Abstract
Genetic markers identifying women at an increased risk of developing breast cancer exist, yet the majority of inherited risk remains elusive. While numerous BRCA1 coding sequence mutations are associated with breast cancer risk, BRCA1 mutations account for less then 5% of breast cancer risk. Since 3' untranslated region (3'UTR) polymorphisms disrupting microRNA (miRNA) binding can be functional and can act as genetic markers of cancer risk, we tested the hypothesis that such polymorphisms in the 3'UTR of BRCA1 and haplotypes containing these functional polymorphisms may be associated with breast cancer risk. We sequenced the BRCA1 3'UTR from breast cancer patients to identify miRNA disrupting polymorphisms. We further evaluated haplotypes of this region including the identified 3'UTR variants in a large population of controls and breast cancer patients (n=221) with known breast cancer subtypes and ethnicities. We identified three 3'UTR variants in BRCA1 that are polymorphic in breast cancer populations, and haplotype analysis including these variants revealed that breast cancer patients harbor five rare haplotypes not generally found among controls (9.50% for breast cancer chromosomes, 0.11% for control chromosomes, p=0.0001). Three of these rare haplotypes contain the rs8176318 BRCA1 3'UTR functional variant. These haplotypes are not biomarkers for BRCA1 coding mutations, as they are found rarely in BRCA1 mutant breast cancer patients (1/129 patients= 0.78%). These rare BRCA1 haplotypes and 3'UTR SNPs may represent new genetic markers of breast cancer risk.
Introduction
Breast cancer is the most frequently diagnosed cancer, and one of the leading causes of cancer death in women today.Citation1,Citation2 Clinical and molecular classification has successfully clustered breast cancer into subgroups and shown unique gene expression in categories that have prognostic significance. Among the categories emerging from these studies are estrogen receptor (ER) or progesterone receptor (PR) positive, HER2 receptor gene-amplified tumors and triple negative ([TN] ER/PR/HER2−) tumors.Citation3 The ER/PR+ and HER2+ tumors together are most prevalent (80%), with basal-like or TN tumors accounting for approximately 15–20% of breast cancers.Citation4 The TN phenotype represents an aggressive and poorly understood subclass of cancer that is most prevalent among younger women and in African American women.Citation5
BRCA1 coding sequence mutations are a well-known risk factor for breast cancer, yet account for less then 5% of all breast cancer cases yearly.Citation6 Overall, breast tumors resulting from BRCA1 mutations are most frequently TN (57%),Citation7 or ER+ breast cancers (34%),Citation8 and are rarely HER2+ breast cancers (about 3%).Citation9 While TN tumors are often characterized by low expression of BRCA1,Citation10 because BRCA1 mutations are quite rare, they only account for approximately 10–20% of the TN tumors.Citation11–Citation13 These findings suggest that there may be additional inherited genetic factors associated with BRCA1 misexpression that could predispose individuals to breast cancer.
Haplotypes are patterns of several SNPs that are in linkage disequilibrium (LD) with one another within a gene or segment of DNA and are thus inherited as a unit.Citation14,Citation15 As haplotypes serve as markers for all measured and unmeasured alleles within a population,Citation14 a study of haplotypes of a region of interest can narrow the search for causal SNPs.Citation14,Citation15 Previous studies of the association of BRCA1 haplotypes with breast cancer have yielded conflicting results. Cox et al. identified five common haplotypes (≥5%) that could be predicted by four tagging SNPs. Testing of these SNPs showed that one of the haplotypes predicted a 20% increased risk (odds ratio 1.18, 95% confidence interval 1.02–1.37) of sporadic breast cancer in Caucasian women in the Nurses' Health Study.Citation16 There was significant interaction (p = 0.05) between this haplotype, positive family history and breast cancer risk.Citation16 In contrast, Freedman et al. tested common variation across the BRCA1 locus in a cohort from the Multiethnic Cohort Study. This group was not able to show that common variants in BRCA1 substantially influence sporadic breast cancer risk.Citation17 These haplotype studies focused primarily on variation at SNPs in the coding and intronic regions of BRCA1.Citation18,Citation19
