Abstract
Peloruside A is a microtubule-stabilizing macrolide that binds to beta tubulin at a site distinct from the taxol site. The site was previously identified by H-D exchange mapping and molecular docking as a region close to the outer surface of the microtubule and confined in a cavity surrounded by a continuous loop of protein folded so as to center on Y340. We have isolated a series of peloruside A-resistant lines of the human ovarian carcinoma cell line A2780(1A9) to better characterize this binding site and the consequences of altering residues in it. Four resistant lines (Pel A-D) are described with single-base mutations in class I β-tubulin that result in the following substitutions: R306H, Y340S, N337D, and A296S in various combinations. The mutations are localized to peptides previously identified by Hydrogen-Deuterium exchange mapping, and center on a cleft in which the drug side chain appears to dock. The Pel lines are 10-15-fold resistant to peloruside A and show cross resistance to laulimalide but not to any other microtubule stabilizers. They show no cross-sensitivity to any microtubule destabilizers, nor to two drugs with targets unrelated to microtubules. Peloruside A induces G2/M arrest in the Pel cell lines at concentrations 10-15 times that required in the parental line. The cells show notable changes in morphology compared to the parental line.
Introduction
Small molecules that alter microtubule (MT) dynamics are usually categorized as MT-destabilizing or MT-stabilizing. All affect microtubules, and therefore all can alter MT-based cellular activities throughout the cell cycle. Perhaps the most dramatic of these drug effects is mitotic arrest. These drugs have had multiple successes in clinical applications, notably cancer chemotherapy, but also including inflammatory conditions such as gout and Familial Mediterranean Fever and parasite infections especially in veterinary contexts. Clinical efficacy, especially in cancer chemotherapy, is usually attributed to the ability of these drugs to arrest mitosis,Citation1 though it may not be the case that all such clinical success is due to mitotic inhibition.Citation2
MT-active compounds bind to one of four major drug binding sites on β-tubulin: two for destabilizing drugs, and two for stabilizing ones.Citation3 Many hundreds of MT-active compounds have been described, but only a handful have found acceptance in human clinical or veterinary use. These include colchicine, several vinca alkaloids, several benzimidazoles, taxanes and epothilones. The destabilizing compounds mostly bind to either the colchicine site or the vinca domain—a series of related and overlapping sites. A number of “secondary” binding sites for destabilizing agents have been described on tubulin, including certain sulfhydryl residues and other sites.Citation4,Citation5 Most stabilizing compounds bind to the paclitaxel site, including the MT stabilizing drugs in clinical use (taxanes and epothilones). The fourth site, and the second site for stabilizing drugs, is the binding site for peloruside A and laulimalide, and is the subject of this paper.
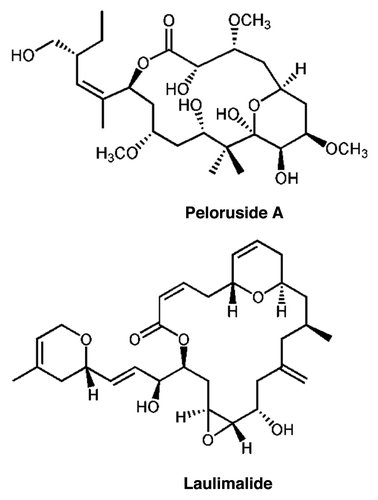
Peloruside A is a macrolide originally isolated from a marine sponge found off the coast of New Zealand.Citation6,Citation7 While it shares with paclitaxel (Taxol®) the ability to stabilize MT in cells and induce polymerization of purified tubulin, it does not bind to the paclitaxel site. This conclusion was reached due to evidence that peloruside A does not displace paclitaxel from MT, and does act synergistically with paclitaxel in cell-based assays.Citation8–Citation11 The macrolide laulimalide shares these properties with peloruside A.Citation9,Citation12 In addition, laulimalide can displace bound peloruside A,Citation8 and laulimalide and peloruside A are not synergistic in combination with each other, but only with paclitaxel binding site ligands.Citation11 These results indicated that peloruside A and laulimalide likely share a new binding site, distinct from the paclitaxel site, and possibly act via mechanisms of stabilization distinct from paclitaxel. This binding site is the second for stabilizing drugs, and the fourth of the four mentioned above. We previously located this site by hydrogen-deuterium exchange mapping (HDX), and showed it to be on β-tubulin, well-removed from the paclitaxel site, and exposed to the outer surface of the MT.Citation13 The structures of peloruside A and laulimalide are shown in . In this paper we describe several mutations in β-tubulin which confer resistance to the action of peloruside A and laulimalide, and which are all located in a crevice in the binding site which we previously identified.
