Abstract
The ubiquitin conjugation system regulates a wide variety of biological phenomena, including protein degradation and signal transduction, by regulating protein function via polyubiquitin conjugation in most cases. Several types of polyubiquitin chains exist in cells, and the type of polyubiquitin chain conjugated to a protein seems to determine how that protein is regulated. We identified a novel linear polyubiquitin chain and the ubiquitin-protein ligase complex that assembles it, designated LUBAC. Both were shown to have crucial roles in the canonical NFκB activation pathway. This year, three groups, including our laboratory, identified SHARPIN as a new subunit of LUBAC. Of great interest, Sharpin was identified as a causative gene of chronic proliferative dermatitis in mice (cpdm), which is characterized by numerous inflammatory symptoms including chronic dermatitis, arthritis and immune disorders. Deletion of SHARPIN drastically reduces the amount of LUBAC and attenuates signal-induced NFκB activation. The pleomorphic symptoms of cpdm mice suggest that LUBAC-mediated NFκB activation may play critical roles in mammals and be involved in various disorders. A forward look into the linear polyubiquitin research is also discussed.
Introduction
In multicellular organisms, cells communicate with each other and function coordinately to maintain homeostasis. Thus, cells receive signals from other cells or from the extracellular environment and must respond to those stimuli appropriately. When they encounter agents that induce tissue damages, including infectious organisms or DNA damaging agents, organisms operate inflammatory responses to remove those agents and repair their own tissues. Nuclear factor κB (NFκB) is one of the transcription factors that plays a central role in inflammatory responses induced by infectious agents, UV or inflammatory cytokines. NFκB induces the expression of proinflammmatory molecules.Citation1 Besides inflammation, NFκB is also involved in many biological phenomena, including cell survival. Abnormal activation of NFκB is observed in many pathological conditions, such as allergic and autoinflammatory diseases and malignancies.Citation2–Citation5 Therefore, the signal-induced NFκB activation pathway has been extensively studied.Citation1 NFκB is a dimeric transcription factor composed of Rel proteins. Two activation pathways exist, the canonical and non-canonical pathways.Citation6
In this manuscript, the pathophysiological function of the novel linear polyubiquitin chain is discussed. Since linear polyubiquitin chains are mainly involved in the canonical pathway,Citation7 this short article will focus on that pathway. NFκB is inactive in resting cells, as it resides in the cytoplasm bound to inhibitor proteins called inhibitors of κBs (IκBs). Upon stimuli by inflammatory cytokines or Toll-like receptor ligands, the IKK (IκB kinase) complex, composed of IKKα, IKKβ and NFκB essential modulator (NEMO), which is also called IKKγ, is activated and phosphorylates specific Ser residues in IκBs. Phosphorylated IκBs are degraded in a ubiquitin-dependent manner, which releases NFκB and allows it to translocate into the nucleus to induce the transcription of target genes ().Citation1
The Ubiquitin Conjugation System
The NFκB activation pathway is tightly linked to the ubiquitin conjugation pathway.Citation8 Although the ubiquitin sysytem was identified as part of an energy-dependent degradation system,Citation9 non-degradation roles were subsequently recognized and are now widely accepted. Most of the non-degradation roles of the conjugation system are mediated by different types of polyubiquitin chains, polymers of ubiquitin that are distinct from those used for protein degradation.Citation10 Polyubiquitin chains are believed to be generated by the repetition of the cascade of reactions catalyzed by three enzyme classes: E1 ubiquitin-activating enzymes, E2 ubiquitin-conjugating enzymes and E3 ubiquitin-protein ligases. Target proteins are specifically recognized by E3 enzymes ().Citation11 Polyubiquitin chains are thought to be generated via Lys residues of ubiquitin. Polyubiquitin chains that function as degradation signals are generated via ubiquitin Lys 48 (K48-linked chains) ().Citation12 Indeed, phosphorylated IκBα is recognized by the SCFβTrCP ubiquitin ligase and targeted for degradation by conjugation to K48-linked chains.Citation13–Citation15
The ubiquitin proteolytic pathway plays crucial roles in cell regulation by recognizing and conjugating polyubiquitin chains to specific substrates in a timely and selective manner.Citation11 Timely and selective protein modification is desirable beyond protein degradation: it is a crucial feature of other modes of protein regulation; for example, signal-induced protein activation. Indeed, Lys 63-linked (K63-linked) polyubiquitin chains are involved in signal transduction and DNA repair ().Citation16,Citation17 The existence of approximately 100 human deubiquitinating enzymes has been suggested ().Citation18 Thus, the ubiquitin conjugation system is now regarded as a reversible post-translational protein modification system that regulates protein function in a wide variety of ways. In addition to K48- and K63-linked polyubiquitin chains, mass spectrometry analyses revealed that inter-ubiquitin linkages via all seven Lys residues of ubiquitin exist in eukaryotic cells.Citation19 Since the type of polyubiquitin chain has been hypothesized to determine the mode of regulation of the conjugated protein,Citation7 the ubiquitin conjugation system may play a much greater role in biology than initially anticipated.Citation7 In that context, a new type of polyubiquitin chain, a linear polyubiquitin chain in which the carboxyl group of a ubiquitin monomer is bound to the α-amino group of another monomer, was identified by our laboratory in 2006.Citation20 Further analysis revealed that linear polyubiquitination is involved in NFκB activation.Citation21 NEMO, a component of the IKK complex, is specifically conjugated with linear polyubiquitin chains in a signal-dependent manner, leading to activation of the IKK complex ().Citation21