MiRNAs are a class of 22-nucleotide non-coding RNAs that are evolutionarily conserved and are aberrantly expressed in virtually all cancers, where they function as a novel class of oncogenes or tumor suppressors.Citation20 The ability of miRNAs to bind to messenger (mRNA) in the 3′UTR is critical for regulating mRNA level and protein expression, binding which can be affected by single nucleotide polymorphisms.Citation21 Data from our group and others indicates that variants in the 3′UTR of cancer genes are strong genetic markers of cancer risk.Citation22–Citation24
The BRCA1 3′UTR has been recently studied for such miRNA binding site SNPs, and the derived (and less frequent) alleles at rs12516 and rs8176318 showed a positive association with familial breast and ovarian cancer in Thai women. The study found that homozygosity for the derived alleles, A, at both SNP sites are found in cancer patients at triple the frequency as seen in unaffected Thais, yielding a significant cancer association (p = 0.007). Analysis showed reduced translation of BRCA1 with the derived alleles at both sites when present on the same chromosome, i.e., in cis, with the greatest reduction seen with the derived allele at rs8176318.Citation24 This study additionally found that the 3′UTR variants were not associated with known BRCA1 mutations. In addition, a study in 1998 reported an allele at a third SNP in the BRCA1 3′UTR, rs3092995, as being associated with increased risk of breast cancer in African American women. The rarer, derived G allele was found to be more common in African American breast cancer cases than African American controls. The age-adjusted OR for breast cancer among African American women and the G allele was 3.5 (95% CI, 1.2–10).Citation25
We hypothesized that studying haplotypes that included functional 3′UTR variants would better identify BRCA1 haplotypes associated with sporadic breast cancer risk. Furthermore, because BRCA1 dysfunction varies by breast cancer subtype we evaluated these haplotypes by breast cancer subtype. We identified 3′UTR SNPs in our breast cancer patients, and then performed haplotype analysis with these variants and five SNPs surrounding the BRCA1 3′UTR to determine the association of these haplotypes with breast cancer. Doing so we identified five haplotypes common in our breast cancer patients but rare in non-cancerous populations. These rare BRCA1 haplotypes may represent new genetic markers of BRCA1 dysfunction associated with breast cancer risk.
Results
Identifying SNPs in the BRCA1 3′UTR.
There are numerous known BRCA1 3′UTR SNPs (Sup. Table 1). To identify the frequency of these known polymorphisms and/or to identify novel SNPs in sporadic breast cancer patients, we sequenced the entire 3′UTR of BRCA1 in breast cancer patients with the three known breast cancer subtypes (TN = 7, HER2+ = 18 and ER/PR+/HER2+ = 14). The initial screen of the entire BRCA1 3′UTR in these patients identified variation at only the three previously reported functional SNPs: rs12516, rs8176318 and rs3092995 (Sup. Table 2). Additionally, we identified a novel SNP in the BRCA1 3′UTR. The novel SNP in BRCA1 is 6824G/A or 5711 + 1113G/A. This SNP was identified as heterozygous in a 61 year old African American HER2+ patient for the previously unseen A allele.
To better evaluate the frequency of these variants across populations we performed population specific genotyping in 2,250 non-cancerous individuals making up 46 populations worldwide (). The three identified BRCA1 3′UTR SNPs, rs12516, rs8176318 and rs3092995 are in strong linkage disequilibrium in populations and vary by ethnicity.
Since we saw significant variation in the identified 3′UTR SNPs by ethnicity in our control populations, we next determined the variation of these SNPs in breast cancer patients of different ethnicity. We genotyped these SNPs in 130 breast cancer European American patients and 38 breast cancer African American patients, and saw variation across these groups (). To determine the association of these SNPs with tumor risk we compared the frequency of these SNPs between breast cancer patients and ethnicity matched controls. We found that the rare variant at rs8176318 in the homozygous form (A/A) is significantly associated with breast cancer for African Americans [Odds ratio (OR), 9.48; 95% confidence interval (CI), 1.01–88.80; p = 0.04]. We did not see any tumor association between breast cancer in European Americans and the rs8176318 SNP ().
Because BRCA1 dysfunction varies among the breast cancer subtypes we next evaluated the three 3′UTR SNPs by ethnicity and breast cancer subtype (Sup. Fig. 1). We found that the homozygous variant form of rs8176318 was significantly associated with risk for TN breast cancer among African American women [OR, 12.19; 95% CI, 1.29–115.21, p = 0.02). We did not see an association for any of the other SNPs or for ER/PR+ or HER2+ breast cancer subtypes (Sup. Table 3).
BRCA1 haplotype evolution and frequencies.
