSCs and cytokines.
Introduction
Multiple myeloma (MM), characterized by increased blood calcium level, renal insufficiency, anemia, and bone lesions (CRAB), is a malignant plasma cell proliferative disorder with about 22 000 new cases annually and about 20% of deaths from all hematological malignancies in the US.Citation1 With the advent of new drugs, such as bortezomib, lenalidomide, pomalidomide, perifosine, etc., 5-y survival rate of MM increased to more than 40%.Citation1 However, MM is still regarded as an incurable disease in spite of new treatment regimens.Citation2 Interaction of MM cells and cells in the bone marrow (such as mesenchymal stem cells, osteoblasts, osteoclasts, endothelial cells, immunocytes, and adipocytes) constitutes a complex tumor microenvironment that support proliferation, growth, chemoresistance, invasion, and metastasis of MM cells.Citation3 Bone marrow mesenchymal stem cells (BMMSCs) are important cells in MM microenvironment. It is now becoming apparent that BMMSCs mediate chemoresistance, angiogenesis, and progression of MM cells though either direct contact or secreted cytokines, chemokines and exosomes.Citation4-Citation6 In solid tumors, previous studies have realized that tumor microenvironment has become a coconspirator in process of carcinogenesis.Citation7 Tumor-stroma interaction modifies malignant cells and “nonmalignant” cells leading them to reciprocal activation and coevolution and turned tumor microenvironment into a fertile soil to support the growth and chemoresistance of tumor cells.Citation8,Citation9 Cancer-associated fibroblasts (CAFs) have been considered as important component in tumor microenvironment and play vital roles in tumor behaviors.Citation8,Citation10,Citation11 However, the precise origin of CAFs is still less known.Citation11 As for the current knowledge, BMMSCs was recognized as one of its origins.Citation12-Citation14 Studies have shown that tumor cells recruit BMMSCs through secreted cytokines or chemokines and educate BMMSCs to become CAFs to support the growth, proliferation, angiogenesis, invasion, and metastasis of malignant cells.Citation12-Citation14 Current knowledge has revealed that CAFs express surface markers such as fibroblast activation protein (FAP), fibroblast surface protein (FSP), α-smooth muscle actin (α-SMA), platelet derived growth factor receptor-α/β (PDGFRα/β), tenascin-C (TN-C), vimentin, and fibronectin.Citation11,Citation14,Citation15 FAP, a member of serine protease family possessing of gelatinase activity and type I collagenase activity, is a vital type II trans membrane protein which is expressed in more than 90% epithelial tumor stromal cells but not normal cells.Citation16 Researches have shown that FAP plays an important roles in tumor proliferation,Citation17 angiogenesis,Citation18,Citation19 immune escape,Citation20 invasion and metastasis,Citation21,Citation22 and tissue remolding.Citation23,Citation24 Considering its important roles in tumor progression and rare expression in normal tissue, FAP is becoming an important therapy target in tumors.Citation25 It is still less known whether or not BMMSCs from MM patients or normal donors had a different expression of FAP and whether or not FAP had important role in mediating MM cells behavior. In this study, we mainly focus on the expression of FAP on BMMSCs and the role of FAP in BMMSCs mediated protection on MM cell lines.
Results and Discussion
Identification of BMMSCs
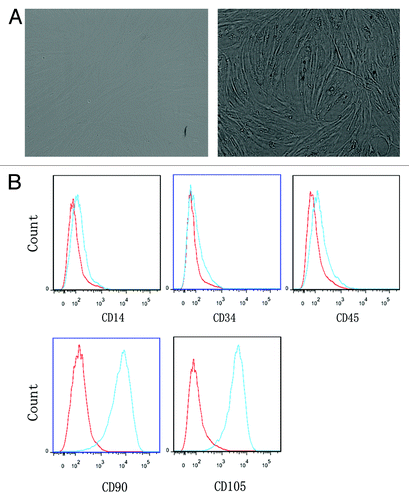
BMMSCs were first observed by the German pathologist Cohnheimin 140 y ago.Citation26 Dramatic interest in MSCs has risen in last two decades. In terms of the current knowledge, there is now a general consensus that three criteria are applied to define MSCs.Citation27-Citation30 First, cultured MSCs are fibroblast-like cells and adherence to plastic. Second, cultured MSCs express CD105 (SH2), CD73 (SH3/4), and CD90 (Thy-1), but are negative for the expression of hematopoietic markers, such as CD34, CD45, CD14 or CD11b, CD19 or CD79α, and HLA-II. Third, MSCs are capable of differentiating into osteoblasts, adipocytes, chondroblasts in vitro. MSCs in bone marrow are very rare with only 1 MSC every 10 000 nucleated cells.Citation26,Citation30 In our experiments, cultured BMMSCs were adherent to the flasks and fibroblast-like in morphology (). BMMSCs of p2 had a low expression of CD14 (1.31 ± 0.37%), CD34 (0.23 ± 0.05%), CD45 (1.20 ± 0.73%), and high expression of CD90 (94.65 ± 1.53%) and CD105 (97.82 ± 0.76%) in three independent experiments. FACS analysis of these CD molecular on BMMSCs is shown in . Taken together, these results met the minimal criteria for defining MSCs.Citation29
Expression of FAP in BMMSCs
FAP was first discovered in the late 1980s with monoclonal antibody (mAb) F19.Citation31,Citation32 In 1990, a dimeric 170-kDa membrane-bound gelatinase was identified in aggressive malignant human melanoma cell line, LOX and named “seprase” in 1994.Citation33,Citation34 Subsequently, FAP and seprase were proved to be the same transmembrane protease with cloning and sequence analysis.Citation35,Citation36 FAP was expressed in 90% tumor stroma of epithelia cancer,Citation16 malignant melanoma cells, invasive ductal carcinoma cells of breast cancer, cancer cells of colorectum, stomach, uterine cervix, and glioma cells,Citation33,Citation37-Citation41 fibroblast foci, and fibrotic interstitium of idiopathic pulmonary fibrosis, fibroblast-like synoviocytes of rheumatoid arthritis, and stellate cells of cirrhosis,Citation42-Citation44 BMMSCs, and cultured fibroblasts.Citation38,Citation45 FAP had important role in tumor progression,Citation17 invasion,Citation22 metastasis,Citation21 and immune escape.Citation20 HEK293 cells and human mammary adenocarcinoma cells MDA-MB-231 cotransfect with FAP cDNA could more easily to form subcutaneous tumors.Citation17,Citation19 Researches have shown that BMMSCs can be educated by TCCM to become CAFsCitation12,Citation46,Citation47 and to promote tumor progression.Citation13 Studies have revealed that BMMSCs from MM patients display distinct profile as compared with those from normal donors.Citation5,Citation48-Citation50 So, we presumed that MM microenvironment may also educate BMMSCs to become CAFs and have an elevated expression of FAP than normal BMMSCs.
