Abstract
Ubiquitination is a post-translational modification that generally directs proteins for degradation by the proteasome or by lysosomes. However, ubiquitination has been implicated in many other cellular processes, including transcriptional regulation, DNA repair, regulation of protein-protein interactions and association with ubiquitin-binding scaffolds. Ubiquitination is a dynamic process. Ubiquitin is added to proteins by E3 ubiquitin ligases as a covalent modification to one or multiple lysine residues as well as non-lysine amino acids. Ubiquitin itself contains seven lysines, each of which can also be ubiquitinated, leading to polyubiquitin chains that are best characterized for linkages occurring through K48 and K63. Ubiquitination can also be reversed by the action of deubiquitination enzymes (DUbs). Like E3 ligases, DUbs play diverse and critical roles in cells.Citation1 Ubiquitin is expressed as a fusion protein, as a linear repeat or as a fusion to ribosomal subunits, and DUbs are necessary to liberate free ubiquitin, making them the first enzyme of the ubiquitin cascade. Proteins destined for degradation by the proteasome or by lysosomes are deubiquitinated prior to their degradation, which allows ubiquitin to be recycled by the cell, contributing to the steady-state pool of free ubiquitin. Proteins destined for degradation by lysosomes are also acted upon by both ligases and DUbs. Deubiquitination can also act as a means to prevent protein degradation, and many proteins are thought to undergo rounds of ubiquitination and deubiquitination, ultimately resulting in either the degradation or stabilization of those proteins. Despite years of study, examining the effects of the ubiquitination of proteins remains quite challenging. This is because the methods that are currently being employed to study ubiquitination are limiting. Here, we briefly examine current strategies to study the effects of ubiquitination and describe an additional novel approach that we have developed.
Keywords: :
One way to determine the role that ubiquitintion of a particular protein plays is to prevent the ubiquitination of that protein and see what happens. The most common way to do this is to mutate lysine residues within a given protein of interest. However, this approach has several caveats. At first glance, it can difficult to predict which lysine(s) within a given protein of interest are susceptible to ubiquitination. Even using a very precise approach, such as mass spectrometry, to determine which residues are ubiquitinated can be problematic. For instance, mutating a lysine that is ubiquitinated may result in another lysine residue within the protein of interest getting ubiquitinated. Also, many commonly used epitopes, such as GFP, myc and GST, contain lysine residues that are susceptible to ubiquitination. Perhaps even more troubling is the observation that serine, threonine, cysteine and the N terminus of some proteins can undergo ubiquitination.Citation2-Citation5 Of course, lysines aren’t just for ubiquitination and may be critical for protein folding, protein/protein interactions or as a target for other post-translational modifications. Together, these caveats make it virtually impossible to generate a mutant protein that is completely resistant to ubiquitination. An alternative approach is to knock down, knock out, chemically inhibit or express dominant-negative forms of an E3 ligase thought to ubiquitinate a particular protein of interest. This approach is limited in that many proteins, including ubiquitin-binding proteins, are ubiquitinated by multiple ubiquitin ligases, leading to the possibility that the reduced activity of one ligase will simply be compensated for by another ligase. For instance, the MATalpha2 transcription factor is ubiquitinated by two Ub-ligases, each of them sufficient for a2 degradation but each using distinct degrons and each likely conveying distinct regulatory pathways.Citation6 Other proteins appear to have the capacity to be targeted by a wide range of E3 enzymes, at least in vitro, underscoring the problem of redundancy with various experimental approaches.Citation7 Moreover, inhibiting a ubiquitin ligase potentially changes the ubiquitination status not only of the protein of interest but of every protein recognized by that ligase. Thus, any resulting phenotype may be an indirect effect independent of the ubiquitinated protein of interest. Specificity is frequently a concern for other approaches as well. For instance, using a chemical inhibitor of the proteasome may block the degradation of a protein of interest but also blocks the degradation of many other proteins, making it difficult to draw conclusions about the specific protein in question. Our recent report in the Journal of Cell Biology focuses on two critical aspects of ubiquitin-dependent trafficking to lysosomes. During this study, we encountered many of these problems when examining the role of ubiquitination within the endocytic pathway. To circumvent many of these issues, we devised a novel method to immunize proteins from ubiquitination.Citation8