To better evaluate the BRCA1 region, we added five additional previously reported tagging SNPs shown to best represent BRCA1 evolutionCitation26 surrounding the three 3′UTR SNPs we identified in our sporadic breast cancer patients. The eight SNPs in total span 267 kb (). This entire region has high LDCitation26–Citation28 and heterozygosities among all eight SNPs composing our haplotype are generally high (30–50%) (http://alfred.med.yale.edu).Citation26,Citation29
These eight SNPs were used to generate global haplotype frequencies (). All of the common haplotypes observed can be explained by accumulation of variation on the ancestral haplotype (). Most of the directly observed haplotypes can be ordered, differing by one derived nucleotide change; in one case two changes are required and in another case a recombination is observed. Collectively, these generate three branches, each starting with a single nucleotide change from the ancestral haplotype. Of note, we found that haplotype diversity is much higher in Africa (with 6–9 haplotypes represented) versus outside of Africa (with 3–5 haplotypes).
BRCA1 haplotypes in breast cancer patients.
We next studied haplotypes consisting of our eight SNPs in our breast cancer patients to determine if there were differences in BRCA1 haplotypes between non-cancerous patients and breast cancer patients. We identified five haplotypes [GGC CGC TA (hot pink, #1), GGC CGC TG (light blue, #2), GGA CGC TA (teal, #3), GGA CGC TG (tan, #4) and GAA CGT TG (violet, #5)], which were enriched in our breast cancer populations (42/442 total breast cancer chromosomes evaluated), but extremely rare in global control populations. For example, in the global sample of 4,500 non-cancerous chromosomes, the teal haplotype (#3) was observed on 3 chromosomes and the tan haplotype (#4) was present on 2 chromosomes, while the hot pink (#1), light blue (#2) and violet (#5) haplotypes were never seen (<0.1%). This represents an overall global frequency of 0.1% for these haplotypes in non-cancerous controls versus a frequency of 9.50% for breast cancer patient chromosomes (p < 0.0001) (). Two haplotypes (Teal and Tan, #3 and #4, respectively) are characterized by the derived allele A within the 3′UTR at SNP rs8176318. A third rare haplotype (violet, #5) has derived alleles (A) at two of the 3′UTR polymorphisms, rs8176318 and rs12516.
Because we had shown that our haplotypes varied by ethnicity, to better compare these rare breast cancer haplotypes with the appropriate ethnic populations we evaluated breast cancer patients and controls matched by ethnicity. Our ethnicity-matched controls were composed of a total of 194 individuals (102 African Americans and 92 European Americans, including a cohort of Yale control Caucasian Americans and African Americans). We found that 8.84% of Caucasian American breast cancer patients and 11.84% of African American breast cancer patients contain our rare haplotypes, and again these haplotypes were rarely found in ethnicity matched controls, with only the teal haplotype found on one European American control chromosome (0.26%, 1/388 chromosomes, p < 0.0001, and Sup. Table 4).
BRCA1 haplotypes in breast cancer patients by breast cancer subtype.
Since known BRCA1 coding sequence mutations vary with breast cancer subtype, we wanted to determine how our rare haplotypes were distributed amongst breast cancer subtypes. We found the prevalence of our rare haplotypes varied significantly between the TN, ER/PR+ and HER2+ subtypes, with the TN subgroup harboring these rare haplotypes at the highest rate, at 14.85% (30/202 chromosomes, p = 0.014 compared to the others), the ER/PR+ breast cancer subtype next at 8.09% (11/136 ER/PR+ chromosomes), and the HER2+ subtype the least at 1% (1/104), ( and Sup. Table 5). It is interesting to note that the tan haplotype (#4) was only identified in TN tumors and not in the other tumor subtypes. We then evaluated the rare haplotypes by both ethnicity and breast tumor subtype (). Two haplotypes (light blue and violet, #2 and 5, respectively) were unique to European American breast cancer patients.
BRCA1 haplotypes and BRCA mutation status.
We wanted to also determine if the rare BRCA1 haplotypes identified in this study were associated with BRCA1 coding sequence mutations, yet we did not have BRCA1 mutation status on most of the patients tested in this study. Therefore, we tested a separate cohort of 129 unrelated European breast cancer patients heterozygous for BRCA1 coding region mutations for the presence of our rare BRCA1 haplotypes. We found that only one BRCA1 coding sequence mutant patient had a rare haplotype (0.8%, violet, #5). The remaining four rare haplotypes were not found in this cohort of BRCA1 mutant patients, suggesting that these rare BRCA1 haplotypes are not surrogate markers of common BRCA1 coding sequence mutations, but are unique and novel biomarkers of BRCA1 alterations associated with breast cancer.