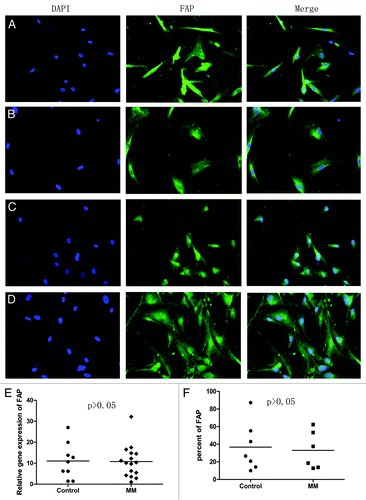
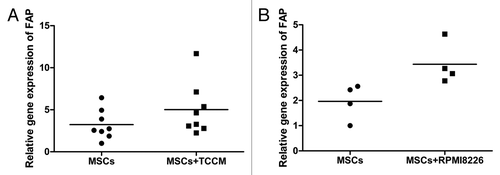
First, we checked the expression of FAP on BMMSCs between MM patients and normal donors. As shown in , there were no differences on the expression of FAP between MM patients and normal donors detected by qRT-PCR (P > 0.05), FCM (P > 0.05), and immunofluorescence. We speculated that two reasons maybe contributed to this result. First, cultured BMMSCs, like MSCs involving in healing wounds,Citation16,Citation37,Citation51 had already activated in the process of culture to express FAP. Second, BMMSCs from MM patients may lack of stimulation by MM cells and/or secreted factors because of long-term in vitro culture. So, next, we added TCCM from myeloma cells or cocultre with RPMI8226 cells to stimulate BMMSCs for 7 d and detected the expression of FAP by qRT-PCR. As shown in , expression of FAP in BMMSCs had a moderate increase in TCCM stimulated group (n = 8) and cocultured group (n = 4) than non-stimulated group and non-cocultured group (P = 0.0256 and P = 0.035, respectively).
FAP protect RPMI8226 and CAG cells from apoptosis induced by bortezomib
It is well known that the crosstalk of MM cells with BMMSCs through direct contact or secreted cytokines can facilitate MM progression,Citation4,Citation52 survival and growth,Citation53,Citation54 chemotherapy resistance,Citation55-Citation57 and angiogenesis.Citation58,Citation59 But the function of FAP, expressed on activated BMMSCs, in MM cells behavior is still less known and need to be further discussed. As shown in , BMMSCs were able to protect bortezomib (20 nM) induced RPMI8226 (n = 3) and CAG (n = 2) cells apoptosis in cocultured system detected by FCM (). This result was consistent with previous study.Citation53 Knockdown of FAP with siRNA weakened this protective effect. The apoptotic cells in FAP-siRNA group increased from about 6% to 16% (FAP-siRNA vs NC-siRNA, 50.94 ± 13.13% vs 42.76 ± 12.34%, P = 0.0014, n = 5) (). A representative apoptosis analysis of CAG and a representative knockdown effect analysis of FAP on BMMSCs detected by FCM were shown in respectively. Next, we detected the apoptosis related protein with western blot in RPMI8226 and CAG cells in coculture system with either FAP knockdown or not in the presence of bortezomib. Our result demonstrated that in cocultured system knockdown of FAP in BMMSCs with siRNA activated caspase 3 and PARP-1 of RPMI8226 and CAG cells. Pro-apoptotic protein Bad and Bak () had a moderate elevate and anti-apoptotic protein survivin () had a slight repression. These results further supported the result of FCM.
Figure 4. The protective effect of FAP on MM cell lines. (A) A representative apoptosis analysis of CAG in coculture system with FAP knockdown or not in the presence of 20 nM bortezomib detected by FCM. Cells of CD138-positive and annexin V-positive were considered as apoptotic cells. (B) FAP could protect RPMI8226 (n = 3) and CAG (n = 2) cells from bortezomib induced apoptosis (FAP-siRNA vs NC siRNA, P = 0.0014, n = 5). (C) A representative knockdown effect of FAP detected by FCM.
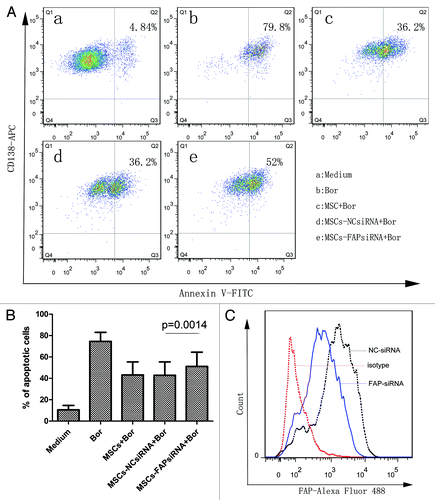
Figure 5. The probable role of FAP in BMMSCs mediated protection of RPMI8226 and CAG cells. BMMSCs could protect MM cell lines from apoptosis induced by 20 nM bortezomib through reducing the activation of caspase 3 and PARP-1 and the expression of pro-apoptotic proteins, such as Bad and Bak. Knockdown FAP could weaken this protection effect.
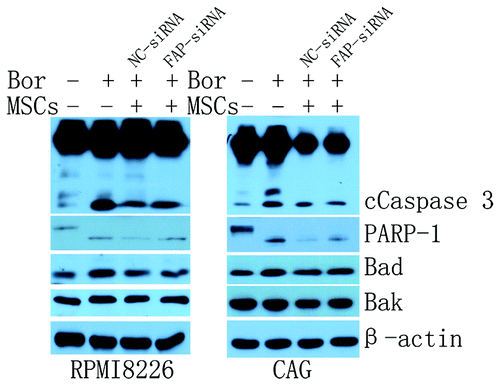
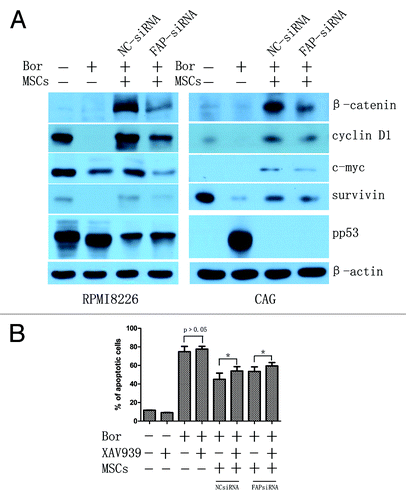
Figure 6. The probable role of FAP in BMMSCs mediated protection of MM cell lines. (A) β-catenin of MM cell lines was significantly activated when cocultured with BMMSCs. Knockdown of FAP in BMMSCs could decrease the expression of β-catenin and its downstream proteins such as c-myc, cyclin D1, and survivin and increase the expression of pp53 in RPMI8226 and CAG cells. (B) RPMI8226 cells pretreated with 40 μM XAV939 for 24 h, and then cocultured with BMMSCs (knockdown FAP or not). As a result, XAV939 could enhance bortezomib induced RPMI8226 cells apoptosis. (P = 0.017 and 0.031 in BMMSCs-NCsiRNA and BMMSCs-FAPsiRNA groups, respectively).