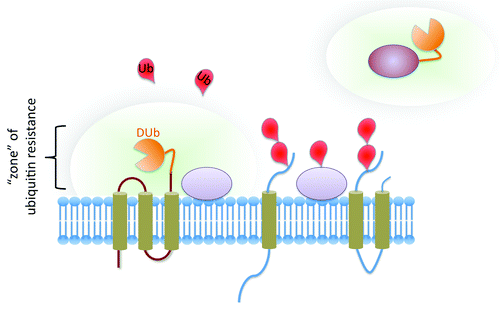
To make proteins resistant to ubiquitination, we covalently fused the catalytic domain of a deubiquitinating enzyme to a protein of interest. The rationale was that this modification would rapidly reverse the ubiquitination of the protein of interest and proteins that closely interacted with that protein of interest, essentially creating a zone around the protein that is not susceptible to ubiquitination (). We reasoned that this approach would have several advantages over conventional methods. By simply fusing a DUb to a protein of interest, ubiquitination is potentially reversed no matter which residues are ubiquitinated, including any potentially ubiquitinatable residues on epitopes. This allows for a relatively rapid assessment as to whether ubiquitination affects a protein or process of interest without knowing every potential site of ubiquitination or worrying about artifacts generated by the ubiquitination of epitopes. Also, DUbs rely on specific residues for their catalytic activity. Thus, an inactive DUb with point mutations of catalytically conserved residues could be used as a negative control. This is a major advantage over the conventional method of mutating lysines, for which there really is no specificity control to demonstrate that the effect of lysine mutation is due solely to loss of ubiquitination. By mutating the DUb and not the protein itself, it becomes easier to draw conclusions. Indeed, in our experiments, fusion of a variety of DUb catalytic domains blocked ubiquitination and Ub-dependent lysosomal trafficking of a wide variety of membrane proteins, and both ubiquitination and ubiquitin-dependent trafficking were restored by mutating the catalytic site of the DUb domain. We also found that DUb fusions provided strong specificity, in that they did not reverse the ubiquitin-dependent trafficking of other endogenous proteins.
Figure 1. Ubiquitin-resistant proteins. The fusion of a catalytic domain from a DUb to an integral membrane protein of interest (left) or a soluble protein of interest (right) renders the entire protein resistant to the effects of ubiquitination. The DUb activity essentially creates a “zone” where the protein and other closely associated proteins within the “zone” are rapidly deubiquitinated. This approach provides a powerful way to completely prevent the ubiquitination of a protein of interest without changing the “ubiquitome” of the entire cell or modifying every potential ubiquitinatable residue.
We employed this approach to examine specific questions related to ubiquitin-dependent trafficking to lysosomes. Proteins that contain ubiquitin-binding domains carry out ubiquitin-dependent processes in cells. Integral membrane proteins destined for degradation by lysosomes are ubiquitinated and recognized by a set of protein complexes known as the ESCRTs.Citation9,Citation10 The ESCRTs shuttle ubiquitinated cargo into intralumenal vesicles that bud inward from the limiting membrane of endosomes. The accumulation of these vesicles, termed multivesicular bodies (MVBs), eventually leads to the degradation of the proteins within MVBs by lysosomes. Several ESCRT subunits contain ubiquitin-binding domains that are crucial to this process. Interestingly, many ubiquitin-binding proteins, including several ESCRT subunits, are themselves ubiquitinated. Previous work has implicated this effect, known as “coupled ubiquitination,” in the modulation of the activity of ubiquitin-binding proteins. While much is known about how ubiquitin-binding proteins are ubiquitinated, it is still unclear whether the ubiquitination of ubiquitin-binding proteins provides a physiological mechanism to regulate their activity. Perturbation of the ubiquitin-binding domains of ubiquitin-binding proteins diminishes their ubiquitination but also renders them incapable of binding ubiquitin. Several ESCRT proteins involved in the sorting of ubiquitinated cargo proteins to lysosomes undergo coupled ubiquitination. Since the ubiquitin-dependent sorting events performed by ESCRTs is via their ability to bind ubiquitin, we could not perturb the ubiquitin-binding domains to inhibit their ubiquitination. However, by fusing a DUb to several of the ESCRTs, we were able to uncouple ubiquitin-binding from ubiquitination. DUb fusions prevented the ubiquitination of the ESCRTs but still allowed for ubiquitin-dependent trafficking to the vacuole, the yeast equivalent of lysosomes. By generating in-frame fusions of ubiquitin to cargo proteins that were not susceptible to the activity of DUbs, we were able to demonstrate that ubiquitin-dependent sorting by ubiquitin-binding proteins was possible in the absence of their ubiquitination. If coupled ubiquitination were a critical part of protein sorting mediated by ESCRTs, DUb fusions should have blocked all ubiquitin-dependent trafficking. Our data suggest that coupled ubiquitination is not a critical mechanism required for protein trafficking to lysosomes.