Results
The 1A9 ovarian carcinoma cell line (a subclone of the A2780 cell line) was chosen for this study because we have already isolated from this line multiple lines resistant to microtubule-stabilizing and destabilizing drugs.Citation14,Citation15 Initial testing with the parental cells showed that the growth inhibitory IC50 for peloruside A was comparable to that of paclitaxel, but slightly higher (peloruside A IC50 ∼ 6 nM, paclitaxel IC50 ∼ 2 nM). Resistant lines were selected by prolonged exposure to peloruside A, as described in Methods, and similar to the protocol that we have used to isolate cell lines resistant to paclitaxelCitation14 and hemiasterlin.Citation15 Resistant cells were selected in an initial single step exposure, then divided into four populations (flasks) and exposed to increasing concentrations of peloruside A. Resistant lines were named with a letter for the flask and a number for the line split from that flask (hence Pel B1 and B2 represent two lines split from flask B early in secondary selection). The resistant lines are collectively referred to as Pelr lines.
Drug resistance of selected lines.
Four resistant lines, each from a different selection flask, were selected for further analysis. In addition, in some cases a second line from the same original flask was retained in the study (e.g., B1 and B2 will be discussed). Typical dose-dependent growth inhibition results are presented in and tabulated in . As can be seen in , the Pel lines exhibit >10-fold resistance to peloruside A compared with the parental line. These lines do not show any cross-resistance to the microtubule-stabilizing drug paclitaxel, as shown in . Similarly, the lines do not show cross-sensitivity to combretastatin A4 a microtubule-destabilizing drug ().
The resistant lines Pel A1, Pel B2, Pel C and Pel D1 were tested for sensitivity to a number of drugs in paired experiments with the parental line. These results are shown in as the fold resistance (IC50 of resistant line divided by the IC50 of the parental line). Results of multiple tests are shown for each resistant line to a range of drugs. The drugs tested include peloruside A and five other microtubule stabilizing drugs (laulimalide, paclitaxel, epothilone A, dictyostatin and discodermolide), four microtubule destabilizing drugs (combretastatin A4, Nocodazole, Vinblastine and D24851) and two drugs with nonmicrotubule targets [etoposide (VP-16) and romidepsin (depsipeptide FK228)].
The results shown in show that the only drug to which these resistant lines possess cross resistance is laulimalide, a compound that is structurally related to peloruside A (see ) and which is known to bind to the same site on tubulin as peloruside A. All lines show lower fold-resistance to laulimalide compared with peloruside A, but Pel D1 shows the lowest. None of the resistant lines show any cross-resistance to any of the other microtubule-stabilizing agents. Similarly, none of the lines show any cross-sensitivity or resistance to any of the microtubule destabilizing agents. Finally, the resistant lines show little resistance or sensitivity to non-microtubule targeting drugs, aside from a weak resistance to etoposide. All of these results are presented in detail in Table S1. In summary, these lines appear to be resistant to agents that bind at the peloruside A/laulimalide binding site on tubulin and not to drugs that target other binding sites.
Cell lines resistant to other microtubule-targeting agents may show alterations in the cell cycle or cycle time in the absence of drug compared with the parental cells.Citation15,Citation17,Citation18 To evaluate this, we examined the growth characteristics of the Pelr lines in the presence and absence of added peloruside A. Doubling times were determined by quantitation of DNA at various times after plating the cells (). The doubling time of the Pelr lines is not significantly different from that of the parental line and is not affected by 25 nM peloruside A added to the growth media. The small increase in DNA seen in the parental line in the presence of 25 nM peloruside A is surprising (since this is significantly above the growth IC50) and highly variable; it may be due to variable DNA synthesis after cell division blockage. The point we would emphasize is that, in the absence or presence of 25 nM peloruside A, the doubling time of the Pelr lines is not longer nor more variable than the parental cells in the absence of drug.
