Abstract
Recent reports have better elucidated the components of the Polycomb Repressive Complex 2 (PRC2) and its functional role in embryonic stem cells (ESCs) and their differentiated derivatives. The depletion of a newly described mammalian PRC2 associated protein, PCL2, leads to an increase in ESC self-renewal and a delay in differentiation, a phenotype similar to knockouts of the core PRC2 members. Genomic and cell biology data suggest that PCL2 is important in cell fate decisions and may play a role in recruitment of PRC2 to target genes and histone methylation. Importantly, depletion of PCL2 in ESCs leads to a decrease in 3meH3K27 at the proximal promoter regions of pluripotency transcription factors Tbx3, Klf4, Foxd3 and a concomitant increase in gene expression. These proteins subsequently activate expression of Oct4, Nanog and Sox2 through a feed-forward gene regulatory circuit, altering the core pluripotency network and driving cell fate decisions towards self-renewal. We propose a model whereby alteration of the epigenetic state of Tbx3, Klf4, and Foxd3 results in the enforced expression of the pluripotency network, preventing differentiation.
Introduction
Polycomb group (PcG) proteins are known epigenetic transcriptional repressors that play critical roles maintaining cellular memory of fate decisions made during development. PcG proteins exist in two main complexes, and act to repress the expression of various target genes, including transcription factors (TFs). The Polycomb Repressive Complex 2 (PRC2) is comprised of a core group of proteins, EZH2, SUZ12 and EEDCitation1–Citation3 and executes its repressive role by modifying chromatin structure via histone methylation. Specifically, EZH2 is a histone methyltransferase, and is able to catalyze the di- and tri-methylation at H3K27. Enzymatic action by EZH2 at target genes requires the binding of SUZ12 and EED.Citation3 Recent developments have expanded the concept of a simple three member PRC2 complex to one that includes multiple associated proteins that may interact with the PRC2 complex to modify its enzymatic function, influence its stability or contribute to its targeting.
The embryonic stem cell (ESC) provides an excellent model to study the molecular mechanisms controlling cell fate. Response to internal or external stimuli leads to changes in molecular, genetic and epigenetic events which provide tightly-regulated control during self-renewal and differentiation. By incorporating genomic and proteomic datasets with cell biological events such as cell signaling or self-renewal, integrated gene regulatory networks (GRNs) may be drafted, enabling the identification of novel interactions as well as identification of novel regulators (or nodes).
To better understand the molecular underpinnings of self-renewal and commitment of ESCs, we drafted and tested GRN modules.Citation4 Through this analysis, we identified Polycomb-like 2 (Pcl2, also known as Mtf2) as a candidate regulator of ESC fate. Pcl2 is highly expressed in ESCs and its promoter is bound by OCT4 and NANOG, suggesting that it is regulated by key pluripotency factors. Further, upon commitment to differentiation, we found that Pcl2 is downregulated in a similar fashion as Oct4, Sox2 and Nanog.Citation4 We now show in our recent study that PCL2 is critical for modulating GRNs involved in both ESC self-renewal and commitment. Our analyses show that PCL2 associates with the core components of the PRC2 complex and plays a critical role in ESC differentiation and self-renewal.Citation5 In this Extra Views article, we highlight our previous findings and propose a model of epigenetic modification by PRC2 and its associated proteins.
Association with the PRC2 Complex
It has been shown that PCL2 can associate with EZH2 in chick,Citation6 and that PCL associates with E(Z) in Drosophila.Citation7 PHF1, another member of the Polycomblike family was also shown to associate with PRC2 in fly and human.Citation8–Citation11 Similarly, our lab and others recently demonstrated by co-immunoprecipitation and mass spectrometry that mammalian PCL2 interacts with PRC2 core members SUZ12, EZH2 and EED.Citation5,Citation12,Citation13 The discovery of accessory proteins, such as PCL2 and JARID2, has expanded the original notion of a simple three member PRC2 complex to one that is much more complex and dynamic. While an association between JARID2 and PCL2 has been shown through mass spectrometry,Citation12,Citation14 there are conditions where this interaction failed to be captured,Citation13 allowing for the possibility that JARID2 and PCL2 may or may not coexist in the same PRC2 complex at any given time.