This approach also allowed us to examine whether polyubiquitination is a necessary signal for trafficking to lysosomes. Many studies reveal that several yeast cargo proteins destined for degradation in the vacuole are modified by K63 polyubiquitination. Several studies showed that cells with a K63R ubiquitin as the sole source of ubiquitin have severe defects in sending proteins into the MVB sorting pathway.Citation11 This finding raises the possibility that K63 polyubiquitin may be a necessary signal on cargo that must be recognized by the endosomal sorting machinery for delivery into lysosomes. However, making firm conclusions from cells unable to make K63-linked chains is confounded by several caveats stemming from the fact that a wide variety of biological processes may be partially compromised. Our approach circumvented these limitations, allowing a more precise examination of what type of ubiquitin signal on cargo was required for entry into MVBs. ESCRT-DUb fusions were sufficient to block the ubiquitin-dependent sorting of every cargo protein that we tested, suggesting that the DUb was capable of removing every ubiquitin moiety from the cargo. However, the addition of a single fused ubiquitin that was not susceptible to DUb activity was sufficient to bypass the DUb block. In previous studies, a single ubiquitin fused to cargo might have acted as a ubiquitin acceptor for subsequent polyubiquitination. Here, that is not a possibility, since any further ubiquitination is susceptible to deubiquitination by the ESCRT-DUb fusions. These data, in combination with other data from our study, suggest that a single ubiquitin is sufficient for cargo trafficking into MVBs.
An additional approach we employed was to fuse a DUb to the E3 ligase Rsp5, which essentially turned the ligase into a dominant-negative protein that could seek out Rsp5 substrates and reverse the work of endogenous Rsp5. The potential of converting an E3 ligase of interest into a specific dominant “anti-ligase” presents several future experimental possibilities, including new strategies to find E3 substrates. There are many orphan ubiquitin ligases, which have no known targets; even well-characterized ligases are likely to have a plethora of undiscovered substrates. At least one experimental difficulty is that many substrates are ubiquitinated by multiple ubiquitin ligases. Hence, inhibiting one ubiquitin ligase many not necessarily correlate with a reduction in the ubiquitination of its substrates, at least not to detectable levels using common biochemical techniques. Fusion of a DUb to an E3 would circumvent this problem, because the targets would be deubiquitinated, even if the targets were ubiquitinated by other ligases. Thus, fusion of a DUb to ubiquitin-ligase could be used in combination with proteomics to map the targets of ubiquitin-ligases by looking for which targets lose ubiquitination when E3-DUb fusions are expressed in cells. One area where this approach might be particularly helpful is the study of ER-associated degradation. Numerous studies characterizing two E3 ligases, gp78 and Hrd1, have revealed many ERAD substrates for these ligases.Citation12 However, while many other ligases have been implicated in ERAD, including RNF5, TEB4, RFP2, Kf-1, Trc8, RMA1 and Parkin,Citation13-Citation16 few substrates for these ligases have been described. This is likely due to redundancy within the ERAD ubiquitination system. Fusion of a DUb to these poorly characterized ligases, would essentially transform these ligases into E3 “reversal enzymes” that would deubiquitinated and stabilize the targets of these ligases. Furthermore, some E3s are destabilized by autoubiquitination or by ubiquitination from other E3s. DUb fusions would prevent the ubiquitination of these E3s, which may help in the further characterization of E3s that are rapidly turned over.Citation17,Citation18