Cell cycle analysis.
Flow cytometry studies were performed in order to examine the possibility that the cell cycle is altered, and to examine the mechanism of toxicity of peloruside A to the Pelr lines at the higher concentrations required for growth arrest. It is possible that at these concentrations, the drug may cause growth arrest by a non-mitotic mechanism, and we wished to demonstrate the actual mechanism. The flow cytometry studies () show that in the absence of drug, the cell cycles of parental and Pel lines are essentially the same. Addition of paclitaxel induces G2/M accumulation as efficiently in the Pelr lines as in the parental line, as might be expected from the data in and . Low concentrations of peloruside A do induce G2/M accumulation in the parental line, but not in the Pelr lines. Higher concentrations of peloruside A, however, do induce G2/M accumulation in the Pelr lines, to a similar extent as peloruside A does in the parental line or as paclitaxel does in all lines. This result demonstrates that the drug is likely still able to bind to its binding site on tubulin in the Pelr cells, with the same functional result as in the parental line, but requiring a higher concentration of drug.
Cell morphology.
In the absence of peloruside A, the Pelr lines seem remarkably similar to the parental cells in terms of growth, cell cycle, and sensitivity to drugs that do not bind to the peloruside A/laulimalide binding site. However, with regard to shape, the different cell lines look different from each other and mostly different from the parental cells. This is shown in the photomicrographs in Figure S1. This result is different from the paclitaxel-resistant Ptx lines described previously in reference Citation15, which do not show the distinctive morphological changes seen with the Pelr lines (data not shown).
Mutations in β-tubulin in Pel lines.
All of the results thus far combine to suggest that the resistance to peloruside A in the Pel lines is due to an alteration in the binding site on tubulin. Our previous study used H-D exchange-mass spectrometry to track the peloruside A binding site to β-tubulin and to a defined locale.Citation13 Hence we expected to find a mutation in β-tubulin. The TUBB (also known as M40) gene codes for the major β-tubulin (class I) in these cells,Citation14 and therefore we sequenced the mRNA for this protein in the parental and Pel lines. Sequencing of the 1A9 parental cells confirmed that β-tubulin was wild type. Sequencing of the 1A9 Pelr cell lines revealed β-tubulin mutations in every population. 1A9 Pel A1 had two heterozygous mutations in β-tubulin; the first mutation changing the codon for amino acid 138 from a Serine to a stop codon (TCA→TAA) and the second converting amino acid 306 from an Arginine to a Histidine (CGC→CAC). 1A9 Pel B1 had a single mutation in β-tubulin which was the same R306H seen in 1A9 Pel A1 however it was homozygous in expression: parental sequence mRNA was not found. 1A9 Pel B2 had two heterozygous mutations in β-tubulin; the R306H mutation and one converting amino acid 340 from a Tyrosine to a Serine (TAC→TCC). 1A9 Pel C had two heterozygous mutations, the R306H mutation and another changing amino acid 337 from an Asparagine to an Aspartic Acid (AAC→GAC). 1A9 Pel D1 had a single mutation converting amino acid 296 from an Alanine to a Serine (GCC→TCC), and homozygous in expression: no parental sequence mRNA was found. Sequencing from cell clones of each Pel line (see Methods) with two heterozygous mutations found that all clones had both mutations. Thus lines that contain two mutations are heterozygous at two sites. These results are summarized in .
Location of mutations in tubulin.
The binding site for peloruside A was previously localized to a region of β-tubulin exposed on the outer surface of the microtubule and near the intradimer contact with β-tubulin.Citation13 All of the mutations found in the Pelr lines map in this site, shown in . Notably, all four mutations map to the cavity that binds the side chains of peloruside A and laulimalide (). The respective binding modes of both peloruside A and laulimalide are indicated in the figure.
