Abstract
Musashi-mediated mRNA translational control has been implicated in the promotion of physiological and pathological stem cell proliferation. During self-renewal of mammalian stem cells, Musashi has been proposed to act to repress the translation of mRNAs encoding inhibitors of cell cycle progression. By contrast, in maturing Xenopus oocytes Musashi activates translation of target mRNAs that encode proteins promoting cell cycle progression. The mechanisms directing Musashi to differentially control mRNA translation in mammalian stem cells and Xenopus oocytes is unknown. In this study, we demonstrate that the mechanisms defining Musashi function lie within the cellular context. Specifically, we show that murine Musashi acts as an activator of translation in maturing Xenopus oocytes while Xenopus Musashi functions as a repressor of target mRNA translation in mammalian cells. We further demonstrate that within the context of a primary mammalian neural stem/progenitor cell, Musashi can be converted from a repressor of mRNA translation to an activator of translation in response to extracellular stimuli. We present current models of Musashi-mediated mRNA translational control and discuss possible mechanisms for regulating Musashi function. An understanding of these mechanisms presents exciting possibilities for development of therapeutic targets to control physiological and pathological stem cell proliferation.
Musashi Promotes Stem Cell Self Renewal
The Musashi mRNA-binding protein plays a critical role in the promotion of stem cell self-renewal by repressing the translation of mRNAs encoding proteins that inhibit cell cycle progression.Citation1,Citation2 Musashi was originally identified as a critical regulator of asymmetric cell division in Drosophila sensory organ precursor cells and subsequently, mammalian Musashi isoforms have been implicated in the self-renewal of neural, epithelial and hematopoietic stem and progenitor cells.Citation2–Citation8 Musashi has been also implicated in proliferative pathologies in various tissues where Musashi may be acting to promote self-renewal of tumor cells with stem cell-like properties.Citation2,Citation7,Citation9–Citation19 Despite indications of a pivotal role in physiological and pathological stem cell proliferation, little is known about the mechanisms by which Musashi regulates mRNA translation or how Musashi function is regulated. Recent findings have demonstrated that Musashi can unexpectedly act as an mRNA translational activator, rather than a translational repressor, in promotion of cell cycle progression in Xenopus oocytes.Citation20,Citation21 This study attempts to reconcile these apparently opposing roles for Musashi function in mRNA translational control during cell cycle progression and previews areas of promising research into the mechanism and regulation of Musashi function.
Musashi Cell Context-Specific mRNA Targets
Two Musashi isoforms are present in vertebrates, Musashi1 and Musashi2. While Musashi2 is ubiquitously expressed, Musashi1 shows more restricted expression with particular enrichment in embryonic and adult neural stem cells.Citation1 The Musashi proteins contain two N-terminal RNA recognition motifs (RRMs) that bind to target mRNAs through a Musashi binding element (MBE, (G/A)U1–3AGU) in the mRNA 3′ untranslated region.Citation22,Citation23 The Musashi1 and Musashi2 isoforms are highly related in sequence within the RRM domains (>90% similarity at the amino acid level), suggesting they may interact with the same target mRNAs. Although very few Musashi-target mRNA have been definitively identified, a recent microarray analysis of co-associated mRNAs indicate that Musashi may regulate a range of mRNAs encoding proteins involved in cell proliferation, cell differentiation and apoptosis.Citation24 Musashi2 has been shown to repress translation of the mRNA encoding the Notch signaling inhibitor Numb, in malignant hematopoietic cells and Musashi1 can repress Numb in NIH3T3 cells.Citation18,Citation22 In P19 embryonal carcinoma cells Musashi1 represses translation of the mRNA encoding the cyclin-dependent kinase (CDK) inhibitor, p21WAF-1.Citation25 Both Numb and p21WAF-1 enforce inhibition of cell cycle progression, promote cell cycle exit and commitment of progenitor cells to differentiate.Citation26,Citation27 By repressing the translation of the mRNAs encoding these inhibitory proteins, Musashi functions to maintain cells in an undifferentiated state capable of self-renewal ().