Discussion
We found that sporadic breast cancer patients harbor five rare BRCA1 haplotypes not generally found in control populations. These haplotypes include three BRCA1 3′UTR SNPs, one of which (rs8176318) shows a significant association with breast cancer among African Americans (p = 0.04) and appears to be a genetic marker of risk most strongly for triple negative breast cancer (p = 0.02) as compared to their ethnicity matched controls. These haplotypes are not associated with common BRCA1 coding mutations. Our findings support the hypothesis that these identified rare BRCA1 haplotypes may represent new genetic markers of an increased risk of developing breast cancer, and likely represent non-coding sequence variations in BRCA1 that impact BRCA1 function and lead to increased breast cancer risk.
There have been previous studies conducting haplotype analysis in the BRCA1 region to determine their association with sporadic breast cancer, with little success.Citation16,Citation17 This is the first BRCA1 haplotype study of sporadic breast cancer to our knowledge that includes rare functional variants in the 3′UTR noncoding regulatory regions of BRCA1 as part of the haplotype analysis. Evidence is fast becoming available to support the theory that variants within the 3′UTR increase susceptibility to cancer through gene expression control.Citation22,Citation23 While we are unable to determine if in our rare haplotypes the increased breast cancer risk is one single variant within the haplotype or a combination of alleles, we hypothesize that it is the combination of the functional 3′UTR variants with the other variants comprising each haplotype that are predictive of meaningful BRCA1 dysfunction. Studies to better understand the function of each of these SNPs in breast cancer are ongoing.
We furthermore analyzed sporadic breast cancer by subtype in our haplotype analysis. Our finding that the rare haplotypes are primarily in TN and ER+ breast cancer is consistent with the published literature showing that BRCA1 coding sequence mutations are most frequently associated with TN (57%),Citation7 and ER+ breast cancers (34%),Citation8 and are rarely found in HER2+ breast cancers (about 3%).Citation9 We hypothesize that the significant association of the rare haplotypes with TN and ER+ breast cancer indicates that these haplotypes (and/or functional SNPs within the haplotypes) are associated with true BRCA1 dysfunction and studies to better prove this are ongoing.
The enrichment of the rare haplotypes in the TN subtype of breast cancer is especially striking. Not only does this subtype statistically associate with our rare haplotypes as compared to controls (p < 0.0001), but TN breast cancer is also the most common subtype associated with our rare haplotypes. Because TN breast cancers have the worst outcome,Citation30 it is perhaps most important to identify those at risk of developing this subtype of breast cancer.
Limitations of our studies are perhaps the small number of patients harboring the rare haplotypes, preventing potential significant associations with age and race to be uncovered. Additionally, our cohort of European breast cancer patients heterozygous for BRCA1 coding region mutations, are mostly western European Caucasian, with a small percentage possibly of mixed European descent. The ethnically narrow group may have limited the findings of our rare haplotypes among BRCA1 mutation carriers. In addition, we did not have tumor and germline DNA from the same individuals, making it impossible to know definitively if the rare haplotypes could be tumor acquired or if they were germline. However, identification of the rare haplotypes in DNA from both germline DNA as well as tumor DNA as well as the high association of our rare haplotypes with breast cancer makes our findings strongly statistically significant. This study provides evidence that the 3′UTR variant, rs8176318, is a marker for breast cancer risk for AA women, and is a marker for TN breast cancer, and that rare haplotypes including this and other 3′UTR variants may be genetic markers of an increased risk of developing breast cancer.
Materials and Methods
Ethics statement/study populations.
After approval from the Human Investigation Committee at Yale, samples from patients with breast cancer receiving treatment at Yale/New Haven Hospital (New Haven, CT) were collected from a total of 221 consenting individuals and samples consisted of 180 tumor FFPE and 41 germline DNA sources (81.4% and 18.6%, respectively) on HIC protocol # 0805003789. FFPE DNA was extracted from fresh frozen tumor biopsies containing over 80% tumor tissue. Germline DNA samples were collected from 22 blood and 19 saliva sources (53.7% and 46.3%, respectively). Patient data were collected including age, ethnicity and family history of cancer. Breast cancer subtypes were established by pathologic classification. Full receptor status (ER, PR and HER2) was documented on all samples using standard histopathology techniques. Women were excluded from studies evaluating breast cancer by subtype if there was insufficient documentation of the pathologic classification. An additional group of 27 control individuals (11 European Americans and 16 African Americans) were recruited from Yale/New Haven Hospital for BRCA1 haplotype analysis and included people without any personal history of cancer except non-melanoma skin cancer. All samples were saliva samples. Information including age, ethnicity and family history was recorded. For BRCA1 3′UTR analysis of genotype and cancer association we used 194 germline DNA controls (92 European Americans and 102 African Americans) and 205 tumor FFPE and germline DNA samples from breast cancer patients with known tumor subtype and ethnicity. 129 unrelated BRCA1 mutation carriers with breast cancer were ascertained at Erasmus University Medical Center through the Rotterdam Family Cancer Clinic and DNA was isolated from peripheral blood samples as described below.