The original docked model of peloruside ACitation13 retained some ambiguity in the orientation of the molecule within the binding site. Our recent efforts with laulimalide model building,Citation16 together with NMR data,Citation19 analog data,Citation20 and recent modeling results,Citation21 suggest an alternative binding mode. In this model peloruside A is reoriented by approximately 180 degrees, such that the side chain is buried in a cavity in the “back” of the binding site. This orientation is shown in . It represents a model based upon a significantly populated low-energy cluster from our previous study.Citation13 This ambiguity resonates with our more extensive analysis of the laulimalide binding mode where both “forward and reverse” orientations are nearly equivalently populated in docking exercises. However, binding free energy analysis indicated a strong preference for a binding mode with the side chain penetrating into a deep, narrow cavity within the site. By extension, we therefore favor this pose for peloruside A. Our model development does not involve a manual rebuilding of loop regions within the site, thus favoring a more extended geometry relative to that presented by Nguyen, et al.
The mass shift analyses based on H/D exchange methods favor overlapping binding sites for both peloruside A and laulimalide.Citation13,Citation16 A quantitative analysis of the shift data for each was performed under conditions where macrolide binding is saturated (Fig. S2). It highlights drug-induced mass shifts in all four peptides spanning the region, although one shows a reduced mass shift for laulimalide relative to peloruside (β294–301). This peptide spans the mutation found in Pel D1 (A296S), which confers the lowest resistance to laulimalide, while still providing resistance to peloruside A.
The residues mutated in the Pelr lines are conserved in evolution, as shown in . In human β isotypes are aligned, and only two differences are seen in the residues affected by Pelr mutations. In βII (TUBB2A) Ala 296 is changed to a Ser, as occurs in Pel D1, and in ?VI (TUBB1) Tyr 340 is changed to a cysteine. In other residues, the few changes that occur between isotypes I–V are fairly conservative. The hematopoietic isotype βVI is the most divergent, differing at multiple positions. This may underlie the unusual binding behavior of βVI-rich tubulin.Citation22 The entire region in which the Pel mutations occur, residues 296–345 is fairly conserved throughout animals, plants and protists, as brief inspection of the alignment in indicates. The four residues identified in the binding site mutants are nearly unchanged. “Nearly” unchanged since some isotypes of β, for example in Gallus, contain a Ser instead of Ala at residue 296, as does human βII. These results raise the possibility that peloruside A may be active in more species than just mammals, and may be differentially active against different isotypes in one species.
Discussion
It has been clear for some time that the binding sites for peloruside A and laulimalide are the same, yet different from the paclitaxel site, which is also the site for all other known microtubule stabilizing agents. This is known from studies that indicate synergy between peloruside A or laulimalide and other microtubulestabilizing drugs, all of which bind at the paclitaxel binding site,Citation9–Citation11,Citation23 as well as from direct binding studies that have shown that microtubules formed in vitro and taxol-saturated can still bind peloruside A and vice-versa.Citation8,Citation21 These results demonstrated that the binding site for peloruside A and laulimalide was a new drug site on tubulin, but did not indicate where on tubulin it might be. For example, it need not even be located on β-tubulin, as the taxol site is, and indeed early computational docking results favored a site on α tubulin.Citation24,Citation25
Our study with hydrogen-deuterium exchange mapping by mass spectrometry, coupled with guided docking indicated that the binding site is on β, not on α.Citation13 These results refined the binding site to a surface cavity on β-tubulin, near the β-α interface. The results of the current study, based on the unrelated approach of mapping resistance-conferring mutations, confirm this assignment of the binding site, and emphasize the importance of the side-chain binding cavity.