In oocytes of the frog Xenopus laevis, Musashi has been shown to direct the activation of the mRNAs encoding the MAP kinase kinase, Mos and cyclin B5 resulting in MAP kinase- and CDK-mediated promotion of cell cycle progression.Citation20,Citation21 Inhibition of Musashi function in oocytes demonstrates that Musashi is a master regulator of oocyte maternal mRNA translational recruitment and functions upstream of subsequent mRNA translation controlled by cytoplasmic polyadenylation elements (CPEs) and the CPE binding protein, CPEB.Citation28 Thus, in stem cells and oocytes Musashi acts by exerting opposite control on target mRNA translation to promote cell cycle progression. It is unclear at this time whether Musashi functions in mRNA translational control in differentiating stem cells or in immature oocytes ().
Musashi-Directed Repression versus Activation of Target mRNA Translation
Very little is known concerning the mechanisms by which Musashi exerts mRNA translational control. A recent breakthrough demonstrated that Musashi exerts mRNA translational repression through association with the poly [A] binding protein (PABP) to preclude PABP interaction with the eIF4G initiation factor.Citation29 As a consequence, Musashi is thought to disrupt recruitment of the large ribosomal subunit and prevent target mRNA translation. Consistent with this inhibitory role, a Musashi mutant protein deleted for the identified PABP interaction domain was shown to be compromised for mRNA translational repression in a rabbit reticulocyte lysate reporter assay.Citation29 Future studies will be necessary to access the contribution of this mechanism to control of Musashi target mRNAs in stem cells.
The mechanism by which Musashi directs mRNA translational activation, rather than translational repression, in Xenopus oocytes is currently unknown. The Xenopus and mammalian Musashi1 protein sequences are highly conserved overall (94% amino acid identity), suggesting that the differential activity is not likely to be due to species-specific differences in the proteins. We sought to verify the functional conservation of Musashi protein from Xenopus and mouse by experimentally reconstituting Musashi activity in the appropriate heterologous system. In Xenopus oocytes, endogenous Musashi function was knocked down through injection of antisense oligonucleotides targeting the mRNAs encoding both the Musashi1 and Musashi2 protein isoforms as described in reference Citation20. We have previously demonstrated that this knockdown strategy caused failure to polyadenylate and subsequently translate Musashi target mRNAs and that both target mRNA polyadenylation (a requirement for translational activation) and cell cycle progression can be rescued by ectopic expression of wild-type Xenopus Musashi1 protein.Citation20 Expression of mouse Musashi1 rescued cell cycle progression () and polyadenylation of the Musashi-regulated cyclin B5 mRNA () and Mos mRNA (data not shown) in Musashi antisense oligonucleotide knock-down Xenopus oocytes. These findings indicate that the mammalian Musashi1 protein can function to activate mRNA translation in Xenopus oocytes. The rescue of activation of mRNA translation by ectopic mammalian Musashi1 required progesterone stimulation indicating that the mammalian Musashi1 protein is regulated through the same progesterone-stimulated mechanisms that control Xenopus Musashi.
In a complementary experiment, we sought to determine whether the Xenopus Musashi1 protein could function to repress translation of a target mRNA in mammalian NIH3T3 cells. NIH3T3 cells do not express endogenous Musashi and thus offer a null background to reconstitute and test Musashi-dependent mRNA translational regulation.Citation22 To address the role of Musashi-mediated translational control in mammalian cells, we have developed a mRNA reporter by linking a β-globin 3′ UTR containing the Xenopus Mos mRNA MBECitation21 to the Firefly luciferase coding sequence within the mammalian pcDNA expression vector (designated Fluc-MBE). As specificity controls, a reporter containing mutational disruption of the consensus Musashi binding element (AUAGU mutated to AUccU; Fluc-MBE mtCitation21,Citation22) or the 3′ UTR lacking the MBE (Fluc-Con) were utilized. NIH3T3 cells were transiently co-transfected with a plasmid expressing one of the Fluc reporter mRNAs, a plasmid expressing either GST tagged murine Musashi1 (mMsi) or GST tagged Xenopus Musashi1 (xMsi), as well as a plasmid encoding Renilla luciferase under the control of the unregulated SV40 3′ UTR (designated Rluc). Following 24 hours of culture, cells were lysed and assayed for Firefly and Renilla luciferase activity using the Dual-luciferase assay kit (Promega) with a Turner Biosytems 20/20 luminometer. Measurement of Renilla luciferase activity allows normalization of transfection efficiency (Fluc/Rluc) between different samples. Expression of Xenopus Musashi resulted in a selective inhibition of translation of the Fluc-MBE reporter compared to the translation of the control Firefly luciferase reporter (; 9% of control reporter ±2% SEM, n = 3, p < 0.001). Under the same experimental conditions expression of murine Musashi1 in NIH3T3 cells also resulted in efficient repression of the MBE reporter (8% of control reporter ±1% SEM, n = 3, p < 0.001), consistent with prior findings.Citation22 No significant repression of the Fluc-MBE mt was observed with either mMsi or xMsi, demonstrating the requirement for interaction of Musashi with the MBE in the 3′ UTR. Semi-quantitative PCR analysis of the samples confirmed that the different Fluc reporter mRNAs were expressed at equivalent levels () and western blotting with GST antibody confirmed equivalent expression of the mMsi and xMsi proteins (data not shown). We conclude that the differential activity of Musashi in Xenopus oocytes and in mammalian cells is not due to species-specific differences in the protein. Moreover, these findings reveal that the mammalian Musashi protein is capable of translational activation in the correct cellular context.