To determine the frequency of the SNP alleles, we used our resource at Yale University of 2,250 unrelated global control individuals representing 46 populations from around the world. This resource is well documented among genetic studies and details can be found there.Citation22,Citation31–Citation33 All subjects gave informed consent under protocols approved by the committees governing human subjects research relevant to each of the population samples. Sample descriptions and sample sizes can be found in the Allele Frequency Database by searching for the population names (alfred.med.yale.edu) and in a previous publication.Citation29 DNA samples were extracted from lymphoblastoid cell lines established and/or grown in the Yale University Laboratory of Kenneth K. Kidd and J.R.K. The methods of transformation, cell culture and DNA purification have been described in reference Citation34. All volunteers were apparently normal and otherwise healthy adult males or females and samples were collected after receipt of appropriate informed consent under protocols approved by all relevant institutional review boards.
Evaluation of 3′UTR sequences.
DNA was isolated from frozen and FFPE tumor breast tissue (4 patients) using RecoverAll Total Nucleic Acid Isolation Kit (Ambion), and from blood and saliva (35 patients) using the DNeasy Blood and Tissue kit (Qiagen). The whole 3′UTR of BRCA1 (1,381 base pairs) was amplified in 39 breast cancer cases using KOD Hot Start DNA polymerase (Novagen) and DNA primers specific to this sequence: BRCA1: 5′-GAG CTG GAC ACC TAC CTG AT-3′ and 5′-GAG AAA GTC GGC TGG CCT A-3′. PCR products were purified using the QIAquick PCR purification kit (Quiagen) and sequenced using nested primers: BRCA1: 5′-CCT ACC TGA TAC CCC AGA TC-3′ and 5′-GGC CTA AGT CTC AAG AAC AGT C-3′.
Marker typing.
For high throughput genotyping, TaqMan 5′ nuclease assays (Applied Biosystems) were designed specifically to identify alleles at each SNP location. We determined the ancestral states of the 8 SNPs employed (rs9911630, rs12516, rs8176318, rs3092995, rs1060915, rs799912, rs9908805, rs17599948) by using the same TaqMan assays to genotype genomic DNA for non-human primates—3 bonobos (Pan paniscus), 3 chimpanzees (Pan troglodytes), 3 gibbons (Hylobates), 3 gorillas (Gorilla gorilla) and 3 orangutans (Pongo pygmaeus).
Statistics.
Frequencies of genotypes across populations were compared using Chi-Square Test of Association and Fisher Exact probability test. Significance of haplotype data was evaluated using Chi-Square Test of Association. p values were considered statistically significant if p > 0.05. All sites within the haplotype are in accordance with Hardy-Weinberg equilibrium among controls within each ethnic group. We used PHASE (software for haplotype reconstruction and recombination rate estimation from population data) to infer haplotypes of patients and control individualsCitation35,Citation36 without subpopulation information. PHASE software provides estimates of the certainty of haplotype assignment. In view of the fairly simple haplotype structure of the BRCA1 gene, the PHASE algorithm was extremely accurate. Of the haplotypes that did need to be estimated, PHASE estimated our cohort with 99% absolute certainty.
Conflict of Interest
Dr. Weidhaas and Dr. Slack have founded a company, Mira Dx, which has licensed IP related to findings reported in this manuscript.