Unique Character of Linear Polyubiquitin Chains
Although the mechanism generating polyubiquitin chains has not yet been conclusively resolved, it is thought that E2 enzymes determine the type of polyubiquitin chain generated.Citation12 For example, E2 complexes containing Ubc13 (Ubc13-Uev1a and Ubc13-MMS2) generate K63-linked chains exclusively,Citation22 while Ube2S generates K11-linked chains,Citation23 and CDC34 and E2-25K generate K48-linked chains.Citation24,Citation25 However, some E2 enzymes, such as UbcH5s, can generate several types of polyubiquitin chains. In contrast to Lys-linked polyubiquitin chains, linear chains are determined by E3.Citation20 The linear ubiquitin chain assembly complex (LUBAC) is the only E3 enzyme to generate linear polyubiquitin chains, together with several E2s, including UbcH5s and E2-25K, the latter of which was shown to generate K48-linked chains specifically.Citation20,Citation25 Since E3s determines the substrate for ubiquitination,Citation26 LUBAC determines the specificity of both the substrate and the type of polyubiquitin chain.Citation26 Thus, the number of linear polyubiquitination substrates seems to be limited. Consequently, the physiological functions of linear polyubiquitin chains must be limited. Any E2 enzyme can bind to a number of E3 enzymes and conjugate polyubiquitin chains to substrates specifically recognized by those E3s.Citation26 Thus, the E2 complex containing Ubc13, for example, can conjugate K63-linked chains to numerous substrates. Moreover, it has been suggested that K63 chains may be generated by other E2 enzymes, such as UbcH5s, even in the absence of E2 complexes containing Ubc13.Citation27 Ubc13 KO is embryonic lethal in mice.Citation28 Therefore, genetic analysis cannot readily be applied to probe the function of specific Lys-linked ubiquitin chains in organisms. However, that is not the case with linear polyubiquitin chains. The unique feature of linear polyubiquitination discussed above enabled us to probe the function of linear chains using genetic analysis, because LUBAC is the only reported E3 known to date to specifically generate linear chains, and no other E2 or E3 enzymes are known to generate the unique chains.Citation7 Indeed, LUBAC component gene knockout provided solid evidence of the involvement of LUBAC-mediated linear polyubiquitination in NFκB signaling.Citation21,Citation29–Citation31
Mechanism Underlying LUBAC-Mediated NFκB Activation
HOIL-1L and HOIP were first identified as components of LUBAC.Citation20 Biochemical analysis subsequently revealed that LUBAC-mediated linear polyubiquitination is involved in NFκB activation.Citation21 Primary hepatocytes isolated from HOIL-1L-KO mice generated in our laboratory have severely impaired TNFα-induced NFκB activation.Citation21 However, TNFα-induced NFκB activation is not completely abolished in HOIL-1L-KO mice.Citation21 Knocking-out of molecules essential for NFκB activation, such as NEMO or IKKβ, is embryonic lethal in mice,Citation32–Citation34 but HOIL-1L KO is not.Citation21 The expression of HOIP, the catalytic center of LUBAC, is drastically decreased but not completely absent in HOIL-1L-KO cells.Citation29 This observation led to the hypothesis that HOIP may have another binding partner besides HOIL-1L and SHARPIN was identified.Citation29 The C terminus of SHARPIN exhibits significant homology with the N-terminal half of HOIL-1L that is essential for binding to HOIP.Citation29–Citation31 Although SHARPIN was isolated as a SHANK-binding protein in 2001,Citation35 SHARPIN was also identified as a causative gene in the cpdm mouse phenotype in 2007.Citation36,Citation37 Cpdm mice are spontaneous mutant mice exhibiting pleomorphic phenotypes, including chronic dermatitis, arthritis and immune disorders. However, the precise mechanism by which loss of SHARPIN provokes these phenotypes in cpdm mice has not been identified. Further analysis showed that SHARPIN forms a complex, not only with HOIP, but also HOIL-1L; namely, SHARPIN formed the tertiary complex with HOIL-1L and HOIP. The complex composed of HOIL-1L, HOIP and SHARPIN conjugates to linear polyubiquitin chains.Citation29–Citation31 Using genetic analysis, the lack of SHARPIN was found to drastically reduce the amount of the other components of LUBAC, HOIL-1L and HOIP, by destabilizing them, thereby attenuating NFκB activation induced by TNFα, CD40 or LT-βR.Citation29–Citation31 Thus, we hypothesized that the complex phenotype of cpdm mice may be induced by severely attenuated but not completely abolished signal-induced NFκB activation, since residual LUBAC, composed of HOIL-1L and HOIP, possesses linear polyubiquitination and NFκB activation activity. This issue will be discussed later in the article.
The molecular mechanism underlying LUBAC-mediated NFκB activation was also analyzed. LUBAC was found to form a complex with NEMO in a signal-dependent manner, such as upon stimulation with TNFα, and to conjugate linear polyubiquitin to NEMO.Citation21 Genetic analysis demonstrated that both linear polyubiquitination of NEMO and activation of IKK were severely impaired in cells lacking HOIL-1L or SHARPIN.Citation21,Citation29–Citation31 Since the introduction of NEMO conjugated to uncleavable linear polyubiquitin can activate NFκB but introduction of GFP conjugated to linear polyubiquitin cannot, linearly polyubiquitinated NEMO seems critical for activation of IKK.Citation21 Thus, the current concept for LUBAC-mediated NFκB activation is as follows: upon stimulation by inflammatory cytokines such as TNFα and IL-1β and by the ligands of some Toll-like receptors, LUBAC recognizes and linearly polyubiquitinates NEMO, which induces IKK activation and subsequent degradation of IκBα. Free NFκB translocates into the nucleus and activates the transcription of target genes ().Citation7 In cpdm mice, the linear polyubiquitination of NEMO is attenuated because of the drastic reduction in the amount of LUBAC composed of HOIL-1L and HOIP due to lack of SHARPIN, resulting in attenuated NFκB activation ().