Flipping the Switch: Mammalian Musashi can Convert from a Repressor to an Activator of Target mRNA Translation during Differentiation of Neural Stem/progenitor Cells
The available evidence suggests that the Musashi target mRNAs p21 and Numb are de-repressed in differentiating neurons.Citation25,Citation30 However, the mechanism by which Musashi function may be altered in response to extracellular cues is unknown. The observations that Musashi1 protein levels are downregulated during neuronal differentiation and absent in mature neuronsCitation31 suggested that de-repression of target mRNAs could occur simply through degradation of Musashi protein. However, there is evidence that ELAV proteins stabilize the Musashi1 mRNA resulting in increased Musashi1 protein levels during early neuronal differentiation of human neuroblastoma cells.Citation32 Further, neuronal differentiation of mouse P19 embryonal carcinoma cells is accompanied by an initial transient increase in Musashi1 protein at a time coincident with endogenous p21WAF-1 mRNA translation.Citation25 The observation that knockout of Musashi1 influences the ratio of differentiated neural cell types from multipotent progenitors also supports an active role for Musashi in differentiation.Citation4 In addition, Musashi-dependent activation of Robo3/Rig-1 mRNA translation during axonal midline crossing in precerebellar neurons has recently been reported.Citation33 These observations suggest that translation of Musashi target mRNAs can occur via modification of Musashi function, independently of Musashi protein degradation.
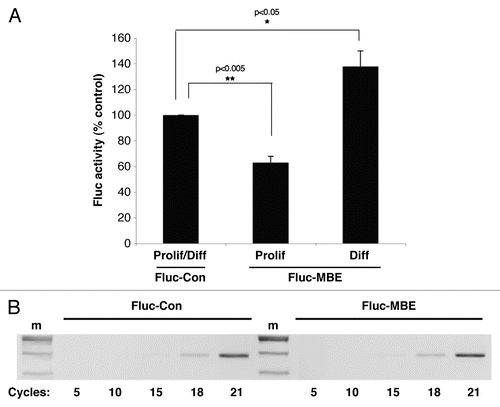
We sought to directly assess Musashi function during early phases of neuronal differentiation. Primary rat neural stem/progenitor cells (NSPCs) were transiently co-transfected with a plasmid encoding the Renilla luciferase reporter and one the Firefly luciferase reporters described for . In cells maintained in proliferative media for 24 hours, we observed that translation of the Fluc-MBE reporter was significantly repressed (63% compared to translation of the control reporter, ±5% SEM n = 3, p < 0.005, ). This observation indicates that endogenous Musashi represses mRNA translation in proliferating primary NSPCs, similar to what has been observed in proliferating P19 embryonal carcinoma cells.Citation25 By contrast, in NSPCs exposed to differentiation conditions for 24 hours, translation of the Fluc-MBE reporter was not repressed but rather was activated above the levels of the control (138% of control reporter ±12.4 SEM, n = 3, p < 0.05, ). Thus, during early phases of neuronal differentiation, Musashi exerts stimulus-dependent activation of target mRNAs, similar to the translational activation of Musashi target mRNAs during Xenopus oocyte maturation.Citation20,Citation21 The Fluc reporter mRNAs were expressed at equivalent levels ().