The probable mechanism for FAP to protect MM cells
Researches have demonstrated that BMMSCs secreted cytokines (IL-6, TNF-α, IGF-I, VEGF, CD44, ICAM-1, etc.) or adhesion of MM cell lines with BMMSCs could activate multitude of signaling pathways such as PI3K/Akt/mTOR, NFκB, JAK-STAT, MEK/ERK, and Notch pathways to support survival, proliferation, and migration as well as angiogenesis of MM cells.Citation60,Citation61 However, BMMSCs-mediated Wnt/β-catenin signaling pathway activation is still less known. Our group demonstrated that Wnt/β-catenin signaling pathway is involved in the protection of BMMSCs in acute lymphoblastic leukemia (ALL) cells in previous study.Citation62 Reports have shown that the activation of canonical Wnt/β-catenin signaling pathway in MM is ubiquitous.Citation63,Citation64 Wnt/β-catenin signaling pathway has an important role in MM proliferation,Citation63 growth,Citation65 chemoresistance,Citation66 MM-related bone disease,Citation65 migration, and invasion.Citation67 Upregulated β-catenin can cause the activation of cyclin D1, c-myc, and survivin and repress the activation of p53.Citation63,Citation68 A previous paper has hypothesized that bone marrow stromal cells-derived Wnt is essential for the activation of Wnt/β-catenin signaling in myeloma cells according to three observations.Citation69 The relationship of FAP and Wnt/β-catenin for MM cells behavior is still unknown. Here, we demonstrated that the expression of β-catenin in RPMI8226 and CAG cells were significantly increased when cocultured with BMMSCs for 48 h even in the presence of bortezomib. This result supported that β-catenin likely plays an important role in BMMSCs mediated chemoresistance including bortezomib and other novel chemotherapeutic drugs. Knockdown of FAP with siRNA could obviously suppress the expression of β-catenin (). Subsequently, we detected the major downstream protein of β-catenin, such as survivin, cyclin D1, and c-myc and regulatory protein of β-catenin, such as pp53 by western blot. As shown in , knockdown of FAP suppressed the downstream proteins of c-myc, survivin, and cyclin D1 and upregulate the expression of pp53. These results showed that FAP may play an important role in participating BMMSCs mediated protection of bortezomib-induced MM cell lines apoptosis and this protection effect may likely through β-catenin parthway.
To verify β-catenin signaling pathway participating BMMSCs- and FAP-mediated protection effect. We inhibited β-catenin signaling pathway with XAV939.Citation70 As shown in , 40 μM XAV939 could enhance cocultured RPMI8226 cells apoptosis. In BMMSCs group without FAP knockdown, the apoptotic cells of RPMI8226 was increased about 9% after treated with bortezomib and XAV939 compared with bortezomib single for 48 h (45.01% ± 6.65% vs 54.08% ± 4.67%, P = 0.017). In BMMSCs group with FAP knockdown, the apoptotic cells of RPMI8226 was increased about 6% after treated with bortezomib and XAV939 compared with bortezomib single for 48 h (53.52% ± 4.90% vs 59.44% ± 3.71%, P = 0.031).
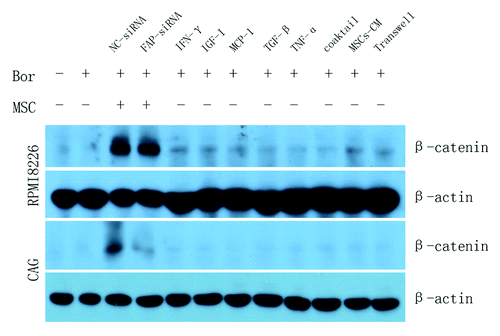
Researches have demonstrated that cytokines,Citation71 such as IGF-I,Citation72 MCP-1,Citation73 IFN-γ,Citation74,Citation75 TNF-α,Citation76 TGF-β,Citation77 etc. could active the expression of β-catenin. In order to verify whether the activation of β-catenin was dependent on direct contact with BMMSCs, we added cytokines mentioned above, conditioned medium (CM) derived from BMMSCs and cocultured RPMI8226 or CAG cells with BMMSCs using transwell chamber. As shown in , β-catenin in RPMI8226 and CAG cells were not activated when separated by transwell chamber or by cytokines and CM derived from BMMSCs. This result demonstrated that the activation of β-catenin was dependent on direct contact with BMMSCs.
Figure 7. MM cell lines treated with additional cytokines, BMMSCs-derived conditioned medium or MM-BMMSCs cocultured system separated with transwell chamber could not activate the expression of β-catenin in MM cell lines demonstrating that the activation of β-catenin dependent on direct contact. Coaktail means the mixture of cytokines.
Taken together, our data indicated that there is no difference on the expression of FAP in the BMMSCs isolated from MM patients and normal donors. The expression of FAP can be increased under TCCM stimulation or coculture with MM cells lines. Further study demonstrated that BMMSCs and FAP can protect MM cells from apoptosis induced by bortezomib, which is likely through β-catenin signaling pathway.
Materials and Methods
Cell cultures
Human multiple myeloma cell lines RPMI8226, MM.1R, CAG, and the cell line of human bone marrow mesenchymal stem cells (BMMSCs) were kindly provided by Dr Qing Yi (Department of Cancer Biology, Lerner Research Institute, Cleveland Clinic). Cells were cultured in RPMI1640 (Thermo Scientific) medium containing 10% heat-inactivated fetal bovine serum (FBS) (Life Technologies), 1% l-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin at 37 °C in 5% CO2. Culture of BMMSCs were according this paper.Citation78 Human bone marrow mononuclear cells (BMMNCs) from normal donors, obtained after informed consent in keeping with ethical guidelines declared by the First Affiliated Hospital of Zhejiang University and the Declaration of Helsinki, were isolated by lymphocyte separation medium and cultured in flasks, half of the medium was replaced every 3–7 d. After being cultured for 2 to 4 wk, cells became adherent and fibroblast-like cells, reaching to about >90% confluency. Cells were digested with trypsin-EDTA and subcultured. Passage 2 to 6 MSCs were used for the experiments.
Identification of BMMSCs
AccutaseTM solution (Merck Millipore) was used to detach the cells from culture flasks. Cells were washed twice with phosphate-buffered saline (PBS). Then cells were incubated with anti-CD14-FITC, anti-CD34-FITC, anti-CD90-APC, anti-CD105-PE antibodies (BD PharMingen), and anti-CD45-PE-Cy7 antibody (Biolegend) for 15 min according to the manufacturer’s instructions. Mouse IgG1, κ (Biolegend) were used as isotype control. Fluorescence staining was performed using a FACScan (Becton Dickinson).