Protein Domains
The mechanism of PRC2 recruitment to its target genes is currently unknown and none of the core components have DNA-binding capability. Based on their protein domains, both PCL2 and JARID2 are candidates for such a role. Three distinct isoforms of PCL2 are found in ESCs, each of which shows conservation of two PHD (plant homeodomain type zinc finger) domains from Drosophila PCL (). JARID2 also contains a PHD domain along with an evolutionarily conserved ARID/BRIGHT domain, both of which have DNA-binding capabilities.Citation15
While the PHD protein domain is generally considered to convey DNA binding function, it has also been shown to bind RNA, proteins and lipid domains.Citation16,Citation17 The protein-protein interactions between PCL2 and the other PRC2 members identified by co-immunoprecipitation may be mediated through the PHD domains, although this will need to be confirmed using binding assays with deletion mutants. The PHD domain has also been associated with a chromatin binding, as it can specifically recognize methylated lysine residues, as observed in the NuRF protein targeting H3K4 areas.Citation18,Citation19 This feature may also aid in the targeting of PCL2 to its targets, since PRC2 is known to bind at highly methylated regions.
In addition to its 2 PHD domains, the two longest isoforms of PCL2 also contain a TUDOR domain (), which is known to aid in targeting proteins to methylated sites on the genome. TUDOR domains are thought to target proteins to methylated lysines during spliceosome assembly or recognition of histone tails. It may be the combination of PHD and TUDOR domains of PCL2 that function to associate with members of PRC2 and recruitment the complex to specific promoter targets based on sequence or histone modifications. Thus, each isoform of PCL2 may have specific protein- or DNA-binding targets. Each isoform has yet to be studied independently in detail, but it is likely that the shortest 56 kDa isoform plays a distinct role or has a unique set of targets, based on its lack of the TUDOR domain. The presence of a TUDOR domain is not found in other PRC2-interacting proteins such as JARID2, AEBP2, YY1 or RBBP4, and this may confer a unique role to PCL2.
The recruitment process has been well studied in Drosophila and Polycomb Response Elements (PREs) have been identified as locations on DNA to which PRC2 members are recruited. Invertebrate PREs are not simply one conserved sequence, but contain multiple PHO, Zeste and GAGA binding sites (reviewed in ref. Citation20). Despite the abundance of genome-wide promoter occupancy data for each of the PRC2 proteins, identification of analogous PREs in mammalian cells has proven unsuccessful. One potential mammalian targeting factor is YY1 (PHO homolog) and two groups have recently reported the identification of regions of mammalian DNA containing multiple YY1 binding sites which are capable of regulating PcG complex binding and gene repression.Citation21,Citation22 Only two such instances have been identified; thus, it is difficult to decipher any rules for mammalian PREs or conclude whether this is a general mechanism of recruitment. With increased data supporting the association between the core PRC2 complex members and multiple accessory proteins such as PCL2, JARID2, AEBP2, YY1, RBBP4, RBB7 and STK38, the assembly of the complex may be dynamic, and recruitment of PRC2 may be varied depending on the target.
Attempts to characterize a consensus binding sequence for JARID2 revealed only that the binding regions tended to be GC-rich,Citation12,Citation14,Citation23 an observation previously noted based on PRC2 binding profiles.Citation24 Thus, it was suggested that JARID2 binds DNA without clear sequence specificity, an outcome that will not facilitate the prediction or further the mechanistic understanding of PRC2 recruitment and binding to its targets. In-depth data mining of PCL2 targets may indicate whether binding of PCL2 is more restrictive than JARID2.