These findings indicate that Musashi can function both to repress and to activate mRNA translation in NSPCs depending on extracellular cues. The stimulus-dependent mechanism that converts Musashi from a repressor of target mRNA translation to an activator of mRNA translation during differentiation of NSPCs is not known. Musashi may switch from a repressor to an activator through post-translational modification of the Musashi protein and/or altered association with protein co-factors by analogy to the well characterized CPEB protein, which converts from a repressor to an activator of mRNA translation in response to extracellular cues in both Xenopus oocytes and mammalian neurons.Citation34 In this regard, it is possible that the switch from translational repressor to translational activator may involve dissociation of Musashi from PABP or modification of PABP function in differentiating NSPCs. Irrespective of the precise underlying molecular regulatory mechanisms, it is clear that the function of Musashi is determined not just by the cell type, but also by exposure to environmental signals.
Conclusions
The emerging mechanistic insights indicate that Musashi is a complex multifunctional protein that is dynamically regulated in response to extracellular stimuli. Musashi can promote cell cycle progression in a diverse range of cell types through various target mRNAs and in a context-dependent manner. A critical avenue for future investigation will be to elucidate the factors that determine whether Musashi acts to repress or to activate target mRNA translation and to determine how hormone and growth factor signals impinge upon the regulation of these factors. Since Musashi-mediated mRNA repression has been implicated in both physiological stem cell and pathological stem cell-like proliferation, an understanding of how Musashi can switch from a repressor to an activator of mRNA translation may allow for development of therapeutic targets to control this regulatory switch in Musashi function.
Abbreviations
| MBE | = | musashi binding element |
| CPE | = | cytoplasmic polyadenylation element |
| CPEB | = | CPE-binding protein |
| CDK | = | cyclin-dependent kinase |
| PABP | = | poly[A] binding protein |
| eIF4G | = | eukaryotic initiation factor 4G |
| 3′ UTR | = | 3′ untranslated region |
| RRM | = | RNA recognition motif |
| SV40 | = | simian virus 40 |
| SEM | = | standard error of the mean |
| NSPC | = | neural stem/progenitor cells |
Figures and Tables
Figure 1 Musashi regulates cell cycle progression through distinct translational control mechanisms in oocytes and in stem/progenitor cells. Musashi promotes cell cycle progression in mammalian stem cells by repression of mRNA translation (top). Similarly, Musashi promotes cell cycle progression in Xenopus oocytes, although by activation rather than repression of mRNA translation (bottom). It is unclear whether Musashi plays a role in during arrest of cell cycle progression in either differentiating stem cells or in immature oocytes.
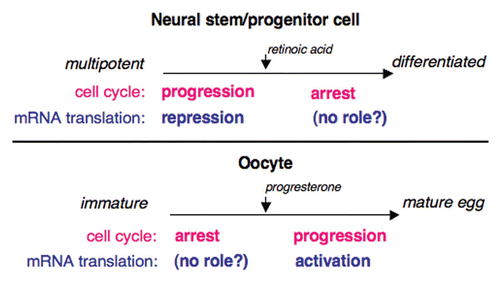
Figure 2 Mammalian Musashi1 functionally compensates for Xenopus Musashi in mediating cell cycle progression in Xenopus oocytes. (A) Progesterone-stimulated cell cycle progression is assayed as percent of oocytes that undergo germinal vesicle breakdown (maturation). Knockdown of Musashi function in Xenopus oocytes, through injection of antisense oligonucleotides targeting the mRNAs encoding endogenous Musashi1 and Musashi2 isoforms, results in inhibition of cell cycle progression (Msi AS). All procedures have been previously described in reference Citation20. Injection of mRNA (23 ng/oocyte) encoding mammalian Musashi1 rescues cell cycle progression in Musashi antisense-treated oocytes (Msi AS + mMsi). Over 50 oocytes were scored for each condition and error bars represent SEM from three independent experiments. Injection of control antisense oligonucleotide did not block cell cycle progression (Con AS). The rescue of cell cycle progression was highly significant (p < 0.005, Student's t-test). (B) Control antisense (Con AS) or Musashi antisense (Msi AS) oligonucleotide injected oocytes were treated with (+) or without (−) progesterone and analyzed for polyadenylation of endogenous cyclin B5 mRNA by RNA ligation-coupled PCR as described in reference Citation20. A progesterone-dependent heterogeneous increase in the size of the mRNA population in Con AS oocytes and Msi AS oocytes injected with mammalian Musashi1 (Msi AS + mMsi) is indicative of polyadenylation and translational activation (bracketed).