An interesting finding came from our study that fusion of a DUb to Gga2, a clathrin adaptor that contains a ubiquitin-binding domain and is thought to facilitate Golgi to endosome trafficking of ubiquitinated cargo, blocked MVB sorting of a subset of cargo.Citation19 Several studies also have suggested that Ggas may play a role in trafficking from the plasma membrane to endosomes; however, evidence for a direct role in this pathway remains elusive, likely due to redundancy among the ubiquitin-binding domain-containing proteins in the endocytic pathway.Citation20-Citation22 Interestingly, the dominant acting Gga2-DUb fusions in our study blocked some but not all of the endosomal MVB cargo we tested, suggesting a role for a subset of cargo proteins as they progress toward the vacuole in the endocytic pathway. Many proteins, including Tom1, Epsins, Eps15, Ggas, Sts1 and Sts2, are all associated with the endocytic pathway and all contain ubiquitin-binding domains.Citation23,Citation24 However, a precise role for these proteins remains elusive, as many of them function in parallel with each other. A DUb fusion of these proteins essentially eliminates their Ub-dependent function and may provide a way to reveal the types of Ub-dependent sorting steps in which these proteins participate.
Major questions persist about the functionality of different polyubiquitin linkages in a variety of processes. Perhaps the most perplexing is the role of polyubiquitin chains linked through K11. Proteomics studies in yeast have revealed that roughly 30% of all polyubiquitin linkages in cells are through K11,Citation25 and distinct enzymes have been discovered that selectively promote formation of K11 chains.Citation26-Citation28 K11 polyubiquitination linkages are thought to promote protein degradation by the proteasome and have been implicated in several processes, including cell cycle and ERAD.Citation29 Structural data indicate that K11 chains are distinct from K48. Why do cells utilize two distinct polyubiquitination mechanisms leading to the same degradative fate by proteasomal degradation? It seems unlikely that cells would have two independent mechanisms for ubiquitination merely for redundancy within the proteasomal degradation pathway. One idea is that different types of polyubiquitination may establish a hierarchy for proteasomal degradation. During times of cells stress or rapid protein turnover, the cell may selectively degrade some substrates faster then others. Fusion of a K11-specific DUb to a protein of interest might be helpful in uncovering a distinct role for K11 polyubiquitination. Indeed, the DUb Cezanne, an OTU family DUb, has been shown to have a strong preference for K11-linked chains. This DUb and perhaps others could be used to understand the role K11-linked polyubiquitination plays in cells.Citation27
A more precise understanding of how proteins are ubiquitinated and what ubiquitination of proteins does is important, since ubiquitination is frequently discussed as a potential target of therapeutics. The fusion of a DUb to a protein of interest is potentially a powerful tool for future studies. The general approach of using DUb fusion proteins could be finer tuned with a number of straightforward improvements that would yield greater control, specificity and accuracy. The linker between the DUb and the protein of interest could be engineered to containing a protease cleavage site so that the DUb could be easily removed in vivo upon introduction of a protease. Also, the linker could be engineered with protein domains that dimerize, such as FKBP/FRB, so that addition of a DUb to a protein of interest could be induced by the addition dimerization drug. Perhaps the most promising application of this technique is as a discovery tool. Rapid assessment of whether ubiquitination is involved in a particular cellular process would provide the impetus for more robust investigation. In combination with more conventional techniques, this novel approach could be a very powerful tool for future discovery.
