Abstract
Comment on: Miyamoto K, et al. Genes Dev 2011; 25:946-58.
Differentiated cells can be experimentally reprogrammed to an embryonic state by different approaches.Citation1 Nuclear reprogramming entails reactivation of silenced embryonic genes, such as Oct4 (Pou5f1). The most efficient method for reprogramming is regarded as nuclear transfer to eggs/oocytes.Citation2 However, oocyte factors and mechanisms participating in this efficient reprogramming have not been elucidated. We have recently identified actin in a nucleus (nuclear actin) as one such factor by using the unique nuclear transfer system into Xenopus oocytes.Citation3
Actin is a major constituent of the cytoskeleton. Growing evidence suggests that actin plays an important role in many nuclear events, including transcription, RNA processing, gene movement and chromatin remodeling.Citation4 In our recent study, we took advantage of the giant nucleus of the Xenopus oocyte, referred to as the germinal vesicle (GV), in order to analyze functions of nuclear actin in transcriptional reprogramming (). Hundreds of somatic nuclei can be injected into one GV, and almost all transplanted nuclei are efficiently reprogrammed to express many embryonic genes.Citation5 It is noteworthy that GVs can be isolated in nonaqueous oil without losing their reprogramming activity, so that reprogramming can be driven exclusively by nuclear contents. This is especially valuable for revealing functions of nuclear actin, because investigations on nuclear actin have been constrained by involvement and contamination of cytoplasmic actin. Indeed, this oil GV method has been used to visualize polymerized nuclear actin by live cell imaging and to show a requirement of nuclear actin polymerization transcription.Citation3
In the cytoplasm, actin dynamically changes its polymerized states with the help of actin nucleators and actin signaling proteins.Citation6 The existence of functional actin nucleators in nuclei has also been reported, such as N-WASP,Citation7,Citation8 Arp2/3 Citation9 and JMY.Citation10 Actin nucleators in nuclei seem to be capable of accelerating actin polymerization, since N-WASP regulates de novo actin polymerization in the in vitro actin assembly assay using nuclear extracts from mammalian cultured cells.Citation7 Apart from actin polymerization, nucleators in nuclei also play an important role in transcription. N-WASP and Arp2/3 are bound to RNA polymerase II (Pol II) and are required for efficient transcription driven by Pol II.Citation7,Citation9 Recently, cofilin-1, which severs polymerized actin, was shown to be necessary for Pol II transcription elongation.Citation11 These results suggest that the regulation of actin polymerization by nuclear actin-binding proteins is a key determinant of Pol II transcription. WASP, an actin nucleator, is also identified as an important molecule for RBBP5, a histone H3K4 trimethyltransferase, and JMJD2A, a H3K9/H3K36 tridemethylase, to execute its full activity on target genes in T cells during transcriptional activation.Citation8 Together, actin nucleators are involved in transcriptional activation and ongoing transcription. Our study has shown that the overexpression of Toca-1, an actin signaling protein and an upstream regulator of N-WASP, enhances nuclear actin polymerization and Oct4 reactivation.Citation3 We favor the idea that Toca-1 overexpression facilitates the transcriptional activation/derepression process by activating downstream targets and thereby increasing chromatin remodeling rather than by accelerating transcription itself.Citation3 However, it is still not clear whether enhanced gene activation by Toca-1 overexpression results from enhanced actin polymerization per se and/or from other actions by downstream actin nucleators of Toca-1, such as histone-modifying activities by WASP. This question needs to be answered in future work so as to reveal nuclear functions of actin nucleators.
We have revealed a novel function of nuclear actin in transcriptional reprogramming of developmentally silenced genes and have shown a requirement for actin polymerization. However, we still do not know how nuclear actin polymerization is involved in transcriptional reprogramming. Given the fact that actin polymerization is regulated by actin nucleators, one clue to this question might be provided by pursuing mechanistically the functions of nucleators in transcriptional reprogramming. Unraveling regulatory machineries of nuclear actin toward transcriptional reprogramming may lead to a better understanding of nuclear reprogramming by oocytes.
Figures and Tables
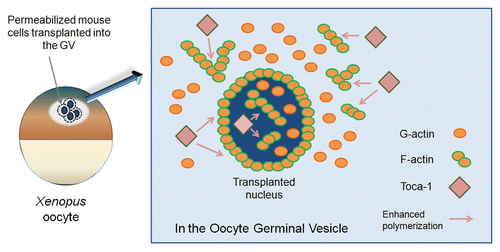
Figure 1 Polymerized actin regulated by an actin signaling protein Toca-1 is present in the oocyte germinal vesicle (GV) and in transplanted nuclei. Permeabilized mouse cells were injected into the GV of Xenopus oocytes. After nuclear transfer, injected GVs were collected in oil and subjected to live cell imaging. Transplanted nuclei were surrounded by polymerized actin and actin filaments were also observed in the nuclei. Toca-1 is present in the GV and in transplanted nuclei and enhances nuclear actin polymerization. G-actin, monomeric actin; F-actin, filamentous actin (polymerized actin).
Comment on: Miyamoto K, et al. Genes Dev 2011; 25:946 - 958
References
- Gurdon JB, et al. Science 2008; 322:1811 - 1815; PMID: 19095934; http://dx.doi.org/10.1126/science.1160810
- Kim K, et al. Nature 2010; 467:285 - 290; PMID: 20644535; http://dx.doi.org/10.1038/nature09342
- Miyamoto K, et al. Genes Dev 2011; 25:946 - 958; PMID: 21536734; http://dx.doi.org/10.1101/gad.615211
- Visa N, et al. Cold Spring Harb Perspect Biol 2010; 2:620; PMID: 20452941; http://dx.doi.org/10.1101/cshperspect.a000620
- Jullien J, et al. Proc Natl Acad Sci USA 2010; 107:5483 - 5488; PMID: 20212135; http://dx.doi.org/10.1073/pnas.1000599107
- Campellone KG, et al. Nat Rev Mol Cell Biol 2010; 11:237 - 251; PMID: 20237478; http://dx.doi.org/10.1038/nrm2867
- Wu X, et al. Nat Cell Biol 2006; 8:756 - 763; PMID: 16767080; http://dx.doi.org/10.1038/ncb1433
- Taylor MD, et al. Sci Transl Med 2010; 2:37 - 44; PMID: 20574068; http://dx.doi.org/10.1126/scitranslmed.3000813
- Yoo Y, et al. J Biol Chem 2007; 282:7616 - 7623; PMID: 17220302; http://dx.doi.org/10.1074/jbc.M607596200
- Zuchero JB, et al. Nat Cell Biol 2009; 11:451 - 459; PMID: 19287377; http://dx.doi.org/10.1038/ncb1852
- Obrdlik A, et al. Nucleus 2011; 2:72 - 79; PMID: 21647301