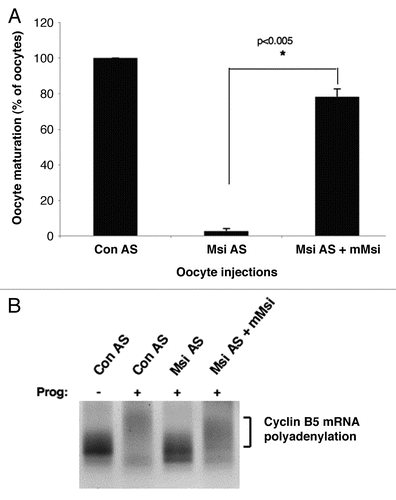
Figure 3 Xenopus Musashi1 represses target mRNA translation in mammalian cells. (A) Mammalian NIH3T3 cells were transiently co-transfected with plasmids encoding Renilla luciferase; a Firefly luciferase fused to either a control 3′ UTR (Con), a 3′ UTR containing a MBE or a 3′ UTR containing a mutant MBE (MBEmt); and either a GST tagged mammalian Musashi1 (mMsi) or GST tagged Xenopus Musashi1 (xMsi) protein. 24 hours after transfection, the cells were lysed and samples prepared for both protein and total RNA analyses. For each protein sample, luciferase activity was measured in triplicate with the Dual-Luciferase assay system and the values normalized as previously described in reference Citation20. Values are shown relative to the Firefly luciferase fused to the control 3′ UTR, arbitrarily set to 100%. Error bars represent SEM from three independent experiments. Repression of the Fluc-MBE reporter by mMsi and xMsi was significant (p < 0.001, Student's t-test). (B) The levels of Firefly luciferase reporter mRNA were determined using semi-quantitative PCR. Total RNA was prepared from the same samples used in (A) and Firefly reporter mRNA was PCR-amplified for different cycle numbers as indicated. The PCR products were visualized after separation through a 2% agarose gel. No significant differences in stability of the Fluc-Con, -MBE or -MBEmt constructs were detected in either mMsi (left) or xMsi (right) co-transfected cells. m, indicates DNA marker lane.
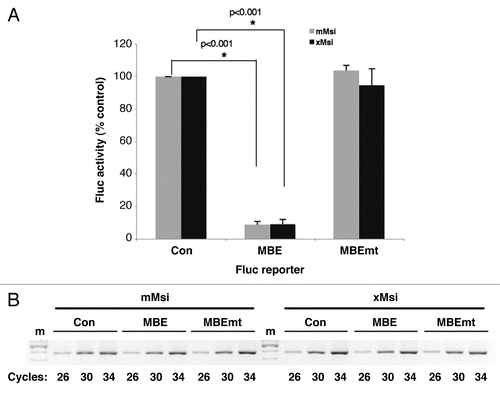
Figure 4 Musashi activates mRNA translation in differentiating neural stem/progenitor cells. Primary neural stem/progenitor cells (NSPCs) were cultured from embryonic rat hippocampal/cortical tissue (Genlantis, San Diego) and proliferation of the NSPCs established through assay of neurosphere formation.Citation35 Differentiation was induced by plating enzymatically and mechanically dispersed single cells on poly-ornithine/fibronectin-coated dishes in the absence of bFGF and EGF and in the presence of retinoic acid.Citation36 Dispersed single NSPCs were co-transfected with plasmids encoding Renilla luciferase and either a Firefly luciferase fused to either a control 3′ UTR (Con) or a 3′ UTR containing a MBE (MBE). Cells were then cultured in either proliferative (Prolif) or differentiation-inducing (Diff) media. 24 hours after the transfection, the cells were lysed and samples prepared for both luciferase activity and reporter mRNA expression as described in the legend to . (A) Luciferase values are shown relative to the Firefly luciferase fused to the control 3′ UTR, arbitrarily set to 100% for both proliferation and differentiation conditions (Prolif/Diff). Error bars represent SEM from three independent experiments. Repression of the Fluc-MBE reporter in proliferating NSPCs was significant (p < 0.005, Student's t-test) as was the activation of the Fluc-MBE reporter in differentiating NSPCs (p < 0.05, Student's t-test). (B) The levels of Firefly reporter mRNA were determined using semi-quantitative PCR and no significant differences in stability of the Fluc-Con or Fluc-MBE reporter mRNAs were detected in NSPCs. The Fluc-MBEmt reporter mRNA gave results indistinguishable from the Fluc-Con reporter (data not shown). m, indicates DNA marker lane.