References
- Hussain S, Zhang Y, Galardy PJ. DUBs and cancer: the role of deubiquitinating enzymes as oncogenes, non-oncogenes and tumor suppressors. Cell Cycle 2009; 8:1688 - 97; http://dx.doi.org/10.4161/cc.8.11.8739; PMID: 19448430
- Shimizu Y, Okuda-Shimizu Y, Hendershot LM. Ubiquitylation of an ERAD substrate occurs on multiple types of amino acids. Mol Cell 2010; 40:917 - 26; http://dx.doi.org/10.1016/j.molcel.2010.11.033; PMID: 21172657
- Kim HC, Huibregtse JM. Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol Cell Biol 2009; 29:3307 - 18; http://dx.doi.org/10.1128/MCB.00240-09; PMID: 19364824
- Wang X, Herr RA, Chua WJ, Lybarger L, Wiertz EJ, Hansen TH. Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J Cell Biol 2007; 177:613 - 24; http://dx.doi.org/10.1083/jcb.200611063; PMID: 17502423
- Cadwell K, Coscoy L. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science 2005; 309:127 - 30; http://dx.doi.org/10.1126/science.1110340; PMID: 15994556
- Rubenstein EM, Hochstrasser M. Redundancy and variation in the ubiquitin-mediated proteolytic targeting of a transcription factor. Cell Cycle 2010; 9:4282 - 5; http://dx.doi.org/10.4161/cc.9.21.13741; PMID: 20980825
- Uchiki T, Kim HT, Zhai B, Gygi SP, Johnston JA, O’Bryan JP, et al. The ubiquitin-interacting motif protein, S5a, is ubiquitinated by all types of ubiquitin ligases by a mechanism different from typical substrate recognition. J Biol Chem 2009; 284:12622 - 32; http://dx.doi.org/10.1074/jbc.M900556200; PMID: 19240029
- Stringer DK, Piper RC. A single ubiquitin is sufficient for cargo protein entry into MVBs in the absence of ESCRT ubiquitination. J Cell Biol 2011; 192:229 - 42; http://dx.doi.org/10.1083/jcb.201008121; PMID: 21242292
- Wollert T, Yang D, Ren X, Lee HH, Im YJ, Hurley JH. The ESCRT machinery at a glance. J Cell Sci 2009; 122:2163 - 6; http://dx.doi.org/10.1242/jcs.029884; PMID: 19535731
- Piper RC, Katzmann DJ. Biogenesis and function of multivesicular bodies. Annu Rev Cell Dev Biol 2007; 23:519 - 47; http://dx.doi.org/10.1146/annurev.cellbio.23.090506.123319; PMID: 17506697
- Lauwers E, Erpapazoglou Z, Haguenauer-Tsapis R, André B. The ubiquitin code of yeast permease trafficking. Trends Cell Biol 2010; 20:196 - 204; http://dx.doi.org/10.1016/j.tcb.2010.01.004; PMID: 20138522
- Kostova Z, Tsai YC, Weissman AM. Ubiquitin ligases, critical mediators of endoplasmic reticulum-associated degradation. Semin Cell Dev Biol 2007; 18:770 - 9; http://dx.doi.org/10.1016/j.semcdb.2007.09.002; PMID: 17950636
- Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T. The ubiquitylation machinery of the endoplasmic reticulum. Nature 2009; 458:453 - 60; http://dx.doi.org/10.1038/nature07962; PMID: 19325625
- Wang L, Dong H, Soroka CJ, Wei N, Boyer JL, Hochstrasser M. Degradation of the bile salt export pump at endoplasmic reticulum in progressive familial intrahepatic cholestasis type II. Hepatology 2008; 48:1558 - 69; http://dx.doi.org/10.1002/hep.22499; PMID: 18798335
- Ron I, Rapaport D, Horowitz M. Interaction between parkin and mutant glucocerebrosidase variants: a possible link between Parkinson disease and Gaucher disease. Hum Mol Genet 2010; 19:3771 - 81; http://dx.doi.org/10.1093/hmg/ddq292; PMID: 20643691