The Pelr resistant lines described here all contain mutations in the TUBB gene. Pel D1 and Pel B1 are homozygous in expression for their mutations. Pel A1 is effectively homozygous in expression, since the second copy has a mutation that truncates the protein at residue 138. The other two lines, Pel B2 and Pel C, are heterozygous for two mutations. All of the lines contain R306H, except for Pel D1, which is homozygous in expression for A296S. We consider it likely that the initial exposure to peloruside A selected for two mutations that were present in the parental cell population that was exposed, and these are likely R306H (in Pel A, B, C) and A296S (in Pel D). The further steps of escalating drug dose following division of the culture resulted in selection for the second mutations that occurred during that culture (Pel B2 and Pel C) or in selection for silencing of the unmutated, parental sequence gene copy (Pel B1 and Pel D1). This is similar to the development of resistance previously observed in paclitaxel-resistant Ptx cell lines.Citation14,Citation26 In those cases and in the Pelr mutants described here, the initially selected, single mutations in one copy of the gene for a target protein probably does not confer substantial resistance. This is both because drug binding requires the participation of multiple residues on the protein, not just one, and because the resulting reduced affinity for drug binding is a recessive phenotype.Citation27–Citation29
The mutations do appear to affect resistance to peloruside A by reducing drug binding affinity, since merely increasing the drug concentration to which the Pelr cells are exposed results in the same pattern of arrest as seen with the parental cells at lower drug concentrations. Other cell lines resistant to microtubule-active drugs have shown different resistant mechanisms, including change in total tubulin concentrations, alterations in the stability of the microtubules in the absence of drug, shifts in isotype expression and checkpoint protein action.Citation13,Citation25,Citation26,Citation30 All of these will cause either collateral resistance or sensitivity to other microtubule-active drugs or changes in cell cycle (length and/or duration), or both. None of those are observed with these mutations. The Pelr lines show no cross-resistance to microtubule stabilizers that bind at the paclitaxel site, nor do they show collateral sensitivity to any microtubule destabilizing drugs. The doubling times for the Pelr lines is the same as the parental in the absence of drug. Significantly, arrest at G2/M is observed in the Pelr mutants at concentrations of peloruside 10-fold higher than in the parental line (), similar to the ratio in IC50 shown in . Thus we conclude that these mutations confer resistance by lowering the affinity of the binding site for peloruside A and for laulimalide. We further conclude that these mutations do not affect the normal functioning of microtubules nor the ability of tubulin to interact with small drugs at any of the other binding sites. It is difficult at this point to ascribe detailed binding functions for each residue mutated, since different mutations in different combinations afford similar levels of drug resistance. Nonetheless, the picture that emerges from them taken together is clear. The mutations all cluster in a cleft in the binding site that appears to accommodate the drug side chain, and thereby highlights the importance of side chain binding to drug action.
Sequence comparisons () indicate that the peloruside A binding site is conserved between many species, but shows some differences between isotypes in a given species, such as humans (). In particular, the βII isotype has the same residue at position 296 as does Pel D1, raising the possibility that cells expressing high levels of βII may be naturally resistant to peloruside A, but minimally so to laulimalide. This comparison also indicates that some isotypes, especially βVI, are quite divergent in the binding site, and might be expected to have binding properties that differ from those of the others. Indeed, this unusual behavior of βVI is just what was reported in a study of chicken erythrocyte tubulin, which is nearly pure βVI.Citation22 Divergent drug binding behavior has been reported for this tubulin at the colchicine site as well.Citation31 Thus βVI tubulin appears to be pharmacologically distinct from the other β isotype-containing tubulins.
It is notable that all of the mutations affect the region of β-tubulin that appears to bind the side chain of peloruside A and laulimalide. In the model of both peloruside A and laulimalide, R306 is a key residue defining the entrance to a deep/narrow hydrophobic cleft, into which the primary side chain of either macrolactone ring may penetrate. It is embedded within a short helical region that is reduced in labeling, in fact the footprint of both macrolides is virtually identical (Fig. S2). However, some differences emerge between the respective models. Laulimalide appears to induce a reorientation of side chains within the cleftCitation16 and a stabilization of R306 through interactions with Y340. This rearrangement is not evident in the model of the peloruside A binding mode (at this stage of model building), rather R306 interacts in a hydrophobic interaction with A296. This seems consistent with our mutational analysis. Mutating A296 to S induces resistance to peloruside A but notably less so to laulimalide, where the binding mode for the latter highlights a slight expansion of the cleft and the reorientation of R306 away from A296. Overall, the induction of resistance by mutations in the binding site may be understood as either directly influencing macrolide binding or distorting the structure of the deep pocket essential for proper side chain penetration.