Flow cytometry analysis for detection of FAP
Cells were detached from culture flasks with AccutaseTM solution (Merck Millipore). Cells were washed with PBS and non-specific antigens were blocked with 5% goat serum (Beijing Biosynthesis Biotechnology Co., LTD) for 1 h. Then cells were incubated with mouse anti-FAP (Santa Cruz Biotechnology) for 2 h. After that, cells were washed one more time and incubated with Alexa Fluor 488-labeled goat anti-mouse IgG (Invitrogen) for 30 min as secondary antibody. Analyses of ñuorescence staining were performed using a FACScan (Becton Dickinson).
Immunofluorescence
BMMSCs were seeded on the cover glass in 6-well plates for 24 h before being used for experiments. Next day, cells were washed three times with PBS for 5 min every time, and fixed in 4% paraformaldehyde for 30 min in room temperature. Cells were then washed three times with PBS and permeabilized within 0.3% Triton X-100 for 10 min. After being washed three times with PBS, non-specific antigens were blocked with 5% goat serum (Beijing Biosynthesis Biotechnology Co., LTD) for 1 h. After being washed three times with PBS, cells were incubated with anti-fibroblast activation protein (FAP) (Santa Cruz Biotechnology) at 4 °C overnight. After washing, cells were incubated avoiding light with Alexa Fluor 488-labeled goat anti-mouse IgG (Invitrogen) for 30 min. After washing, cells were incubated avoiding light with DAPI (4’,6-diamidino-2-phenylindole, 1:5000; Wako) for 5 min. After that, cells were visualized using fluorescence microscopy (Olympus) and photos were taken.
Quantitative real-time RT-PCR
Total RNA was extracted from BMMSCs using RNAiso™ Plus (TaKaRa) according to the manufacturer’s instructions. cDNA was generated from 1 μg of total RNA with PrimeScriptTM T reagent Kit with DNA Eraser (Perfect Real Time, Takara) according to the manufacturer’s instructions. Semiquantitative real-time PCR (qRT-PCR) was performed with SYBR® Premix Ex TaqTM II (Tli RNaseH Plus) (Takara) following the manufacturers’ instructions using iQ5 Multicolor Real-Time PCR Detection System (Bio-Rad Inc.). Semiquantitative real-time PCR reactions showed no non-specific amplifications. Sequences of primers for FAP (NM_004460) amplification were: forward, 5′-GTATTTGGAG TTGCCACCTC TG; reverse, 5′- GAAGGGCGTA AGACAATGCA C; GAPDH was used as an endogenous control. The sequences for GAPDH amplification were: forward, 5′-AAGGTGAAGG TCGGAGTCAA C; reverse, 5′-GGGGTCATTG ATGGCAACAA TA. The qRT-PCR program of FAP and GAPDH were: initial step at 95 °C for 30 s followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s. The specificity of primers was verified by melt curve. The 2−ΔΔCT method was used to calculate relative quantification of FAP in BMMSCs.
Preparation of tumor cells conditioned medium (TCCM) and stimulation of BMMSCs by TCCM or coculture with RPMI8226 cells
RPMI8226 and MM1.R cells were cultured at a density of 1 × 106/mL in RPMI-1640 containing 10% FBS. Sixteen hours later, culture medium were collected, and stored at 4 °C for up to 1 wk before use.Citation79 To examine the effects of myeloma-derived factors on the expression of FAP in BMMSCs, we added TCCM to the cell cultures of BMMSCs (50% TCCM to 50% complete medium) on day 1 and on day 4 when medium was replaced.Citation79 The division of RPMI8226 cells was blocked by addition of mitomycin C (10 μg/mL) for 2 h. Then BMMSCs were cocultured with pretreated RPMI8226 cells and RPMI8226 cells was changed every 2 d. After 7 d stimulation, BMMSCs were collected, washed with PBS, and cryopreserved with RNAiso™ Plus (TaKaRa) in –80 °C.
Transfection and cell apoptosis analysis by flow cytometry
The day before transfection, BMMSCs were planted in 12-orifice plates. When reaching about 90% confluency, cells were transfected with FAP siRNA (Genepharma Co. Ltd), forward (5′ → 3′): CGCCCUUCAA GAGUUCAUAT T, reverse (5′ → 3′): UAUGAACUCU UGAAGGGCGT T, using lipofectamine™ 2000 (Invitrogen). After 6 h, opti-medium was removed (Invitrogen), RPMI8226 or CAG cells were added to coculture with transfected BMMSCs (MM cells:BMMSCs = 1–2:1). Meanwhile, 20 nM bortezomib (Millennium Pharmaceuticals, Inc.) was added to the well to induce apoptosis in MM cells. After 48 h, cocultured RPMI8226 or CAG cells were separated from the BMMSCs monolayer by gently and careful pipetting with medium and washed twice with PBS. Then, cells were stained with fluorescein isothiocyanate (FITC)-conjugated annexin V (Biouniquer) and APC-conjugated anti-CD138 (Biolegend) following the manufacturers’ instructions. Annexin V-FITC positive cells were regarded as apoptotic cells. The interfering effect was detected by qRT-PCR and flow cytometry.
Western blot analysis
To examine intracellular signaling of cocultured RPMI8226 and CAG cells after FAP knockdown and bortezomib-induced apoptosis, we detect the changes of apoptosis-related proteins and β-catanin-related proteins. BMMSCs (2 × 105) and RPMI8226 or CAG cells (5 × 105) were cocultured in 6-well plates after BMMSCs were transfected with FAP-siRNA. RPMI8226 and CAG cells were also treated with conditioned medium derived from BMMSCs (2 × 105) or 100 ng/mL cytokines (IFN-γ, IGF-I, MCP-1, TGF-β, TNF-α, and mixture of cytokines. All cytokines were from PeproTech), or were cocultured with BMMSCs separated by 0.4 μm transwell chambers (Corning Costar). After 48 h, RPMI8226 and CAG cells were collected and washed twice with PBS. Totol cell lysates were performed using lysis buffer supplemented with 1 mM PMSF (Beyotime). The protein concentration was detected using bicinchoninic acid (BCA, Sangon Biotech) assay according to the protocol. An equal amount of 20–40 µg of protein was separated by 10–12% sodium dodecyl sulfate/PAGE (SDS-PAGE) and transferred to a polyvinylidene difluoride membrane (PVDF) (Merck Millipore) using an electroblot apparatus (Bio-Rad Inc.). After being blocked with 5% non-fat milk for 2 h. PVDF membranes were incubated with the corresponding primary antibodies overnight at 4 °C. Anti-caspase 3, β-catenin, c-Myc, and pp53 antibody were both from Cell Signaling Technology. Anti-PARP-1, survivin, cyclin D1, bak, bad antibodies were from Abcam. Anti-β-actin antibody from Sigma-Aldrich. The membranes were then washed and incubated with the horseradish peroxidase (HRP)-conjugated antibody (Jackson ImmunoResearch Laboratories, Inc.; 1:5000) in TBST at room temperature for 2 h. After being washed, the bands on the membrane were detected on X-ray films using an enhanced chemiluminescence (ECL) detection kit for HRP (Biological Industries, Beit Haemek LTD).