Global and Specific Changes in Histone Methylation
Since PcG proteins are generally thought of as transcriptional repressors, we considered whether PCL2 may have a direct effect on gene expression. To determine whether PCL2 binding results in direct transcriptional repression, we analyzed highly enriched targets of PCL2 that were downregulated in Pcl2 knockdown ESCs. Luciferase-reporter assays for Bmp2, Foxc1, Gsc, Lhx1, Pitx2, Sox1, Tbx3, Wnt3 and Oct4-reporter lines did not show any change in Luciferase level, suggesting that PCL2 does not directly modulate transcription of developmental targets (unpublished). This data led us to postulate that PCL2 may act in a repressive manner through PRC2 recruitment leading to epigenetic modifications.
PRC2 exerts its effect on gene expression through deposition of 3meH3K27, a repressive histone modification, mediated by the enzymatic activity of EZH2. Thus, we asked whether depletion of PCL2 would result in a decrease in 3meH3K27. Although downregulation of Pcl2 in ESCs did not alter global levels of 3meH3K27, when analyzed using ChIP-qPCR we observed a reduction in 3meH3K27 at specific target genes. Importantly, at promoter regions of genes such as Tbx3, Klf4, T and Evx1, there was also a reduction in binding of SUZ12, EZH2 and EED, indicating that PCL2 may be involved in the recruitment or stabilization of PRC2 at particular targets.
In contrast, methylation data JARID2 is more difficult to interpret. It was reported that in Jarid2 null or Jarid2 knockdown ESCs, global levels of 3meH3K27 were unaffected but specific targets showed a slight increaseCitation13 or decreaseCitation12 in 3meH3K27. While many groups have suggested JARID2 acts to fine-tune modulation of 3meH3K27 at PRC2 targets, there has been conflicting data regarding whether it inhibitsCitation13,Citation23 or stimulatesCitation12,Citation25 the methyltransferase activity of PRC2. Thus, several key issues remain unresolved: (1) is JARID2 responsible for inhibition or stimulation of PRC2 function and 3meH3K27? (2) instead, is JARID2 responsible only for the recruitment of PRC2 but not its enzymatic activity? and (3) is 3meH3K27 increased, decreased or unchanged at target promoters following depletion of Jarid2? It is clear that the biochemical role of JARID2 is still under debate and that the methylation pattern at target genes following depletion of a PRC2 accessory protein is still unknown. Further investigation into the function of PCL2 and JARID2 will help to refine any proposed model of PRC2 recruitment and epigenetic modification.
Polycomb complex formation, target recruitment and chromatin modification appear to be dynamic during ESC commitment and early differentiation. If PRC2 targets change during the course of ESC commitment to particular lineages, it is likely that more than one accessory protein, along with other transcription factors and elements such as ncRNA,Citation26 will interact to recruit and stabilize the PRC2 complex at specific locations. It has previously been suggested that a dynamic change in the stoichiometric ratio of accessory proteins in the PRC2 complex may influence the levels of methylation at H3K27.Citation27 This may explain why knockdown of Jarid2 has been shown to both increaseCitation13,Citation23 and decreaseCitation12,Citation25 3meH3K27 at certain targets.
Perturbations of ESC Differentiation
We and others hold the general hypothesis that genes regulated at the onset of commitment to differentiation are critical for regulating ESC fate decisions. PRC2 members are strong candidate regulators as they are all highly expressed in undifferentiated ESCs and are immediately downregulated upon differentiation. Genome-wide ChIP analyses established that PRC2 binds a wide range of developmental TFs.Citation24 It was further noted that many PRC2 targets were commonly bound by OCT4, NANOG and SOX2.Citation28 Together these observations culminated in the proposition that PRC2 cooperated with the ESC GRN to silence developmental targets in undifferentiated ESCs, preventing precocious differentiation. Once given the cue to differentiate, PRC2 would be downregulated, releasing repression of its targets and allowing upregulation of developmental TFs. If this simple explanation of ESC maintenance were true, one would expect that depletion of a PRC2 component would diminish the self-renewing status of the ESC, resulting in upregulation of target gene expression and increased differentiation. To identify the role of PCL2 in ESCs, we performed loss-of-function assays in mouse ESCs. Interestingly, downregulation of Pcl2 resulted in maintenance of high levels of Oct4 throughout the time course of differentiation as measured by high-content imaging.Citation5 Clonogenic assays demonstrated that a higher proportion of single Pcl2 knockdown cells formed undifferentiated colonies compared to wild-type and control knockdown ESCs.