The precise mechanism by which the linear polyubiquitination of NEMO induces IKK activation has not yet been conclusively shown; however, the finding that NEMO binds to linear di-ubiquitin with much higher affinity than to other Lys-linked ubiquitin chains via its ubiquitin-binding motif, called UBAN or CoZi domain, may be insightful.Citation38,Citation39 Since this topic was discussed in our previous review in reference Citation7, it is described only briefly here. Recognition by NEMO of linear polyubiquitin chains conjugated to the NEMO molecules of other IKK complexes brings IKKβs close together and allows IKKβ trans-autophosphorylation, a process similar to that observed with receptor tyrosine kinases upon ligand-mediated dimerization. Alternatively, binding of linear ubiquitin polymers to the UBAN domain of NEMO induces conformational changes in NEMO and triggers changes in the spatial positioning of IKKα and IKKβ, leading to IKKβ activation. However, the mechanism underlying IKK activation is still extensively debated.Citation40,Citation41 Results using HOIL-1L-KO and cpdm mice conclusively show that LUBAC-mediated linear polyubiquitination plays a crucial role in NFκB activation.Citation21,Citation29–Citation31 However, since neither HOIL-1L-KO nor cpdm mice are embryonic lethal,Citation21,Citation29–Citation31 it is not yet known whether linear polyubiquitination is essential for canonical NFκB activation. Knockdown of HOIL-1L in cpdm cells suppresses the expression of HOIL-1L and HOIP and abolishes TNFα- and IL-1β-mediated NFκB activation almost completely.Citation29 Thus, LUBAC-mediated linear polyubiquitination was hypothesized to be indispensable for TNFα- and IL-1β-induced NFκB activation; however, HOIL-1L-KO mice crossed with cpdm mice or HOIP-KO mice will be needed to clarify this issue.
Involvement of Multiple Ubiquitin Chains in NFκB Activation and Their Possible Roles
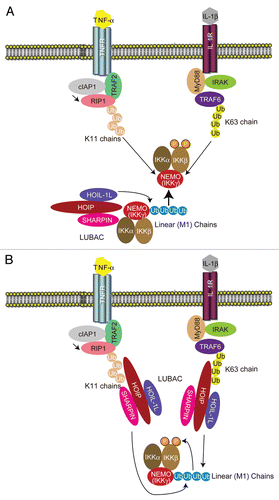
As mentioned above, the ubiquitin conjugation system is heavily involved in NFκB activation.Citation8 It has been well documented that K63-linked chains generated by Ubc13-Uev1a are involved in NFκB activation.Citation16 Extensive work on the role of K63 chains in signaling has been performed. It is currently hypothesized that the Ubc13-Uev1a complex and TRAF6 conjugate K63-linked chains to TRAF6 itself and to RIP1 upon stimulation. This event recruits the TAK1-TAB1-TAB2/3 complex to K63-linked chains through the K63-selective binding of NZFs of TAB2/3.Citation8,Citation42 The IKK complex was also suggested to be recruited to K63-linked chains via ubiquitin-binding domains in NEMO,Citation43 allowing IKK activation by phosphorylation of IKKβ by TAK1Citation44 and leading to the activation of NFκB. However, the involvement of K63-linked chains in NFκB activation has been challenged by the observation that TNFα-mediated NFκB activation is not overtly affected in cells isolated from Ubc13-KO mice, although TNFα-induced JNK activation is severely impaired in those cells.Citation28 Moreover, the finding that NEMO binds preferentially to linear chains also challenges this concept.Citation38,Citation39 Nonetheless, as mentioned previously, the lack of the LUBAC components SHARPIN or HOIL-1L severely impairs TNFα-mediated IKK activation but does not completely abolish it.Citation21,Citation29–Citation31 Although we suspect that residual LUBAC composed of the other two components can activate NFκB in those cells, the existence of other pathways that can activate IKK co-laterally by linear polyubiquitination cannot be excluded. Data suggested that NEMO can bind to tetra-K63-linked ubiquitin with comparable affinity to tetra-linear chains.Citation41,Citation45,Citation46 It has also been reported that c-IAPs are indispensable for TNFα-induced NFκB activation,Citation47,Citation48 and c-IAPs can generate K11-linked chains that can be recognized by NEMO with comparable affinity to K63-linked chains.Citation49 Therefore, K63-linked or K11-linked chains might be recognized by NEMO and can activate IKK (). However, K63-linked chains are indispensable for IL-1β-induced NFκB activation, although K63-linked chains are dispensable for TNFα-mediated NFκB activation.Citation50 Therefore, K63 chains may exert additional roles in NFκB activation besides activation of IKK directly. For example, K63-linked chains may recruit LUBAC to the IL-1 receptor. K11-linked chains generated by cIAPs may recruit LUBAC to TNFR1 (). Since LUBAC exhibits ubiquitin-binding activity,Citation20,Citation30,Citation31 further analysis, including analysis of the ubiquitin-binding domains in LUBAC, will clarify the roles of the different polyubiquitin chains in NFκB activation.
In addition to NFκB, LUBAC-mediated linear polyubiquitination might be involved in other signaling pathways. CD40-mediated activation of JNK is attenuated in splenic B cells from mice lacking SHARPIN or HOIL-1L,Citation29–Citation31 suggesting that LUBAC-mediated linear polyubiquitination is also involved in MAPK activation. It has been hypothesized that the linear polyubiquitination activity of LUBAC stabilizes the complex formed with TNFR1 in TNFα-stimulated cells and leads to the activation of MAPKs. However, TNFα-mediated JNK activation was not downregulated in MEFs from mice lacking HOIL-1L or SHARPIN.Citation21,Citation29 In addition, ABIN-2, which has a ubiquitin-binding domain homologous to the UBAN domain of NEMO,Citation51 is involved in MAPK activation.Citation52 Moreover, the protein substrates of LUBAC, which is involved in the stabilization of the TNFR1 complex, are not yet known. Thus, further analysis is needed to clarify the involvement of LUBAC in MAPK activation.