Cell apoptosis analysis by flow cytometry after inhibition of β-catenin signaling pathway
To verify whether β-catenin signaling pathway participated in BMMSCs- and FAP-mediated protection effect of MM cells or not. We inhibited β-catenin signaling pathway with XAV939 (Sigma-Aldrich). RPMI8226 cells were pretreated with XAV939 (40 μM) for 24 h before cocultured with BMMSCs (knockdown FAP or not) and exposed to 20 nM bortezomib. After 48 h, cocultured RPMI8226 cells were separated from the BMMSCs and washed twice with PBS. Then, cells were stained with APC-conjugated anti-CD138 and FITC-conjugated annexin V. Annexin V-FITC positive cells were regarded as apoptotic cells.
Statistical analysis
Statistical analysis was performed using the Student t test to compare various experimental groups and all tests were considered statistically significant if P < 0.05.
Disclosure of Potential Conflicts of Interest
The authors declare that they have no competing interests.
Acknowledgments
This work was supported by grants from the Major Research Plan of the National Natural Science Foundation of China (91029740) and the National Natural Science Foundation of China (81071936 and 81200385).
References
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin 2013; 63:11 - 30; http://dx.doi.org/10.3322/caac.21166; PMID: 23335087
- Mahindra A, Laubach J, Raje N, Munshi N, Richardson PG, Anderson K. Latest advances and current challenges in the treatment of multiple myeloma. Nature reviews. Clin Oncol 2012; 9:135 - 43
- De Raeve HR, Vanderkerken K. The role of the bone marrow microenvironment in multiple myeloma. Histol Histopathol 2005; 20:1227 - 50; PMID: 16136504
- Roccaro AM, Sacco A, Maiso P, Azab AK, Tai YT, Reagan M, Azab F, Flores LM, Campigotto F, Weller E, et al. BM mesenchymal stromal cell-derived exosomes facilitate multiple myeloma progression. J Clin Invest 2013; 123:1542 - 55; http://dx.doi.org/10.1172/JCI66517; PMID: 23454749
- Corre J, Mahtouk K, Attal M, Gadelorge M, Huynh A, Fleury-Cappellesso S, et al. Bone marrow mesenchymal stem cells are abnormal in multiple myeloma. Leukemia: official journal of the Leukemia Society of America. Leukemia Research Fund, UK 2007; 21:1079 - 88
- Reagan MR, Ghobrial IM. Multiple myeloma mesenchymal stem cells: characterization, origin, and tumor-promoting effects. Clin Cancer Res 2012; 18:342 - 9; http://dx.doi.org/10.1158/1078-0432.CCR-11-2212; PMID: 22065077
- Littlepage LE, Egeblad M, Werb Z. Coevolution of cancer and stromal cellular responses. Cancer Cell 2005; 7:499 - 500; http://dx.doi.org/10.1016/j.ccr.2005.05.019; PMID: 15950897
- Polyak K, Haviv I, Campbell IG. Co-evolution of tumor cells and their microenvironment. Trends Genet 2009; 25:30 - 8; http://dx.doi.org/10.1016/j.tig.2008.10.012; PMID: 19054589
- Weinberg RA. Coevolution in the tumor microenvironment. Nat Genet 2008; 40:494 - 5; http://dx.doi.org/10.1038/ng0508-494; PMID: 18443582
- Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, Richardson AL, Weinberg RA. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 2005; 121:335 - 48; http://dx.doi.org/10.1016/j.cell.2005.02.034; PMID: 15882617
- Mao Y, Keller ET, Garfield DH, Shen K, Wang J. Stromal cells in tumor microenvironment and breast cancer. Cancer Metastasis Rev 2013; 32:303 - 15; http://dx.doi.org/10.1007/s10555-012-9415-3; PMID: 23114846
- Haviv I, Polyak K, Qiu W, Hu M, Campbell I. Origin of carcinoma associated fibroblasts. Cell Cycle 2009; 8:589 - 95; http://dx.doi.org/10.4161/cc.8.4.7669; PMID: 19182519
- Spaeth EL, Dembinski JL, Sasser AK, Watson K, Klopp A, Hall B, Andreeff M, Marini F. Mesenchymal stem cell transition to tumor-associated fibroblasts contributes to fibrovascular network expansion and tumor progression. PLoS One 2009; 4:e4992; http://dx.doi.org/10.1371/journal.pone.0004992; PMID: 19352430
- Mishra PJ, Mishra PJ, Glod JW, Banerjee D. Mesenchymal stem cells: flip side of the coin. Cancer Res 2009; 69:1255 - 8; http://dx.doi.org/10.1158/0008-5472.CAN-08-3562; PMID: 19208837
- Giannoni E, Bianchini F, Masieri L, Serni S, Torre E, Calorini L, Chiarugi P. Reciprocal activation of prostate cancer cells and cancer-associated fibroblasts stimulates epithelial-mesenchymal transition and cancer stemness. Cancer Res 2010; 70:6945 - 56; http://dx.doi.org/10.1158/0008-5472.CAN-10-0785; PMID: 20699369
- Garin-Chesa P, Old LJ, Rettig WJ. Cell surface glycoprotein of reactive stromal fibroblasts as a potential antibody target in human epithelial cancers. Proc Natl Acad Sci U S A 1990; 87:7235 - 9; http://dx.doi.org/10.1073/pnas.87.18.7235; PMID: 2402505
- Cheng JD, Dunbrack RL Jr., Valianou M, Rogatko A, Alpaugh RK, Weiner LM. Promotion of tumor growth by murine fibroblast activation protein, a serine protease, in an animal model. Cancer Res 2002; 62:4767 - 72; PMID: 12183436
- Shi W, Liu J, Li M, Gao H, Wang T. Expression of MMP, HPSE, and FAP in stroma promoted corneal neovascularization induced by different etiological factors. Curr Eye Res 2010; 35:967 - 77; http://dx.doi.org/10.3109/02713683.2010.502294; PMID: 20958185
- Huang Y, Wang S, Kelly T. Seprase promotes rapid tumor growth and increased microvessel density in a mouse model of human breast cancer. Cancer Res 2004; 64:2712 - 6; http://dx.doi.org/10.1158/0008-5472.CAN-03-3184; PMID: 15087384
- Kraman M, Bambrough PJ, Arnold JN, Roberts EW, Magiera L, Jones JO, Gopinathan A, Tuveson DA, Fearon DT. Suppression of antitumor immunity by stromal cells expressing fibroblast activation protein-alpha. Science 2010; 330:827 - 30; http://dx.doi.org/10.1126/science.1195300; PMID: 21051638