Concomitant with increased self-renewal, depletion of PCL2 resulted in impaired differentiation capacity of ESCs. In a short-term monolayer neuroectoderm differentiation assay, Pcl2 knockdown cells failed to express Nestin and maintained high levels of OCT4. To evaluate whether the delay in differentiation is temporal or absolute, we generated embryoid bodies (EBs) from Pcl2 knockdown ESCs. While control EBs formed differentiated cells expressing proteins of all three germ layers, Pcl2 knockdown EBs were small, generated very few differentiated cells, never formed cystic structures, and did not express markers of the endoderm. Pcl2 knockdown EB outgrowths from day 8 cultures continued to express OCT4 robustly and outgrowths from day 25 cultures formed undifferentiated ALP + colonies. Together these phenotypic data suggest that PCL2 is a critical regulator of ESC fate decisions and lack of PCL2 contributes to a delay in differentiation and maintenance of a population of cells with ESC-like characteristics.
A similar phenotype has been observed in knockout or knockdown experiments using other PRC2 complex members, including Suz12,Citation29 Ezh2,Citation30 and Eed.Citation31 JARID2, a member of the Jumonji family of histone methyltransferases and recently identified as a PRC2 accessory protein, has also demonstrated similar defects in differentiation of knockdown ESCs.Citation13,Citation14 In all cases, ESCs exhibited strong self-renewal characteristics and maintained normal levels of Oct4, Nanog and Sox2, demonstrating that PRC2 core and accessory proteins are dispensable for ESC self-renewal. Instead, the importance of these PcG proteins appears to be during exit from the pluripotent state, with absence of any one component resulting in delayed downregulation of Oct4, Nanog and Sox2 during early differentiation. It is this delayed downregulation of the pluripotency network, along with delayed or incomplete upregulation of developmental targets which causes failure to develop into more specialized cell types.
Association with the PRC2 Gene Regulatory Network
To better understand the molecular role of PCL2 in self-renewal and early differentiation, we performed genome-wide transcriptome analyses of Pcl2 knockdown ESCs. We then we performed ChIP-seq to comprehensively map genome-wide PCL2 binding in ESCs. Gene ontology analyses of both our transcriptome and promoter occupancy data demonstrated that proteins regulated by PCL2 function in various developmental processes such as differentiation and pattern specification and include a high proportion of TFs. We compared our PCL2 ChIP-seq data to available PRC2 ChIP-seq datasetsCitation24 and found that many PCL2 targets overlap with those of SUZ12 and EZH2. By combining transcriptome and promoter occupancy data, we were able to draft the PCL2-PRC2 gene regulatory network, which includes many developmental targets controlling differentiation into all three germ layers. Interestingly, approximately 50% of the top 1,000 PCL2 targets are bound by JARID2,Citation12 supporting the possibility that JARID2 and PCL2 may both associate with the PRC2 complex at a subset of specific targets.
A GRN Model of PRC2 Function through Epigenetic Modification
We propose an updated model of PRC2 function in ESCs, incorporating recent data, particularly from analysis of the newly described PRC2 accessory proteins PCL2 and JARID2. A compelling explanation for the gene expression profiles and resulting phenotype of PRC2 member depleted ESCs comes as a result of our work in drafting a PRC2 GRN. By integrating transcriptome and ChIP-seq data, we have determined that PCL2 and other members of core PRC2 are integrated into the pluripotency network.