Mechanism Underlying the Generation of cpdm Phenotypes Induced by Lack of SHARPIN
Chronic dermatitis (), which is the most overt phenotype of cpdm mice, is characterized by epidermal hyperplasia and infiltration of inflammatory cells.Citation53 Three groups, including ours, have shown that TNFα-induced NFκB activation was severely impaired in cpdm cells.Citation29–Citation31 Keratinocyte-specific deletion of IKKβ or NEMO induces dermatitis with epidermal hyperplasia and infiltration of inflammatory cells.Citation54,Citation55 Crossing the conditional KO mice with TNFR1-KO mice remits dermatitis and inflammatory cell infiltra tion.Citation54,Citation55 In cpdm keratinocytes, TNFα-induced NFκB activation is also strongly attenuated. Since dermatitis disappears upon crossing cpdm mice with TNFα-KO mice,Citation31 attenuation of TNFα-mediated NFκB activation may be involved in the pathogenesis of proliferative dermatitis. However, attenuated TNFα-induced NFκB activation per se may not be the only cause of the dermatitis observed in cpdm mice, since HOIL-1L-KO mice do not have dermatitis (data not shown) despite severe attenuation of TNFα-induced NFκB activation.Citation21 Cpdm cells are more sensitive to TNFα-induced apoptosis than HOIL-1L-KO cells. Although the mechanism underlying enhanced sensitivity to TNFα-induced apoptosis in cpdm mice has not been solved, it may be attributed to the fact that SHARPIN plays crucial roles in linear ubiquitination or induction of molecules suppressing apoptosis.Citation56 Since keratinocyte apoptosis may play a crucial role in the dermatitis observed in keratinocyte-specific IKKβ or NEMO-KO mice,Citation57 enhanced TNFα-induced apoptosis in cpdm mice may underlie the pathogenesis of the dermatitis, although the mechanism underlying the phenotypic differences between these two mice has not yet been identified.Citation29–Citation31 In addition to enhanced sensitivity to TNFα-induced apoptosis, cpdm mice exert immunological abnormality. Increased production of type 2 cytokines (IL-4, IL-5 and IL-13) and decreased IFNγ secretion were reported in cpdm mice. Treatment with IL-12, a known inducer of IFNγ, alleviated dermatitis in cpdm mice.Citation58 Therefore, enhanced TNFα-induced apoptosis and reduction of type 1 cytokines in skin might also be involved in the pathogenesis of dermatitis in cpdm, not just attenuation of TNFα-induced NFκB activation. Although crossing cpdm mice with TNFα-KO mice suppresses proliferative dermatitis, the immunological abnormalities were not suppressed.Citation31 Since LUBAC-induced linear polyubiquitination is involved in NFκB activation by several TNFR and Toll-like receptor family members,Citation29–Citation31 attenuation of NFκB activation induced by some of these receptors, except TNFR1, might mediate the immunological abnormalities observed in cpdm mice.
Future Directions
A function of linear polyubiquitin chains has become clear:Citation7 LUBAC-induced linear polyubiquitination is involved in NFκB activation by several stimuli, including UV.Citation59 NEMO has been identified as a substrate of LUBAC, and linearly ubiquitinated NEMO may trigger IKK activation.Citation7 Thus, the analysis of the molecular mechanisms underlying LUBAC-induced NFκB activation is one of the most important issues in linear polyubiquitin and NFκB research, including the identification of deubiquitinating enzymes for linear ubiquitin chains. Several deubiquitinating enzymes, including A20 and CYLD, downregulate NFκB signaling.Citation60–Citation63 CYLD can digest linear chains but A20 cannot.Citation64 Since some reports suggested that deubiquitinating activity of A20 is low, A20 is suggested to exert its NFκB suppression activity in a deubiquitination-independent manner.Citation65 Although CYLD downregulates NFκB activation by degrading K63-linked chains,Citation66 it may function through linear chains. Alternatively, unknown deubiquitinating enzymes may downregulate NFκB signaling by digesting linear chains. In addition, since LUBAC may be involved in the TNFα-stimulated TNF receptor complex assembly, the identification of other LUBAC substrates besides NEMO and RIP1 may help identify the molecular mechanisms underlying complex stabilization.Citation31,Citation67 Another important issue to be addressed is whether linear polyubiquitin chains and/or LUBAC are involved in biological phenomena other than NFκB activation. Recent structural and biochemical analyses of ubiquitin-binding domains revealed the existence of ubiquitin binding domains that discriminate types of polyubiquitin chains.Citation68–Citation70 In the case of linear chains, the UBAN motif of NEMO exhibits a much higher affinity to linear di-ubiquitin than to K63 di-ubiquitin.Citation38,Citation39 The identification of linear chain-specific binding domains and the functional analysis of proteins containing these domains will help elucidate new roles for linear polyubiquitination. It has recently been suggested that linear polyubiquitination is involved in autophagy.Citation71 Optineurin, which possesses a domain homologous to NEMO,Citation72 is involved in autophagy in Salmonella and binds to linear polyubiquitin chains.Citation71 Optineurin was identified as one of the causative genes of amyotrophic lateral sclerosis (ALS), a motor neuron disease,Citation73 and an optineurin mutant found in ALS patients fails to bind linear polyubiquitin and to induce autophagy in Salmonella.Citation71 These reports strongly indicate that linear polyubiquitination is involved in selective autophagy and in the pathogenesis of ALS. Thus, whether LUBAC is involved in selective autophagy is of interest, since, so far, LUBAC is the only known E3 to generate linear chains. Alternatively, other E3 enzymes that specifically generate linear chains may be involved in autophagy. Preliminary analysis revealed that, among the three subunits of LUBAC, HOIP, which is the catalytic center of the complex, plays a crucial role in determining the type of ubiquitin chain generated by the ligase complex (data not shown). Since we have not identified any E3 enzyme that exhibits significant homology with the region of HOIP that seems crucial for linear chain generation so far (data not shown), we cannot suggest the presence of other E3s that generate linear polyubiquitin chains specifically. Linear polyubiquitin chains might conceivably be generated by a special combination of E2(s) and E3(s). The identification of other E3 enzymes or of combinations of E2 and E3 enzymes capable of generating linear polyubiquitin chains and the dissection of their pathophysiological functions might reveal unexpected roles for linear polyubiquitination in biology and medicine.