- Zhang Y, Tang H, Cai J, Zhang T, Guo J, Feng D, Wang Z. Ovarian cancer-associated fibroblasts contribute to epithelial ovarian carcinoma metastasis by promoting angiogenesis, lymphangiogenesis and tumor cell invasion. Cancer Lett 2011; 303:47 - 55; http://dx.doi.org/10.1016/j.canlet.2011.01.011; PMID: 21310528
- Huang Y, Simms AE, Mazur A, Wang S, León NR, Jones B, Aziz N, Kelly T. Fibroblast activation protein-α promotes tumor growth and invasion of breast cancer cells through non-enzymatic functions. Clin Exp Metastasis 2011; 28:567 - 79; http://dx.doi.org/10.1007/s10585-011-9392-x; PMID: 21604185
- Wang XM, Yu DM, McCaughan GW, Gorrell MD. Fibroblast activation protein increases apoptosis, cell adhesion, and migration by the LX-2 human stellate cell line. Hepatology 2005; 42:935 - 45; http://dx.doi.org/10.1002/hep.20853; PMID: 16175601
- Jacob M, Chang L, Puré E. Fibroblast activation protein in remodeling tissues. Curr Mol Med 2012; 12:1220 - 43; http://dx.doi.org/10.2174/156652412803833607; PMID: 22834826
- Fischer E, Chaitanya K, Wüest T, Wadle A, Scott AM, van den Broek M, Schibli R, Bauer S, Renner C. Radioimmunotherapy of fibroblast activation protein positive tumors by rapidly internalizing antibodies. Clin Cancer Res 2012; 18:6208 - 18; http://dx.doi.org/10.1158/1078-0432.CCR-12-0644; PMID: 22992515
- Prockop DJ. Marrow stromal cells as stem cells for nonhematopoietic tissues. Science 1997; 276:71 - 4; http://dx.doi.org/10.1126/science.276.5309.71; PMID: 9082988
- Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science 1999; 284:143 - 7; http://dx.doi.org/10.1126/science.284.5411.143; PMID: 10102814
- Jiang Y, Jahagirdar BN, Reinhardt RL, Schwartz RE, Keene CD, Ortiz-Gonzalez XR, Reyes M, Lenvik T, Lund T, Blackstad M, et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature 2002; 418:41 - 9; http://dx.doi.org/10.1038/nature00870; PMID: 12077603
- Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop Dj, Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006; 8:315 - 7; http://dx.doi.org/10.1080/14653240600855905; PMID: 16923606
- Chamberlain G, Fox J, Ashton B, Middleton J. Concise review: mesenchymal stem cells: their phenotype, differentiation capacity, immunological features, and potential for homing. Stem Cells 2007; 25:2739 - 49; http://dx.doi.org/10.1634/stemcells.2007-0197; PMID: 17656645
- Rettig WJ, Chesa PG, Beresford HR, Feickert HJ, Jennings MT, Cohen J, Oettgen HF, Old LJ. Differential expression of cell surface antigens and glial fibrillary acidic protein in human astrocytoma subsets. Cancer Res 1986; 46:6406 - 12; PMID: 2877731
- Rettig WJ, Spengler BA, Chesa PG, Old LJ, Biedler JL. Coordinate changes in neuronal phenotype and surface antigen expression in human neuroblastoma cell variants. Cancer Res 1987; 47:1383 - 9; PMID: 3028608
- Aoyama A, Chen WT. A 170-kDa membrane-bound protease is associated with the expression of invasiveness by human malignant melanoma cells. Proc Natl Acad Sci U S A 1990; 87:8296 - 300; http://dx.doi.org/10.1073/pnas.87.21.8296; PMID: 2172980
- Monsky WL, Lin CY, Aoyama A, Kelly T, Akiyama SK, Mueller SC, Chen WT. A potential marker protease of invasiveness, seprase, is localized on invadopodia of human malignant melanoma cells. Cancer Res 1994; 54:5702 - 10; PMID: 7923219
- Mathew S, Scanlan MJ, Mohan Raj BK, Murty VV, Garin-Chesa P, Old LJ, Rettig WJ, Chaganti RS. The gene for fibroblast activation protein alpha (FAP), a putative cell surface-bound serine protease expressed in cancer stroma and wound healing, maps to chromosome band 2q23. Genomics 1995; 25:335 - 7; http://dx.doi.org/10.1016/0888-7543(95)80157-H; PMID: 7774951
- Piñeiro-Sánchez ML, Goldstein LA, Dodt J, Howard L, Yeh Y, Tran H, Argraves WS, Chen WT. Identification of the 170-kDa melanoma membrane-bound gelatinase (seprase) as a serine integral membrane protease. J Biol Chem 1997; 272:7595 - 601; http://dx.doi.org/10.1074/jbc.272.12.7595; PMID: 9065413
- Rettig WJ, Garin-Chesa P, Beresford HR, Oettgen HF, Melamed MR, Old LJ. Cell-surface glycoproteins of human sarcomas: differential expression in normal and malignant tissues and cultured cells. Proc Natl Acad Sci U S A 1988; 85:3110 - 4; http://dx.doi.org/10.1073/pnas.85.9.3110; PMID: 2896356
- Rettig WJ, Garin-Chesa P, Healey JH, Su SL, Ozer HL, Schwab M, Albino AP, Old LJ. Regulation and heteromeric structure of the fibroblast activation protein in normal and transformed cells of mesenchymal and neuroectodermal origin. Cancer Res 1993; 53:3327 - 35; PMID: 8391923
- Kelly T, Kechelava S, Rozypal TL, West KW, Korourian S. Seprase, a membrane-bound protease, is overexpressed by invasive ductal carcinoma cells of human breast cancers. Modern pathology: an official journal of the United States and Canadian Academy of Pathology. Inc 1998; 11:855 - 63
- Mori Y, Kono K, Matsumoto Y, Fujii H, Yamane T, Mitsumata M, Chen WT. The expression of a type II transmembrane serine protease (Seprase) in human gastric carcinoma. Oncology 2004; 67:411 - 9; http://dx.doi.org/10.1159/000082926; PMID: 15713998
- Mentlein R, Hattermann K, Hemion C, Jungbluth AA, Held-Feindt J. Expression and role of the cell surface protease seprase/fibroblast activation protein-α (FAP-α) in astroglial tumors. Biol Chem 2011; 392:199 - 207; http://dx.doi.org/10.1515/bc.2010.119; PMID: 20707604
- Acharya PS, Zukas A, Chandan V, Katzenstein AL, Puré E. Fibroblast activation protein: a serine protease expressed at the remodeling interface in idiopathic pulmonary fibrosis. Hum Pathol 2006; 37:352 - 60; http://dx.doi.org/10.1016/j.humpath.2005.11.020; PMID: 16613331