Figures and Tables
Figure 1 (A) Global population-specific frequencies of the derived alleles at three BRCA1 3′UTR SNPs. Populations are listed across the X-axis and the corresponding frequency for the derived alleles at each SNP is shown on the y-axis: rs12516 allele A, rs8176318 allele A, and rs3092995 allele G. The SNPs were genotyped in 2,250 individuals. They represent 46 populations from around the world which are categorized based on geography: Africa; Europe, Southwest Asia and western Siberia; South Central Asia, East Asia, and the Pacific; and the Americas (left to right). (B) BRCA1 3′UTR allele frequencies among breast cancer patients by ethnicity. Populations are listed across the X-axis and the corresponding allele frequency for each genotyped SNP is shown on the y-axis. Both global control and breast cancer populations are listed across the X-axis by ethnicity and the corresponding allele frequency for the derived allele at each SNP is shown on the y-axis: rs12516 allele A, rs8176318 allele A, and rs3092995 allele G. The SNPs were examined in 335 individuals, including control populations and breast cancer populations grouped as European Americans and African Americans (left to right).
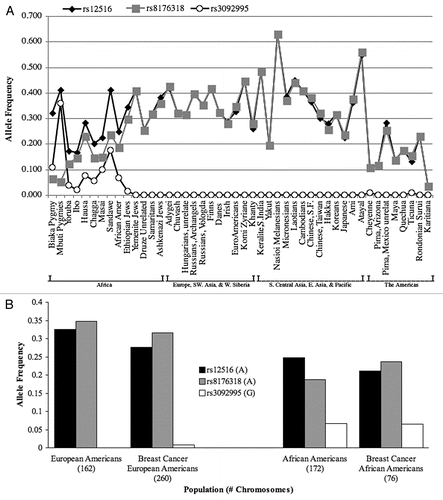
Figure 2 (A) BRCA1 global haplotype frequencies. The frequencies observed are based on 46 populations, 2,250 individuals and 8 SNPs spanning 267 kb. The 11 most common haplotypes are displayed as averages for the populations in each of the major geographic regions. The remaining haplotype frequencies with non-zero estimates are combined into the residual class. The three 3′UTR polymorphisms are displayed in red font and the derived alleles within the 3′UTR are underlined. (B) Proposed evolution of BRCA1 region haplotypes. The ten haplotypes shown are those that occur with a frequency of at least 0.27% of the global population. The grey haplotype is the ancestral state as determined by non-human primate genotypes. The globally most common haplotype is orange. Alleles at positions two through four are highlighted in red and represent the three 3′UTR polymorphisms. The alleles located in positions two through four are underlined if a derived allele is present. The order of the two boxed-in haplotypes represented by pink and red coloring cannot be inferred with the SNPs employed. The two haplotypes represented by the two shades of green are found in all regions of the world. The yellow haplotype is found in regions of the new world only.
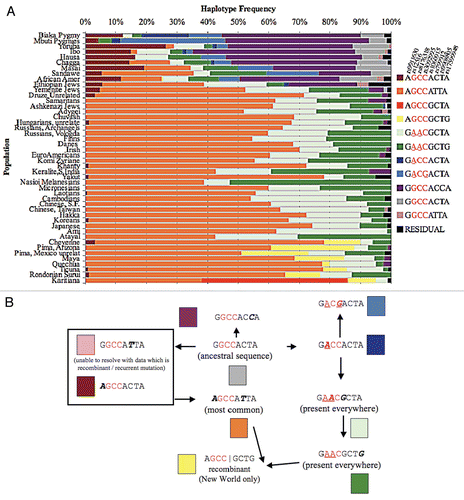
Figure 3 (A) BRCA1 rare haplotype freqeuncies among breast cancer patients. Breast cancer patients were evaluated for haplotypes found to be rare among global control populations but common in breast cancer patients. The five rare haplotype frequencies are displayed along the Y-axis. (B) BRCA1 haplotype freqeuncies among breast cancer by ethnicity. European and African American breast cancer patients were evaluated for haplotype frequencies. Non-cancerous European Americans and African Americans were added as controls. Nine common haplotypes are shown. Five additional rare haplotypes found in breast cancer patients are shown. The remaining haplotype frequencies with non-zero estimates are combined into the residual class. The three 3′UTR polymorphisms are displayed in red font and the derived alleles within the 3′UTR are underlined. Asterisks represent the 5 rare haplotypes in the chart. Additionally, they are boxed in red on the key to the right of the figure.