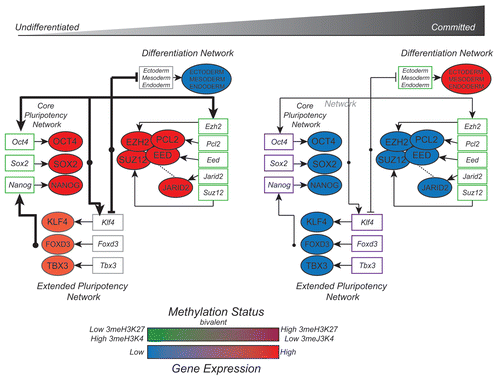
Through our work on the PCL2-PRC2 GRN, we have seen that the depletion of PCL2 results in upregulation of key TF nodes, such as Tbx3, Klf4 and Foxd3, which are known to support self-renewal through the stabilization of OCT4/SOX2/NANOG pluripotency network. This connection is highly significant because each of these genes is activated by OCT4 or NANOG. Even more importantly, TBX3, KLF4 and FOXD3 have been shown to contribute to the stabilization of the self-renewal network through feedback mechanisms.Citation32–Citation34 We propose that in wild type cells, repression of Tbx3, Klf4 and Foxd3 from PCL2-PRC2 tempers the self-renewal network so that cells are able to respond to external stimuli (). As can be seen in the Pcl2 knockdown ESC lines, without this internal control mechanism the increase in TBX3, KLF4 and FOXD3 drives the continued expression of Oct4, Nanog and Sox2, despite the withdrawal of LIF from the media. Consistent with our GRN model, overexpression of TBX3 or KLF4 has been shown to support the pluripotency network even in the absence of LIF.Citation35 Our GRN model suggests that a lack of the PRC2 complex prevents histone modification of the promoters of downstream pluripotency factors (e.g., Tbx3, Klf4, Foxd3). In turn, the increased expression of these pluripotency TFs leads to increased transcriptional activation the core pluripotency network of Oct4, Nanog and Sox2, even in the presence of external stimuli signalling differentiation (). The dominant effect of the core pluripotency GRN nodes, such as Oct4 and Sox2 is evident in current methods of reprogramming. Reactivation of the pluripotency GRN by forced expression of Oct4, Sox2, Klf4 and c-Myc is able to reprogram terminally-differentiated cells into pluripotent cells (i.e., induced pluripotent stem cells or (iPSCs) in both mouse and human.Citation36,Citation37
We propose that the PRC2 complex, along with PCL2 and JARID2, plays a key role during the short time window when ESCs become committed progenitors. To properly transition during differentiation, ESCs must downregulate expression of the pluripotency network, while upregulating expression of developmental TFs. As the cell transitions from pluripotent to committed, Oct4, Nanog and Sox2 may rely on the repressive mark laid out by PRC2 to prevent widespread activation of developmental genes while expression of cell-type specific developmental TFs are established to appropriately and precisely activate/repress the plethora of genes available for expression. Thus, the PRC2 provides a delicate balance to promote self-renewal but enable efficient cell commitment. Knockout or knockdown of PRC2 complex members or accessory proteins tips the scales, preventing the proper downregulation of the extended pluripotency network and maintaining ESC self-renewal characteristics. A time course methylation analysis in Jarid2 ESCs provides insight into what might be happening to the pluripotency network during differentiation.Citation12 Jarid2 knockdown ESCs maintain proper downregulation of the 3meH3K4 activation mark on the promoters of Oct4 and Nanog over a 7 day differentiation time course, but display a delay in upregulation of 3meH3K27 at the same targets. More in-depth analysis of bivalent histone modifications on target genes during a time course of differentiation will help to delineate the role of various PRC2 components, including PCL2, during early commitment.
It has recently been shown that the downregulation of 3meH3K27 by PRC2 at particular sites is followed by an upregulation of acetylation at H3K27, leading to transcriptional activation of target genes.Citation38 Re-expression of SUZ12 in Suz12-/- cells restores 3meH3K27 methylation while decreasing H3K27Ac, supporting the notion that the PRC2 complex may play a role in preventing the accumulation of acetylation marks on particular histones. Histone deacetylation may be mediated by the Mi2/NuRD complex, which contains multiple proteins including RBBP4 and RBBP7.Citation39 In leukemic cells, it has been proposed that the coordinated recruitment of both the Mi-2/NuRD complex and PRC2 is needed for effective chromatin remodelling and transcriptional repression.Citation40 Mass spectrometry data on PCL2, EHZ2, SUZ12 and JARID2,Citation5,Citation12,Citation13,Citation25 has indicated an association of the components with RBBP4 and RBBP7, which suggests a link between PCL2-PRC2 methylation and NuRD deacetylation on chromatin in ESCs.