Acknowledgements
We gratefully acknowledge the excellent technical assistance of Linda Hardy and thank Dr. A. Wilczynska for generation of the Fluc reporter mRNAs. A.M.M. was supported by the National Institutes of Health (HD35688), the American Cancer Society (RPG 101279), and the Arkansas BioSciences Institute; M.C.M. was supported by NIH grant RR020146.
References
- Nishimoto Y, Okano H. New insight into cancer therapeutics: Induction of differentiation by regulating the Musashi/Numb/Notch pathway. Cell Res 2010; 20:1083 - 1085
- Okano H, Kawahara H, Toriya M, Nakao K, Shibata S, Imai T. Function of RNA-binding protein Musashi-1 in stem cells. Exp Cell Res 2005; 306:349 - 356
- Nakamura M, Okano H, Blendy JA, Montell C. Musashi, neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron 1994; 13:67 - 81
- Sakakibara S, Nakamura Y, Yoshida T, Shibata S, Koike M, Takano H, et al. RNA-binding protein Musashi family: Roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proc Natl Acad Sci USA 2002; 99:15194 - 15199
- Asai R, Okano H, Yasugi S. Correlation between Musashi-1 and c-hairy-1 expression and cell proliferation activity in the developing intestine and stomach of both chicken and mouse. Dev Growth Differ 2005; 47:501 - 510
- Wang XY, Yin Y, Yuan H, Sakamaki T, Okano H, Glazer RI. Musashi1 modulates mammary progenitor cell expansion through proliferin-mediated activation of the Wnt and Notch pathways. Mol Cell Biol 2008; 28:3589 - 3599
- Kharas MG, Lengner CJ, Al-Shahrour F, Bullinger L, Ball B, Zaidi S, et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat Med 2010; 16:903 - 908
- Hope KJ, Cellot S, Ting SB, MacRae T, Mayotte N, Iscove NN, et al. An RNAi screen identifies Msi2 and Prox1 as having opposite roles in the regulation of hematopoietic stem cell activity. Cell Stem Cell 7:101 - 113
- Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci USA 2003; 100:15178 - 15183
- Kanemura Y, Mori K, Sakakibara S, Fujikawa H, Hayashi H, Nakano A, et al. Musashi1, an evolutionarily conserved neural RNA-binding protein, is a versatile marker of human glioma cells in determining their cellular origin, malignancy and proliferative activity. Differentiation 2001; 68:141 - 152
- Toda M, Iizuka Y, Yu W, Imai T, Ikeda E, Yoshida K, et al. Expression of the neural RNA-binding protein Musashi1 in human gliomas. Glia 2001; 34:1 - 7
- Yokota N, Mainprize TG, Taylor MD, Kohata T, Loreto M, Ueda S, Dura W, et al. Identification of differentially expressed and developmentally regulated genes in medulloblastoma using suppression subtraction hybridization. Oncogene 2004; 23:3444 - 3453
- Clarke RB, Spence K, Anderson E, Howell A, Okano H, Potten CS. A putative human breast stem cell population is enriched for steroid receptor-positive cells. Dev Biol 2005; 277:443 - 456
- Clarke RB, Anderson E, Howell A, Potten CS. Regulation of human breast epithelial stem cells. Cell Prolif 2003; 36:45 - 58
- Kayahara T, Sawada M, Takaishi S, Fukui H, Seno H, Fukuzawa H, et al. Candidate markers for stem and early progenitor cells, Musashi-1 and Hes1, are expressed in crypt base columnar cells of mouse small intestine. FEBS Lett 2003; 535:131 - 135
- Sanchez-Diaz PC, Burton TL, Burns SC, Hung JY, Penalva LO. Musashi1 modulates cell proliferation genes in the medulloblastoma cell line Daoy. BMC cancer 2008; 8:280
- Sureban SM, May R, George RJ, Dieckgraefe BK, McLeod HL, Ramalingam S, et al. Knockdown of RNA binding protein musashi-1 leads to tumor regression in vivo. Gastroenterology 2008; 134:1448 - 1458