Figures and Tables
Figure 1 The NFκB activation pathway. NFκB (p65-p50 heterodimer) resides in the cytoplasm in resting cells by binding to the inhibitor protein IκBα. Upon activation by various stimuli, IκBα is phosphorylated by the IKK complex. K48-linked polyubiquitination of phosphorylated IκBα leads to its degradation. Subsequently, free NFκB translocates into the nucleus and induces the expression of target genes.
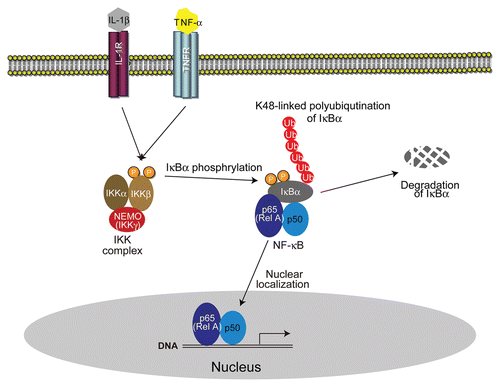
Figure 2 The ubiquitin conjugation system. Ubiquitin conjugation is a reversible post-translational modification that regulates numerous biological phenomena by conjugating ubiquitin polymers to proteins. Polyubiquitin chains are generated by the repetition of the cascade of reactions catalyzed by three enzymes, E1 ubiquitin activating enzymes, E2 ubiquitin conjugating enzymes and E3 ubiquitin-protein ligases, on target proteins specifically recognized by E3s (A). Polyubiquitin chains have been thought to be generated via the Lys residues of ubiquitin. Polyubiquitin chains that function as degradation signals are generated via the Lys 48 of ubiquitin (B). K63-linked chains are involved in DNA repair and signal transduction and do not function as degradation signals (C). A new type of polyubiquitin chain was identified, a linear polyubiquitin (M1-linked) chain in which the C-terminal of ubiquitin is bound to the α-amino group of another ubiquitin. Linear polyubiquitin chains play crucial roles in NFκB activation (D).
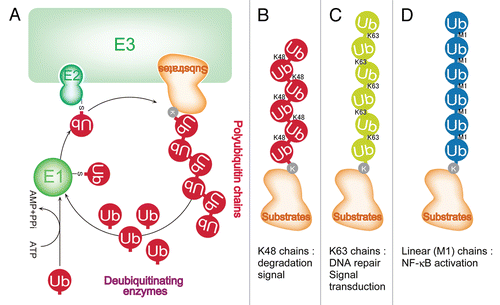
Figure 3 Involvement of LUBAC-mediated linear ubiquitination in NFκB activation and mechanism underlying attenuated NFκB activation by loss of SHARPIN. Upon stimulation by inflammatory cytokines including TNFα and IL-1β, LUBAC, which is composed of HOIL-1L, HOIP and SHARPIN, recognizes and linearly polyubiquitinates NEMO, which induces IKK activation and leads to the degradation of IκBα. Free NFκB translocates into the nucleus and activates the transcription of target genes (A). In cpdm mice, the linear polyubiquitination of NEMO is attenuated because of the drastic reduction in the amount of LUBAC, composed solely of HOIL-1L and HOIP due to lack of SHARPIN, resulting in attenuated linear polyubiquitination of NEMO and attenuated NFκB activation (B).
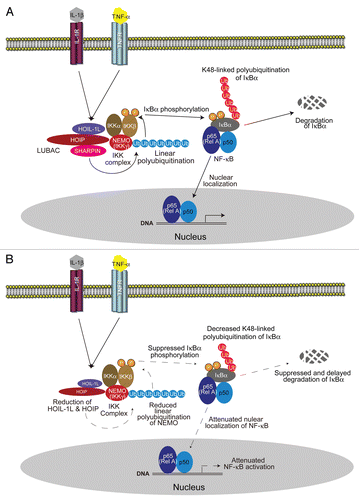
Figure 4 Possible roles of different polyubiquitin chains in NFκB activation. (A) LUBAC-mediated linear polyubiquitinated NEMO can activate IKK. A hypothesis is shown according to which linearly polyubiquitinated NEMO can be recognized by other NEMO molecules, which induces phosphorylation of IKKβ. K63-linked chains or K11-linked chains generated by TRAF6 or cIAP1, respectively, are also recognized by NEMO, which may induce phosphorylation of IKKβ. (B) K63-linked chains generated by TRAF6 and K11-linked chains by cIAP1 may recruit LUBAC to the IL-1 receptor or the TNF receptor, respectively, and induce linear polyubiquitination of NEMO. Linearly polyubiquitinated NEMO induces IKKβ phosphorylation and NFκB activation.
Acknowledgments
Work in my laboratory was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan and the CREST Japan Science Technology Corporation.