- Mehnert M, Sommer T, Jarosch E. ERAD ubiquitin ligases: multifunctional tools for protein quality control and waste disposal in the endoplasmic reticulum. Bioessays 2010; 32:905 - 13; http://dx.doi.org/10.1002/bies.201000046; PMID: 20806269
- Shmueli A, Tsai YC, Yang M, Braun MA, Weissman AM. Targeting of gp78 for ubiquitin-mediated proteasomal degradation by Hrd1: cross-talk between E3s in the endoplasmic reticulum. Biochem Biophys Res Commun 2009; 390:758 - 62; http://dx.doi.org/10.1016/j.bbrc.2009.10.045; PMID: 19835843
- Magnifico A, Ettenberg S, Yang C, Mariano J, Tiwari S, Fang S, et al. WW domain HECT E3s target Cbl RING finger E3s for proteasomal degradation. J Biol Chem 2003; 278:43169 - 77; http://dx.doi.org/10.1074/jbc.M308009200; PMID: 12907674
- Scott PM, Bilodeau PS, Zhdankina O, Winistorfer SC, Hauglund MJ, Allaman MM, et al. GGA proteins bind ubiquitin to facilitate sorting at the trans-Golgi network. Nat Cell Biol 2004; 6:252 - 9; http://dx.doi.org/10.1038/ncb1107; PMID: 15039776
- Erpapazoglou Z, Froissard M, Nondier I, Lesuisse E, Haguenauer-Tsapis R, Belgareh-Touzé N. Substrate- and ubiquitin-dependent trafficking of the yeast siderophore transporter Sit1. Traffic 2008; 9:1372 - 91; http://dx.doi.org/10.1111/j.1600-0854.2008.00766.x; PMID: 18489705
- Deng Y, Guo Y, Watson H, Au WC, Shakoury-Elizeh M, Basrai MA, et al. Gga2 mediates sequential ubiquitin-independent and ubiquitin-dependent steps in the trafficking of ARN1 from the trans-Golgi network to the vacuole. J Biol Chem 2009; 284:23830 - 41; http://dx.doi.org/10.1074/jbc.M109.030015; PMID: 19574226
- Lauwers E, Jacob C, André B. K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J Cell Biol 2009; 185:493 - 502; http://dx.doi.org/10.1083/jcb.200810114; PMID: 19398763
- Puertollano R. Interactions of TOM1L1 with the multivesicular body sorting machinery. J Biol Chem 2005; 280:9258 - 64; http://dx.doi.org/10.1074/jbc.M412481200; PMID: 15611048
- Hoeller D, Crosetto N, Blagoev B, Raiborg C, Tikkanen R, Wagner S, et al. Regulation of ubiquitin-binding proteins by monoubiquitination. Nat Cell Biol 2006; 8:163 - 9; http://dx.doi.org/10.1038/ncb1354; PMID: 16429130
- Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 2009; 137:133 - 45; http://dx.doi.org/10.1016/j.cell.2009.01.041; PMID: 19345192
- Wu T, Merbl Y, Huo Y, Gallop JL, Tzur A, Kirschner MW. UBE2S drives elongation of K11-linked ubiquitin chains by the anaphase-promoting complex. Proc Natl Acad Sci U S A 2010; 107:1355 - 60; http://dx.doi.org/10.1073/pnas.0912802107; PMID: 20080579
- Bremm A, Freund SM, Komander D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat Struct Mol Biol 2010; 17:939 - 47; http://dx.doi.org/10.1038/nsmb.1873; PMID: 20622874
- Bosanac I, Phu L, Pan B, Zilberleyb I, Maurer B, Dixit VM, et al. Modulation of K11-linkage formation by variable loop residues within UbcH5A. J Mol Biol 2011; 408:420 - 31; http://dx.doi.org/10.1016/j.jmb.2011.03.011; PMID: 21396940
- Baboshina OV, Haas AL. Novel multiubiquitin chain linkages catalyzed by the conjugating enzymes E2EPF and RAD6 are recognized by 26 S proteasome subunit 5. J Biol Chem 1996; 271:2823 - 31; http://dx.doi.org/10.1074/jbc.271.5.2823; PMID: 8576261