- Bauer S, Jendro MC, Wadle A, Kleber S, Stenner F, Dinser R, Reich A, Faccin E, Gödde S, Dinges H, et al. Fibroblast activation protein is expressed by rheumatoid myofibroblast-like synoviocytes. Arthritis Res Ther 2006; 8:R171; http://dx.doi.org/10.1186/ar2080; PMID: 17105646
- Levy MT, McCaughan GW, Abbott CA, Park JE, Cunningham AM, Müller E, Rettig WJ, Gorrell MD. Fibroblast activation protein: a cell surface dipeptidyl peptidase and gelatinase expressed by stellate cells at the tissue remodelling interface in human cirrhosis. Hepatology 1999; 29:1768 - 78; http://dx.doi.org/10.1002/hep.510290631; PMID: 10347120
- Bae S, Park CW, Son HK, Ju HK, Paik D, Jeon CJ, Koh GY, Kim J, Kim H. Fibroblast activation protein alpha identifies mesenchymal stromal cells from human bone marrow. Br J Haematol 2008; 142:827 - 30; http://dx.doi.org/10.1111/j.1365-2141.2008.07241.x; PMID: 18510677
- Hall B, Dembinski J, Sasser AK, Studeny M, Andreeff M, Marini F. Mesenchymal stem cells in cancer: tumor-associated fibroblasts and cell-based delivery vehicles. Int J Hematol 2007; 86:8 - 16; http://dx.doi.org/10.1532/IJH97.06230; PMID: 17675260
- Mishra PJ, Mishra PJ, Humeniuk R, Medina DJ, Alexe G, Mesirov JP, Ganesan S, Glod JW, Banerjee D. Carcinoma-associated fibroblast-like differentiation of human mesenchymal stem cells. Cancer Res 2008; 68:4331 - 9; http://dx.doi.org/10.1158/0008-5472.CAN-08-0943; PMID: 18519693
- Garayoa M, Garcia JL, Santamaría C, Garcia-Gomez A, Blanco JF, Pandiella A, et al. Mesenchymal stem cells from multiple myeloma patients display distinct genomic profile as compared with those from normal donors. Leukemia: official journal of the Leukemia Society of America. Leukemia Research Fund, UK 2009; 23:1515 - 27; http://dx.doi.org/10.1038/leu.2009.65
- Arnulf B, Lecourt S, Soulier J, Ternaux B, Lacassagne MN, Crinquette A, et al. Phenotypic and functional characterization of bone marrow mesenchymal stem cells derived from patients with multiple myeloma. Leukemia: official journal of the Leukemia Society of America. Leukemia Research Fund, UK 2007; 21:158 - 63; http://dx.doi.org/10.1038/sj.leu.2404466
- Garayoa M, Garcia JL, Santamaria C, Garcia-Gomez A, Blanco JF, Pandiella A, et al. Mesenchymal stem cells from multiple myeloma patients display distinct genomic profile as compared with those from normal donors. Leukemia: official journal of the Leukemia Society of America. Leukemia Research Fund, UK 2009; 23:1515 - 27; http://dx.doi.org/10.1038/leu.2009.65
- Wu Y, Chen L, Scott PG, Tredget EE. Mesenchymal stem cells enhance wound healing through differentiation and angiogenesis. Stem Cells 2007; 25:2648 - 59; http://dx.doi.org/10.1634/stemcells.2007-0226; PMID: 17615264
- Gunn WG, Conley A, Deininger L, Olson SD, Prockop DJ, Gregory CA. A crosstalk between myeloma cells and marrow stromal cells stimulates production of DKK1 and interleukin-6: a potential role in the development of lytic bone disease and tumor progression in multiple myeloma. Stem Cells 2006; 24:986 - 91; http://dx.doi.org/10.1634/stemcells.2005-0220; PMID: 16293576
- Xu S, Menu E, De Becker A, Van Camp B, Vanderkerken K, Van Riet I. Bone marrow-derived mesenchymal stromal cells are attracted by multiple myeloma cell-produced chemokine CCL25 and favor myeloma cell growth in vitro and in vivo. Stem Cells 2012; 30:266 - 79; http://dx.doi.org/10.1002/stem.787; PMID: 22102554
- Kim J, Denu RA, Dollar BA, Escalante LE, Kuether JP, Callander NS, Asimakopoulos F, Hematti P. Macrophages and mesenchymal stromal cells support survival and proliferation of multiple myeloma cells. Br J Haematol 2012; 158:336 - 46; http://dx.doi.org/10.1111/j.1365-2141.2012.09154.x; PMID: 22583117
- Tassone P, Neri P, Carrasco DR, Burger R, Goldmacher VS, Fram R, Munshi V, Shammas MA, Catley L, Jacob GS, et al. A clinically relevant SCID-hu in vivo model of human multiple myeloma. Blood 2005; 106:713 - 6; http://dx.doi.org/10.1182/blood-2005-01-0373; PMID: 15817674
- Markovina S, Callander NS, O’Connor SL, Xu G, Shi Y, Leith CP, Kim K, Trivedi P, Kim J, Hematti P, et al. Bone marrow stromal cells from multiple myeloma patients uniquely induce bortezomib resistant NF-kappaB activity in myeloma cells. Mol Cancer 2010; 9:176; http://dx.doi.org/10.1186/1476-4598-9-176; PMID: 20604947
- Corre J, Labat E, Espagnolle N, Hébraud B, Avet-Loiseau H, Roussel M, Huynh A, Gadelorge M, Cordelier P, Klein B, et al. Bioactivity and prognostic significance of growth differentiation factor GDF15 secreted by bone marrow mesenchymal stem cells in multiple myeloma. Cancer Res 2012; 72:1395 - 406; http://dx.doi.org/10.1158/0008-5472.CAN-11-0188; PMID: 22301101
- Gupta D, Treon SP, Shima Y, Hideshima T, Podar K, Tai YT, et al. Adherence of multiple myeloma cells to bone marrow stromal cells upregulates vascular endothelial growth factor secretion: therapeutic applications. Leukemia: official journal of the Leukemia Society of America. Leukemia Research Fund, UK 2001; 15:1950 - 61; http://dx.doi.org/10.1038/sj.leu.2402295
- Pan B, Lentzsch S. The application and biology of immunomodulatory drugs (IMiDs) in cancer. Pharmacol Ther 2012; 136:56 - 68; http://dx.doi.org/10.1016/j.pharmthera.2012.07.004; PMID: 22796518
- Hideshima T, Bergsagel PL, Kuehl WM, Anderson KC. Advances in biology of multiple myeloma: clinical applications. Blood 2004; 104:607 - 18; http://dx.doi.org/10.1182/blood-2004-01-0037; PMID: 15090448
- Fulciniti M, Carrasco DR. Role of Wnt Signaling Pathways in Multiple Myeloma Pathogenesis. Advances in Biology and Therapy of Multiple Myeloma: Springer, 2013:85-95.