References
- Hayden MS, Ghosh S. Shared principles in NFκB signaling. Cell 2008; 132:344 - 362; PMID: 18267068; http://dx.doi.org/10.1016/j.cell.2008.01.020
- Staudt LM. Oncogenic activation of NFκB. Cold Spring Harb Perspect Biol 2010; 2:109; PMID: 20516126; http://dx.doi.org/10.1101/cshperspect.a000109
- Kato M, Sanada M, Kato I, Sato Y, Takita J, Takeuchi K, et al. Frequent inactivation of A20 in B-cell lymphomas. Nature 2009; 459:712 - 716; PMID: 19412163; http://dx.doi.org/10.1038/nature07969
- Paradisi A, Mehlen P. Netrin-1, a missing link between chronic inflammation and tumor progression. Cell Cycle 2010; 9:1253 - 1262; PMID: 20305387; http://dx.doi.org/10.4161/cc.9.7.11072
- Demchenko YN, Kuehl WM. A critical role for the NFκB pathway in multiple myeloma. Oncotarget 2010; 1:59 - 68; PMID: 20890394
- Bonizzi G, Karin M. The two NFκB activation pathways and their role in innate and adaptive immunity. Trends Immunol 2004; 25:280 - 288; PMID: 15145317; http://dx.doi.org/10.1016/j.it.2004.03.008
- Iwai K, Tokunaga F. Linear polyubiquitination: a new regulator of N NFκB activation. EMBO Rep 2009; 10:706 - 713; PMID: 19543231; http://dx.doi.org/10.1038/embor.2009.144
- Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NFκB regulatory pathways. Annu Rev Biochem 2009; 78:769 - 796; PMID: 19489733; http://dx.doi.org/10.1146/annurev.biochem.78.070907.102750
- Ciehanover A, Hod Y, Hershko A. A heat-stable polypeptide component of an ATP-dependent proteolytic system from reticulocytes. Biochem Biophys Res Commun 1978; 81:1100 - 1105; PMID: 666810; http://dx.doi.org/10.1016/0006-291X(78)91249-4
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 2002; 82:373 - 428; PMID: 11917093
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem 1998; 67:425 - 479; PMID: 9759494; http://dx.doi.org/10.1146/annurev.biochem.67.1.425
- Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol 2009; 10:755 - 764; PMID: 19851334; http://dx.doi.org/10.1038/nrm2780
- Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning AM, et al. Identification of the receptor component of the IκBα-ubiquitin ligase. Nature 1998; 396:590 - 594; PMID: 9859996; http://dx.doi.org/10.1038/25159
- Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW. The SCFβTRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev 1999; 13:270 - 283; PMID: 9990852; http://dx.doi.org/10.1101/gad.13.3.270
- Spencer E, Jiang J, Chen ZJ. Signal-induced ubiquitination of IκBα by the F-box protein Slimb/β-TrCP. Genes Dev 1999; 13:284 - 294; PMID: 9990853; http://dx.doi.org/10.1101/gad.13.3.284
- Deng L, Wang C, Spencer E, Yang L, Braun A, You J, et al. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 2000; 103:351 - 361; PMID: 11057907; http://dx.doi.org/10.1016/S0092-8674(00)00126-4
- Spence J, Sadis S, Haas AL, Finley D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol 1995; 15:1265 - 1273; PMID: 7862120
- Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem 2009; 78:363 - 397; PMID: 19489724; http://dx.doi.org/10.1146/annurev.biochem.78.082307.091526
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol 2003; 21:921 - 926; PMID: 12872131; http://dx.doi.org/10.1038/nbt849
- Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J 2006; 25:4877 - 4887; PMID: 17006537; http://dx.doi.org/10.1038/sj.emboj.7601360
- Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, et al. Involvement of linear polyubiquitylation of NEMO in NFκB activation. Nat Cell Biol 2009; 11:123 - 132; PMID: 19136968; http://dx.doi.org/10.1038/ncb1821
- Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 1999; 96:645 - 653; PMID: 10089880; http://dx.doi.org/10.1016/S0092-8674(00)80575-9
- Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell 2011; 144:769 - 781; PMID: 21376237; http://dx.doi.org/10.1016/j.cell.2011.01.035
- Pierce NW, Kleiger G, Shan SO, Deshaies RJ. Detection of sequential polyubiquitylation on a millisecond timescale. Nature 2009; 462:615 - 619; PMID: 19956254; http://dx.doi.org/10.1038/nature08595
- Haldeman MT, Xia G, Kasperek EM, Pickart CM. Structure and function of ubiquitin conjugating enzyme E2-25K: the tail is a core-dependent activity element. Biochemistry 1997; 36:10526 - 10537; PMID: 9265633; http://dx.doi.org/10.1021/bi970750u
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem 2009; 78:399 - 434; PMID: 19489725; http://dx.doi.org/10.1146/annurev.biochem.78.101807.093809
- Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, et al. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem 2007; 282:17375 - 17386; PMID: 17426036; http://dx.doi.org/10.1074/jbc.M609659200
- Yamamoto M, Okamoto T, Takeda K, Sato S, Sanjo H, Uematsu S, et al. Key function for the Ubc13 E2 ubiquitin-conjugating enzyme in immune receptor signaling. Nat Immunol 2006; 7:962 - 970; PMID: 16862162; http://dx.doi.org/10.1038/ni1367