- Ito T, Kwon HY, Zimdahl B, Congdon KL, Blum J, Lento WE, et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature 2010; 466:765 - 768
- Bobryshev YV, Freeman AK, Botelho NK, Tran D, Levert-Mignon AJ, Lord RV. Expression of the putative stem cell marker Musashi-1 in Barrett's esophagus and esophageal adenocarcinoma. Dis Esophagus 2010; 23:580 - 589
- Arumugam K, Wang Y, Hardy LL, MacNicol MC, MacNicol AM. Enforcing temporal control of maternal mRNA translation during oocyte cell cycle progression. EMBO J 2010; 29:387 - 397
- Charlesworth A, Wilczynska A, Thampi P, Cox LL, MacNicol AM. Musashi regulates the temporal order of mRNA translation during Xenopus oocyte maturation. EMBO J 2006; 25:2792 - 2801
- Imai T, Tokunaga A, Yoshida T, Hashimoto M, Mikoshiba K, Weinmaster G, et al. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol Cell Biol 2001; 21:3888 - 3900
- Miyanoiri Y, Kobayashi H, Imai T, Watanabe M, Nagata T, Uesugi S, et al. Origin of higher affinity to RNA of the N-terminal RNA-binding domain than that of the C-terminal one of a mouse neural protein, musashi1, as revealed by comparison of their structures, modes of interaction, surface electrostatic potentials and backbone dynamics. J Biol Chem 2003; 278:41309 - 41315
- de Sousa Abreu R, Sanchez-Diaz PC, Vogel C, Burns SC, Ko D, Burton TL, et al. Genomic analyses of musashi1 downstream targets show a strong association with cancer-related processes. J Biol Chem 2009; 284:12125 - 12135
- Battelli C, Nikopoulos GN, Mitchell JG, Verdi JM. The RNA-binding protein Musashi-1 regulates neural development through the translational repression of p21(WAF-1). Mol Cell Neurosci 2006; 31:85 - 96
- Wakamatsu Y, Maynard TM, Jones SU, Weston JA. NUMB localizes in the basal cortex of mitotic avian neuroepithelial cells and modulates neuronal differentiation by binding to NOTCH-1. Neuron 1999; 23:71 - 81
- Kabos P, Kabosova A, Neuman T. Blocking HES1 expression initiates GABAergic differentiation and induces the expression of p21(CIP1/WAF1) in human neural stem cells. J Biol Chem 2002; 277:8763 - 8766
- MacNicol MC, MacNicol AM. Developmental timing of mRNA translation—integration of distinct regulatory elements. Mol Reprod Dev 2010; 77:662 - 669
- Kawahara H, Imai T, Imataka H, Tsujimoto M, Matsumoto K, Okano H. Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP. J Cell Biol 2008; 181:639 - 653
- Okano H, Imai T, Okabe M. Musashi: A translational regulator of cell fate. J Cell Sci 2002; 115:1355 - 1359
- Sakakibara S, Okano H. Expression of neural RNA-binding proteins in the postnatal CNS: implications of their roles in neuronal and glial cell development. J Neurosci 1997; 17:8300 - 8312
- Ratti A, Fallini C, Cova L, Fantozzi R, Calzarossa C, Zennaro E, et al. A role for the ELAV RNA-binding proteins in neural stem cells: stabilization of Msi1 mRNA. J Cell Sci 2006; 119:1442 - 1452
- Kuwako K, Kakumoto K, Imai T, Igarashi M, Hamakubo T, Sakakibara S, et al. Neural RNA-binding protein Musashi1 controls midline crossing of precerebellar neurons through posttranscriptional regulation of Robo3/Rig-1 expression. Neuron 2010; 67:407 - 421
- Richter JD. CPEB: A life in translation. Trends Biochem Sci 2007; 32:279 - 285
- Nakamura Y, Sakakibara S, Miyata T, Ogawa M, Shimazaki T, Weiss S, et al. The bHLH gene hes1 as a repressor of the neuronal commitment of CNS stem cells. J Neurosci 2000; 20:283 - 293
- Cao QL, Howard RM, Dennison JB, Whittemore SR. Differentiation of engrafted neuronal-restricted precursor cells is inhibited in the traumatically injured spinal cord. Experimental neurology 2002; 177:349 - 359