What about the large set of developmental targets bound by PCL2? We observed that despite decreased levels of 3meH3K27 at their promoters in Pcl2 knockdown cells, targets were expressed at lower levels than in wild-type cells and showed a delay in upregulation during differentiation. We propose that this apparent contradiction is also explained by the reinforced activation of the self-renewal network. Many of the developmental targets of PRC2 are also bound by OCT4, NANOG and SOX2. It is believed that these TFs repress such developmental targets either directly or through the extended pluripotency network. Thus, in Pcl2 knockdown cells, the increased repression exerted by the pluripotency network must override any transcriptional activation brought on by a decrease in 3meH3K27.
Thus, it is clear that the role of the PRC2 complex in controlling gene repression is more dynamic than previously thought. With the wealth of recent data regarding new players PCL2 and JARID2, we have gathered much insight into the PRC2 but new questions have also been raised regarding how Polycomb repression of targets may be modulated by accessory proteins during ESC differentiation.
Abbreviations
| PcG | = | polycomb group |
| PRC2 | = | polycomb repressive complex 2 |
| Pcl2 | = | polycomb-like 2 |
| ESC | = | embryonic stem cell |
| GRN | = | gene regulatory network |
| TF | = | transcription factor |
| PRE | = | polycomb repressive element |
Figures and Tables
Figure 1 Clustal W amino acid alignment of three PCL2 isoforms with PHD Zinc Finger (purple) and TUDOR (green) domains highlighted.
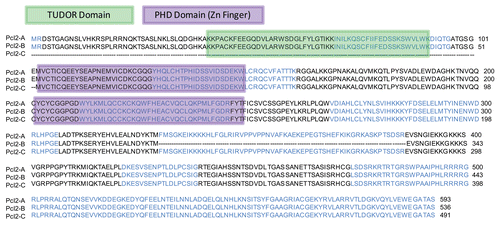
Figure 2 PCL2-PRC2-JARID2 gene regulatory network during early differentiation. In ESCs, promoters of PRC2 genes, including PCL2 and JARID2, carry the 3meH3K4 methylation mark and genes are highly expressed. PRC2 represses bivalent-marked members of the extended pluripotency network, Klf4, Foxd3 and Tbx3 which then act to induce transcription of the core pluripotency network (Oct4, Sox2, Nanog) via feed-forward mechanisms. High expression of the core and extended pluripotency network, along with PRC2 repression of developmental genes, maintains ESC self-renewal and pluripotency. During differentiation, the downregulation of Oct4, Sox2 and Nanog leads to the decreased expression of Klf4, Tbx3, Foxd3 and PRC2 genes, and collapse of the pluripotency network. Developmental genes previously poised for activation, are now de-repressed due to the decreased expression of PRC2. Loss of 3meH3K27 and increased expression of differentiation genes allows cells to become committed to various lineages. Methylation states are reflected in colour of gene boxes. For simplicity, gene expression levels are represented as protein expression levels, although this may not always reflect the true case. Dashed lines indicate potential interactions.
Acknowledgements
This work was supported by an operating grant from the Canadian Cancer Society Research Institute (19122) to W.L.S. E.W. and J.L.M. were both supported by CIHR Banting and Best CGS Doctoral Research Awards; W.Y.C. was supported by a Heart & Stroke Foundation of Canada Postdoctoral Fellowship; and W.L.S. was supported by a Canada Research Chair.