- Tokunaga F, Nakagawa T, Nakahara M, Saeki Y, Taniguchi M, Sakata S, et al. SHARPIN is a component of the NFκB-activating linear ubiquitin chain assembly complex. Nature 2011; 471:633 - 636; PMID: 21455180; http://dx.doi.org/10.1038/nature09815
- Ikeda F, Deribe YL, Skanland SS, Stieglitz B, Grabbe C, Franz-Wachtel M, et al. SHARPIN forms a linear ubiquitin ligase complex regulating NFκB activity and apoptosis. Nature 2011; 471:637 - 641; PMID: 21455181; http://dx.doi.org/10.1038/nature09814
- Gerlach B, Cordier SM, Schmukle AC, Emmerich CH, Rieser E, Haas TL, et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 2011; 471:591 - 596; PMID: 21455173; http://dx.doi.org/10.1038/nature09816
- Schmidt-Supprian M, Bloch W, Courtois G, Addicks K, Israel A, Rajewsky K, et al. NEMO/IKK γ-deficient mice model incontinentia pigmenti. Mol Cell 2000; 5:981 - 992; PMID: 10911992; http://dx.doi.org/10.1016/S1097-2765(00)80263-4
- Rudolph D, Yeh WC, Wakeham A, Rudolph B, Nallainathan D, Potter J, et al. Severe liver degeneration and lack of N NFκB activation in NEMO/IKKγ-deficient mice. Genes Dev 2000; 14:854 - 862; PMID: 10766741
- Li ZW, Chu W, Hu Y, Delhase M, Deerinck T, Ellisman M, et al. The IKKbeta subunit of IκB kinase (IKK) is essential for nuclear factor κB activation and prevention of apoptosis. J Exp Med 1999; 189:1839 - 1845; PMID: 10359587; http://dx.doi.org/10.1084/jem.189.11.1839
- Lim S, Sala C, Yoon J, Park S, Kuroda S, Sheng M, et al. Sharpin, a novel postsynaptic density protein that directly interacts with the shank family of proteins. Mol Cell Neurosci 2001; 17:385 - 397; PMID: 11178875; http://dx.doi.org/10.1006/mcne.2000.0940
- Seymour RE, Hasham MG, Cox GA, Shultz LD, Hogenesch H, Roopenian DC, et al. Spontaneous mutations in the mouse Sharpin gene result in multiorgan inflammation, immune system dysregulation and dermatitis. Genes Immun 2007; 8:416 - 421; PMID: 17538631; http://dx.doi.org/10.1038/sj.gene.6364403
- HogenEsch H, Gijbels MJ, Offerman E, van Hooft J, van Bekkum DW, Zurcher C. A spontaneous mutation characterized by chronic proliferative dermatitis in C57BL mice. Am J Pathol 1993; 143:972 - 982; PMID: 8362989
- Lo YC, Lin SC, Rospigliosi CC, Conze DB, Wu CJ, Ashwell JD, et al. Structural basis for recognition of diubiquitins by NEMO. Mol Cell 2009; 33:602 - 615; PMID: 19185524; http://dx.doi.org/10.1016/j.molcel.2009.01.012
- Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NFκB activation. Cell 2009; 136:1098 - 1109; PMID: 19303852; http://dx.doi.org/10.1016/j.cell.2009.03.007
- Chiu YH, Zhao M, Chen ZJ. Ubiquitin in NFκB signaling. Chem Rev 2009; 109:1549 - 1560; PMID: 19281271; http://dx.doi.org/10.1021/cr800554j
- Laplantine E, Fontan E, Chiaravalli J, Lopez T, Lakisic G, Veron M, et al. NEMO specifically recognizes K63-linked poly-ubiquitin chains through a new bipartite ubiquitin-binding domain. EMBO J 2009; 28:2885 - 2895; PMID: 19763089; http://dx.doi.org/10.1038/emboj.2009.241
- Kanayama A, Seth RB, Sun L, Ea CK, Hong M, Shaito A, et al. TAB2 and TAB3 activate the NFκB pathway through binding to polyubiquitin chains. Mol Cell 2004; 15:535 - 548; PMID: 15327770; http://dx.doi.org/10.1016/j.molcel.2004.08.008
- Chen F, Bhatia D, Chang Q, Castranova V. Finding NEMO by K63-linked polyubiquitin chain. Cell Death Differ 2006; 13:1835 - 1838; PMID: 16858426; http://dx.doi.org/10.1038/sj.cdd.4402014
- Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 2001; 412:346 - 351; PMID: 11460167; http://dx.doi.org/10.1038/35085597
- Hubeau M, Ngadjeua F, Puel A, Israel L, Feinberg J, Chrabieh M, et al. A new mechanism of X-linked anhidrotic ectodermal dysplasia with immunodeficiency: impairment of ubiquitin binding despite normal folding of NEMO protein. Blood 2011;
- Yoshikawa A, Sato Y, Yamashita M, Mimura H, Yamagata A, Fukai S. Crystal structure of the NEMO ubiquitin-binding domain in complex with Lys 63-linked di-ubiquitin. FEBS Lett 2009; 583:3317 - 3322; PMID: 19766637; http://dx.doi.org/10.1016/j.febslet.2009.09.028
- Vince JE, Wong WW, Khan N, Feltham R, Chau D, Ahmed AU, et al. IAP antagonists target cIAP1 to induce TNFα-dependent apoptosis. Cell 2007; 131:682 - 693; PMID: 18022363; http://dx.doi.org/10.1016/j.cell.2007.10.037
- Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, Garg P, et al. IAP antagonists induce autoubiquitination of c-IAPs, NFκB activation and TNFalpha-dependent apoptosis. Cell 2007; 131:669 - 681; PMID: 18022362; http://dx.doi.org/10.1016/j.cell.2007.10.030
- Dynek JN, Goncharov T, Dueber EC, Fedorova AV, Izrael-Tomasevic A, Phu L, et al. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J 2010; 29:4198 - 4209; PMID: 21113135; http://dx.doi.org/10.1038/emboj.2010.300
- Xu M, Skaug B, Zeng W, Chen ZJ. A Ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFα and IL-1β. Mol Cell 2009; 36:302 - 314; PMID: 19854138; http://dx.doi.org/10.1016/j.molcel.2009.10.002