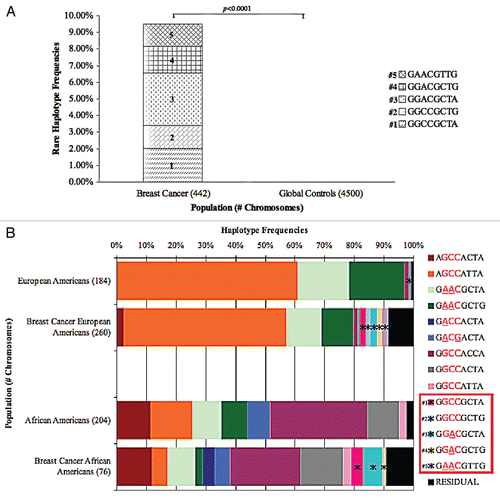
Figure 4 (A) BRCA1 rare haplotype frequencies among breast cancer patients by subtype. Breast cancer patients were grouped by subtype and evaluated for the identified rare haplotypes. The five rare haplotype frequencies are displayed along the Y-axis. (B) Rare Haplotype frequencies by breast cancer subtype and ethnicity. European and African American breast cancer patients were further grouped by breast tumor subtype and evaluated for rare haplotype frequencies. European Americans and African Americans were added as controls.
Table 1 The BRCA1 3′UTR SNP rs8176318 and breast cancer association by ethnicity and breast cancer subtype
Table 2 Eight Polymorphisms studied spanning 267 kb and encompassing BRCA1
Additional material
Download Zip (193.2 KB)Acknowledgements
This work was supported by multiple funding sources. J.B.W. was supported by a National Institute of Health (NIH) K08 grant [CA124484], J.B.W., F.S. and C.P. by a R01 from NIH [CA131301-01A1], and W.C.S. and K.K.K. by a P01 from NIH [GM057672]. The work was supported by the Shannon Family Charitable Fund, and a Clinical and Translational Science Award (CTSA) Grant Number [UL1 RR024139] from the National Center for Research Resources (NCRR), a component of the NIH, and NIH roadmap for Medical Research and the Radiation Therapy Oncology Group (RTOG) Translational Research Program, funded through grant [U10CA21661] by the National Cancer Institute (NCI).
References
- Bradbury AR, Olopade OI. Genetic susceptibility to breast cancer. Rev Endocr Metab Disord 2007; 8:255 - 267
- Edlich RF, Winters KL, Lin KY. Breast cancer and ovarian cancer genetics. J Long Term Eff Med Implants 2005; 15:533 - 545
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumors. Nature 2000; 406:747 - 752
- Irvin WJ Jr, Carey LA. What is triple-negative breast cancer?. Eur J Cancer 2008; 44:2799 - 2805
- Anders C, Carey LA. Understanding and treating triple-negative breast cancer. Oncology (Williston Park) 2008; 22:1233 - 1239
- O'Donovan PJ, Livingston DM. BRCA1 and BRCA2: breast/ovarian cancer susceptibility gene products and participants in DNA double-strand break repair. Carcinogenesis 2010; 31:961 - 967
- Atchley DP, Albarracin CT, Lopez A, Valero V, Amos CI, Gonzalez-Angulo AM, et al. Clinical and pathologic characteristics of patients with BRCA-positive and BRCA-negative breast cancer. J Clin Oncol 2008; 26:4282 - 4288
- Tung N, Wang Y, Collins LC, Kaplan J, Li H, Gelman R, et al. Estrogen receptor positive breast cancers in BRCA1 mutation carriers: clinical risk factors and pathologic features. Breast Cancer Res 2010; 12:12
- Lakhani SR, Van De Vijver MJ, Jacquemier J, Anderson TJ, Osin PP, McGuffog L, et al. The pathology of familial breast cancer: Predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2 and p53 in patients with mutations in BRCA1 and BRCA2. J Clin Oncol 2002; 20:2310 - 2318
- Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nature Rev Cancer 2004; 4:814 - 819
- Young SR, Pilarski RT, Donenberg T, Shapiro C, Hammond LS, Miller J, et al. The prevalence of BRCA1 mutations among young women with triple-negative breast cancer. BMC Cancer 2009; 9:86
- Malone KE, Daling JR, Doody DR, Hsu L, Bernstein L, Coates RJ, et al. Prevalence and predictors of BRCA1 and BRCA2 mutations in a population-based study of breast cancer in white and black American women ages 35 to 64 years. Cancer Res 2006; 66:8297 - 8308
- Nanda R, Schumm LP, Cummings S, Fackenthal JD, Sveen L, Ademuyiwa F, et al. Genetic testing in an ethnically diverse cohort of high-risk women: A comparative analysis of BRCA1 and BRCA2 mutations in American families of European and African ancestry. JAMA 2005; 294:1925 - 1933