References
- Faust C, Schumacher A, Holdener B, Magnuson T. The eed mutation disrupts anterior mesoderm production in mice. Development (Cambridge, England) 1995; 121:273 - 285
- O'Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol 2001; 21:4330 - 4336
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 2004; 23:4061 - 4071
- Walker E, Ohishi M, Davey RE, Zhang W, Cassar PA, Tanaka TS, et al. Prediction and testing of novel transcriptional networks regulating embryonic stem cell self-renewal and commitment. Cell stem cell 2007; 1:71 - 86
- Walker E, Chang WY, Hunkapiller J, Cagney G, Garcha K, Torchia J, et al. Polycomb-like 2 associates with PRC2 and regulates transcriptional networks during mouse embryonic stem cell self-renewal and differentiation. Cell stem cell 2010; 6:153 - 166
- Wang S, Yu X, Zhang T, Zhang X, Zhang Z, Chen Y. Chick Pcl2 regulates the left-right asymmetry by repressing Shh expression in Hensen's node. Development (Cambridge, England) 2004; 131:4381 - 4391
- O'Connell S, Wang L, Robert S, Jones CA, Saint R, Jones RS. Polycomblike PHD fingers mediate conserved interaction with enhancer of zeste protein. J Biol Chem 2001; 276:43065 - 43073
- Cao R, Wang H, He J, Erdjument-Bromage H, Tempst P, Zhang Y. Role of hPHF1 in H3K27 methylation and Hox gene silencing. Mol Cell Biol 2008; 28:1862 - 1872
- Nekrasov M, Klymenko T, Fraterman S, Papp B, Oktaba K, Kocher T, et al. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. EMBO J 2007; 26:4078 - 4088
- Sarma K, Margueron R, Ivanov A, Pirrotta V, Reinberg D. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol Cell Biol 2008; 28:2718 - 2731
- Savla U, Benes J, Zhang J, Jones RS. Recruitment of Drosophila Polycomb-group proteins by Polycomblike, a component of a novel protein complex in larvae. Development (Cambridge, England) 2008; 135:813 - 817
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev 2010; 24:368 - 380
- Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, et al. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell 2009; 139:1303 - 1314
- Landeira D, Sauer S, Poot R, Dvorkina M, Mazzarella L, Jorgensen HF, et al. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat Cell Biol 2010; 12:618 - 624
- Kim TG, Kraus JC, Chen J, Lee Y. JUMONJI, a critical factor for cardiac development, functions as a transcriptional repressor. J Biol Chem 2003; 278:42247 - 42255
- Klug A. Zinc finger peptides for the regulation of gene expression. J Mol Biol 1999; 293:215 - 218
- Matthews JM, Sunde M. Zinc fingers—folds for many occasions. IUBMB Life 2002; 54:351 - 355
- Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 2006; 442:91 - 95
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 2006; 442:86 - 90
- Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development (Cambridge, England) 2009; 136:3531 - 3542
- Sing A, Pannell D, Karaiskakis A, Sturgeon K, Djabali M, Ellis J, et al. A vertebrate Polycomb response element governs segmentation of the posterior hindbrain. Cell 2009; 138:885 - 897
- Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell 2010; 140:99 - 110
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, et al. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 2009; 139:1290 - 1302
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 2008; 4:1000242
- Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, et al. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 2010; 464:306 - 310
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 2008; 322:750 - 756
- Herz HM, Shilatifard A. The JARID2-PRC2 duality. Genes Dev 2010; 24:857 - 861
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 2006; 441:349 - 353
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol 2007; 27:3769 - 3779
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Molecular cell 2008; 32:491 - 502
- Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells 2008; 26:1496 - 505
- Hanna LA, Foreman RK, Tarasenko IA, Kessler DS, Labosky PA. Requirement for Foxd3 in maintaining pluripotent cells of the early mouse embryo. Genes Dev 2002; 16:2650 - 2661
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, et al. Dissecting self-renewal in stem cells with RNA interference. Nature 2006; 442:533 - 538
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell 2008; 132:1049 - 1061
- Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 2009; 460:118 - 122
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007; 131:861 - 872
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126:663 - 676
- Pasini D, Malatesta M, Jung HR, Walfridsson J, Willer A, Olsson L, et al. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res 2010; 38:4958 - 4969
- Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene 2007; 26:5433 - 5438
- Morey L, Brenner C, Fazi F, Villa R, Gutierrez A, Buschbeck M, et al. MBD3, a component of the NuRD complex, facilitates chromatin alteration and deposition of epigenetic marks. Mol Cell Biol 2008; 28:5912 - 5923