- Yang Y, Mallampati S, Sun B, Zhang J, Kim S-B, Lee J-S, Gong Y, Cai Z, Sun X. Wnt pathway contributes to the protection by bone marrow stromal cells of acute lymphoblastic leukemia cells and is a potential therapeutic target. Cancer Lett 2013; 333:9 - 17; http://dx.doi.org/10.1016/j.canlet.2012.11.056; PMID: 23333798
- Derksen PW, Tjin E, Meijer HP, Klok MD, MacGillavry HD, van Oers MH, Lokhorst HM, Bloem AC, Clevers H, Nusse R, et al. Illegitimate WNT signaling promotes proliferation of multiple myeloma cells. Proc Natl Acad Sci U S A 2004; 101:6122 - 7; http://dx.doi.org/10.1073/pnas.0305855101; PMID: 15067127
- Giuliani N, Morandi F, Tagliaferri S, Lazzaretti M, Donofrio G, Bonomini S, Sala R, Mangoni M, Rizzoli V. Production of Wnt inhibitors by myeloma cells: potential effects on canonical Wnt pathway in the bone microenvironment. Cancer Res 2007; 67:7665 - 74; http://dx.doi.org/10.1158/0008-5472.CAN-06-4666; PMID: 17702698
- Qiang YW, Shaughnessy JD Jr., Yaccoby S. Wnt3a signaling within bone inhibits multiple myeloma bone disease and tumor growth. Blood 2008; 112:374 - 82; http://dx.doi.org/10.1182/blood-2007-10-120253; PMID: 18344425
- Bjorklund CC, Ma W, Wang Z-Q, Davis RE, Kuhn DJ, Kornblau SM, Wang M, Shah JJ, Orlowski RZ. Evidence of a role for activation of Wnt/β-catenin signaling in the resistance of plasma cells to lenalidomide. J Biol Chem 2011; 286:11009 - 20; http://dx.doi.org/10.1074/jbc.M110.180208; PMID: 21189262
- Qiang YW, Walsh K, Yao L, Kedei N, Blumberg PM, Rubin JS, Shaughnessy J Jr., Rudikoff S. Wnts induce migration and invasion of myeloma plasma cells. Blood 2005; 106:1786 - 93; http://dx.doi.org/10.1182/blood-2005-01-0049; PMID: 15886323
- Levina E, Oren M, Ben-Ze’ev A. Downregulation of beta-catenin by p53 involves changes in the rate of beta-catenin phosphorylation and Axin dynamics. Oncogene 2004; 23:4444 - 53; http://dx.doi.org/10.1038/sj.onc.1207587; PMID: 15064706
- Bueno C, Lopes LF, Menendez P. Bone marrow stromal cell-derived Wnt signals as a potential underlying mechanism for cyclin D1 deregulation in multiple myeloma lacking t(11;14)(q13;q32). Blood Cells Mol Dis 2007; 39:366 - 8; http://dx.doi.org/10.1016/j.bcmd.2007.06.001; PMID: 17632021
- Yang Y, Mallampati S, Sun B, Zhang J, Kim SB, Lee JS, Gong Y, Cai Z, Sun X. Wnt pathway contributes to the protection by bone marrow stromal cells of acute lymphoblastic leukemia cells and is a potential therapeutic target. Cancer Lett 2013; 333:9 - 17; http://dx.doi.org/10.1016/j.canlet.2012.11.056; PMID: 23333798
- Satoh J, Kuroda Y. Beta-catenin expression in human neural cell lines following exposure to cytokines and growth factors. Neuropathology 2000; 20:113 - 23; http://dx.doi.org/10.1046/j.1440-1789.2000.00293.x; PMID: 10935448
- Playford MP, Bicknell D, Bodmer WF, Macaulay VM. Insulin-like growth factor 1 regulates the location, stability, and transcriptional activity of beta-catenin. Proc Natl Acad Sci U S A 2000; 97:12103 - 8; http://dx.doi.org/10.1073/pnas.210394297; PMID: 11035789
- Speranza FJ, Mahankali M, Gomez-Cambronero J. Macrophage migration arrest due to a winning balance of Rac2/Sp1 repression over β-catenin-induced PLD expression. J Leukoc Biol 2013; 94:953 - 62; http://dx.doi.org/10.1189/jlb.0313174; PMID: 23898047
- Orr SJ, Burg AR, Chan T, Quigley L, Jones GW, Ford JW, Hodge D, Razzook C, Sarhan J, Jones YL, et al. LAB/NTAL facilitates fungal/PAMP-induced IL-12 and IFN-γ production by repressing β-catenin activation in dendritic cells. PLoS Pathog 2013; 9:e1003357; http://dx.doi.org/10.1371/journal.ppat.1003357; PMID: 23675302
- Li W, Henderson LJ, Major EO, Al-Harthi L. IFN-gamma mediates enhancement of HIV replication in astrocytes by inducing an antagonist of the beta-catenin pathway (DKK1) in a STAT 3-dependent manner. J Immunol 2011; 186:6771 - 8; http://dx.doi.org/10.4049/jimmunol.1100099; PMID: 21562161
- Liu Z, Brooks RS, Ciappio ED, Kim SJ, Crott JW, Bennett G, Greenberg AS, Mason JB. Diet-induced obesity elevates colonic TNF-α in mice and is accompanied by an activation of Wnt signaling: a mechanism for obesity-associated colorectal cancer. J Nutr Biochem 2012; 23:1207 - 13; http://dx.doi.org/10.1016/j.jnutbio.2011.07.002; PMID: 22209007
- Dai C, Wen X, He W, Liu Y. Inhibition of proinflammatory RANTES expression by TGF-beta1 is mediated by glycogen synthase kinase-3beta-dependent beta-catenin signaling. J Biol Chem 2011; 286:7052 - 9; http://dx.doi.org/10.1074/jbc.M110.174821; PMID: 21189258
- Pittenger MF. Mesenchymal stem cells from adult bone marrow. Mesenchymal Stem Cells: Springer, 2008:27-44.
- Wang S, Hong S, Yang J, Qian J, Zhang X, Shpall E, Kwak LW, Yi Q. Optimizing immunotherapy in multiple myeloma: Restoring the function of patients’ monocyte-derived dendritic cells by inhibiting p38 or activating MEK/ERK MAPK and neutralizing interleukin-6 in progenitor cells. Blood 2006; 108:4071 - 7; http://dx.doi.org/10.1182/blood-2006-04-016980; PMID: 16917008