- Akey J, Jin L, Xiong M. Haplotypes vs. single marker linkage disequilibrium tests: what do we gain?. Eur J Hum Genet 2001; 9:291 - 300
- Zhang K, Calabrese P, Nordborg M, Sun F. Haplotype block structure and its applications to association studies: Power and study designs. Am J Hum Genet 2002; 71:1386 - 1394
- Cox DG, Kraft P, Hankinson SE, Hunter DJ. Haplotype analysis of common variants in the BRCA1 gene and risk of sporadic breast cancer. Breast Cancer Res 2005; 7:171 - 175
- Freedman ML, Penney KL, Stram DO, Riley S, McKean-Cowdin R, Le Marchand L, et al. A haplo-type-based case-control study of BRCA1 and sporadic breast cancer risk. Cancer Res 2005; 65:7516 - 7522
- Dunning AM, Chiano M, Smith NR, Dearden J, Gore M, Oakes S, et al. Common BRCA1 variants and susceptibility to breast and ovarian cancer in the general population. Human molecular genetics 1997; 6:285 - 289
- Bau DT, Fu YP, Chen ST, Cheng TC, Yu JC, Wu PE, et al. Breast cancer risk and the DNA double-strand break end-joining capacity of nonhomologous end-joining genes are affected by BRCA1. Cancer Res 2004; 64:5013 - 5019
- Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nature reviews 2006; 6:259 - 269
- Chen K, Song F, Calin GA, Wei Q, Hao X, Zhang W. Polymorphisms in microRNA targets: a gold mine for molecular epidemiology. Carcinogenesis 2008; 29:1306 - 1311
- Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res 2008; 68:8535 - 8540
- Landi D, Gemignani F, Naccarati A, Pardini B, Vodicka P, Vodickova L, et al. Polymorphisms within micro-RNA-binding sites and risk of sporadic colorectal cancer. Carcinogenesis 2008; 29:579 - 584
- Pongsavee M, Yamkamon V, Dakeng S, Oc P, Smith DR, Saunders GF, et al. The BRCA1 3′-UTR: 5711 + 421T/T_5711 + 1286T/T genotype is a possible breast and ovarian cancer risk factor. Genet Test Mol Biomarkers 2009; 13:307 - 317
- Newman B, Mu H, Butler LM, Millikan RC, Moorman PG, King MC. Frequency of breast cancer attributable to BRCA1 in a population-based series of American women. JAMA 1998; 279:915 - 921
- Kidd JR, Speed WC, Pakstis AJ, Kidd KK. A 100 kb block encompassing BRCA1. (abstract/program #58) 2003; 53rd Annual Meeting of The American Society of Human Genetics November 4–8th 2003 Los Angeles, California
- Bonnen PE, Wang PJ, Kimmel M, Chakraborty R, Nelson DL. Haplotype and linkage disequilibrium architecture for human cancer-associated genes. Genome Res 2002; 12:1846 - 1853
- Ribas G, Milne RL, Gonzalez-Neira A, Benitez J. Haplotype patterns in cancer-related genes with long-range linkage disequilibrium: No evidence of association with breast cancer or positive selection. Eur J Hum Genet 2008; 16:252 - 260
- Cheung KH, Osier MV, Kidd JR, Pakstis AJ, Miller PL, Kidd KK. ALFRED: An allele frequency database for diverse populations and DNA polymorphisms. Nucleic Acids Res 2000; 28:361 - 363
- Gluz O, Liedtke C, Gottschalk N, Pusztai L, Nitz U, Harbeck N. Triple-negative breast cancer—current status and future directions. Ann Oncol 2009; 20:1913 - 1927
- Speed WC, Kang SP, Tuck DP, Harris LN, Kidd KK. Global variation in CYP2C8-CYP2C9 functional haplotypes. Pharmacogenomics J 2009; 9:283 - 290
- Speed WC, O'Roak BJ, Tarnok Z, Barta C, Pakstis AJ, State MW, et al. Haplotype evolution of SLITRK1, a candidate gene for Gilles de la Tourette syndrome. Am J Med Genet B Neuropsychiatr Genet 2008; 147:463 - 466
- Yamtich J, Speed WC, Straka E, Kidd JR, Sweasy JB, Kidd KK. Population-specific variation in haplotype composition and heterozygosity at the POLB locus. DNA Repair 2009; 8:579 - 584
- Anderson MA, Gusella JF. Use of cyclosporin A in establishing Epstein-Barr virus-transformed human lymphoblastoid cell lines. In vitro 1984; 20:856 - 858
- Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 2003; 73:1162 - 1169
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 2001; 68:978 - 989