- Wagner S, Carpentier I, Rogov V, Kreike M, Ikeda F, Lohr F, et al. Ubiquitin binding mediates the N NFκB inhibitory potential of ABIN proteins. Oncogene 2008; 27:3739 - 3745; PMID: 18212736; http://dx.doi.org/10.1038/sj.onc.1211042
- Papoutsopoulou S, Symons A, Tharmalingham T, Belich MP, Kaiser F, Kioussis D, et al. ABIN-2 is required for optimal activation of Erk MAP kinase in innate immune responses. Nat Immunol 2006; 7:606 - 615; PMID: 16633345; http://dx.doi.org/10.1038/ni1334
- Gijbels MJ, Zurcher C, Kraal G, Elliott GR, HogenEsch H, Schijff G, et al. Pathogenesis of skin lesions in mice with chronic proliferative dermatitis (cpdm/cpdm). Am J Pathol 1996; 148:941 - 950; PMID: 8774148
- Pasparakis M, Courtois G, Hafner M, Schmidt-Supprian M, Nenci A, Toksoy A, et al. TNF-mediated inflammatory skin disease in mice with epidermis-specific deletion of IKK2. Nature 2002; 417:861 - 866; PMID: 12075355; http://dx.doi.org/10.1038/nature00820
- Nenci A, Huth M, Funteh A, Schmidt-Supprian M, Bloch W, Metzger D, et al. Skin lesion development in a mouse model of incontinentia pigmenti is triggered by NEMO deficiency in epidermal keratinocytes and requires TNF signaling. Hum Mol Genet 2006; 15:531 - 542; PMID: 16399796; http://dx.doi.org/10.1093/hmg/ddi470
- Rushworth SA, Zaitseva L, Langa S, Bowles KM, MacEwan DJ. FLIP regulation of HO-1 and TNF signalling in human acute myeloid leukemia provides a unique secondary anti-apoptotic mechanism. Oncotarget 2010; 1:359 - 366; PMID: 21307400
- Stratis A, Pasparakis M, Rupec RA, Markur D, Hartmann K, Scharffetter-Kochanek K, et al. Pathogenic role for skin macrophages in a mouse model of keratinocyte-induced psoriasis-like skin inflammation. J Clin Invest 2006; 116:2094 - 2104; PMID: 16886058; http://dx.doi.org/10.1172/JCI27179
- HogenEsch H, Torregrosa SE, Boggess D, Sundberg BA, Carroll J, Sundberg JP. Increased expression of type 2 cytokines in chronic proliferative dermatitis (cpdm) mutant mice and resolution of inflammation following treatment with IL-12. Eur J Immunol 2001; 31:734 - 742; PMID: 11241277; http://dx.doi.org/10.1002/1521-4141(200103)31:3<734::AIDIMMU734>3.0.CO;2-9
- Niu J, Shi Y, Iwai K, Wu ZH. LUBAC regulates NFκB activation upon genotoxic stress by promoting linear ubiquitination of NEMO. EMBO J In press PMID: 21811235
- Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NFκB signalling. Nature 2004; 430:694 - 699; PMID: 15258597; http://dx.doi.org/10.1038/nature02794
- Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NFκB. Nature 2003; 424:797 - 801; PMID: 12917690; http://dx.doi.org/10.1038/nature01811
- Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NFκB signalling by deubiquitination. Nature 2003; 424:801 - 805; PMID: 12917691; http://dx.doi.org/10.1038/nature01802
- Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NFκB activation by TNFR family members. Nature 2003; 424:793 - 796; PMID: 12917689; http://dx.doi.org/10.1038/nature01803
- Komander D, Reyes-Turcu F, Licchesi JD, Odenwaelder P, Wilkinson KD, Barford D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep 2009; 10:466 - 473; PMID: 19373254; http://dx.doi.org/10.1038/embor.2009.55
- Shembade N, Harhaj E. A20 inhibition of NFκB and inflammation: targeting E2:E3 ubiquitin enzyme complexes. Cell Cycle 2010; 9:2481 - 2482; PMID: 20543575; http://dx.doi.org/10.4161/cc.9.13.12269
- Sun SC. CYLD: a tumor suppressor deubiquitinase regulating NFκB activation and diverse biological processes. Cell Death Differ 2010; 17:25 - 34; PMID: 19373246; http://dx.doi.org/10.1038/cdd.2009.43
- Haas TL, Emmerich CH, Gerlach B, Schmukle AC, Cordier SM, Rieser E, et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol Cell 2009; 36:831 - 844; PMID: 20005846; http://dx.doi.org/10.1016/j.molcel.2009.10.013
- Kulathu Y, Akutsu M, Bremm A, Hofmann K, Komander D. Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat Struct Mol Biol 2009; 16:1328 - 1330; PMID: 19935683; http://dx.doi.org/10.1038/nsmb.1731
- Sims JJ, Cohen RE. Linkage-specific avidity defines the lysine 63-linked polyubiquitinbinding preference of rap80. Mol Cell 2009; 33:775 - 783; PMID: 19328070; http://dx.doi.org/10.1016/j.molcel.2009.02.011
- Sato Y, Yoshikawa A, Mimura H, Yamashita M, Yamagata A, Fukai S. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J 2009; 28:2461 - 2468; PMID: 19536136; http://dx.doi.org/10.1038/emboj.2009.160
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 2011; 333:228 - 233; PMID: 21617041; http://dx.doi.org/10.1126/science.1205405
- Zhu G, Wu CJ, Zhao Y, Ashwell JD. Optineurin negatively regulates TNFα-induced NFκB activation by competing with NEMO for ubiquitinated RIP. Curr Biol 2007; 17:1438 - 1443; PMID: 17702576; http://dx.doi.org/10.1016/j.cub.2007.07.041
- Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature 2010; 465:223 - 226; PMID: 20428114; http://dx.doi.org/10.1038/nature08971