The surface exposure of this cavity suggests the possibility that this site may be a site of binding of some cellular proteins that associate with the MT surface. This suggestion is similar to the proposal by SuffnessCitation32 that the paclitaxel site may be a natural site for binding of microtubule-regulatory proteins. This proposal has been supported by evidence of MAP binding at the paclitaxel site.Citation32–Citation34 It will be interesting to see if the peloruside binding site is also a docking site for protein(s) that bind to the MT outer surface.
Methods
Materials.
Peloruside was synthesized as described by Jin and Taylor.Citation35 Laulimalide, discodermolide and dictyostatin were kind gifts from Dr. Billy Day (University of Pittsburg). Combretastatin A4 was a kind gift from Dr. Karl Werbovetz (Ohio State University). Epothilone A and D24851 were from Calbiochem. Paclitaxel, nocodazole, verapamil, vinblastine and VP16 (Etoposide) were from Sigma Chemical Company.
Cell culture.
1A9 cells, a subclone of the A2780 human ovarian carcinoma cell line,Citation14 was maintained in RPMI medium supplemented with 10% fetal bovine serum and grown in 5% CO2 incubator. Drug resistant cells were maintained in media with drug for one passage followed by three passages without drug, due to the limited quantities of compound. Resistance level was not altered by this procedure or even following much longer periods in the absence of drug.
Cytotoxicity testing.
Drug sensitivity was determined by the Sulforhodamine B (SRB) method using cells that had been grown in drug-free media for 4–7 d prior to plating in 96-well plates. Cells were exposed to serial dilutions of drugs for 4 d, followed by fixation, SRB staining and GI50 determination.Citation14
Selection for peloruside resistance.
Initial selection attempts were made at 30 nM, ∼5 times the 4-d IC50, but there were no survivors. Hence a lower drug concentration of 10 nM peloruside A was used for selection, in the presence of 5 µM Verapamil. Resistant cells were selected by continuous exposure to drug, with several changes of media to remove dead cells. Resistant cells were divided into four separate flasks and exposed to increasing concentrations of peloruside A + 5 µM Verapamil over several months, until the cells survived in culture containing 50 nM peloruside A. Several clones were picked from each flask and maintained as well as frozen.
Determination of cell doubling time.
Cells were plated in two triplicate sets on four 96-well plates, at a density of 2,000 (parental) or 3,000 (Pelr lines) cells per well in 100-uL of complete RPMI media. After 24 h, the media was removed from one plate, which was then stored at −80°C. In each of the three remaining plates, one triplicate set received 25 nM of peloruside A in RMPI while the other set received fresh RPMI media (control). The treated plates were allowed to incubate for an additional 24, 48 and 72 h. Once removed, the plates were stored at −80°C.
After the final plate had been frozen for at least 24 h, the plates were thawed at room temperature. DNA content per well was measured by fluorescence assay using the CyQuant Cell Proliferation Assay kit (Invitrogen), as described by the supplier.
Flow cytometry.
Untreated cells or those treated with peloruside or taxol for 24 h, were trypsinized, washed in DPBS, centrifuged and resuspended in 500 µl of a stain solution containing 0.1 mg/ml propidium iodide and 0.6% Triton-X in PBS. Subsequently, 500 µl of RNase A solution (200 U/ml in PBS) was added and the cells incubated for 30 min at 22°C. Prior to analysis, the cells were passed through nylon mesh to remove clumps and aggregates. The cells were analyzed using a FACSort flow cytometer (Becton Dickinson). Excitation was at 488 nm with a 15 mW argon laser and fluorescence was detected with a 585 nm filter.
β-tubulin sequencing of 1A9 parental and Pel resistant cell lines.
Total RNA was extracted from 1A9 parental and 1A9 Pel cell lines using the RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. RT was performed on the samples using SuperScript III (Invitrogen) according to the manufacturer's instructions. PCR amplification of β-tubulin (NM_178014) was performed with Phusion High-Fidelity PCR Master Mix (New England Biolabs) according to the manufacturer's instructions using the following primers: CTC TGG GGC GCA TTC CAA CCT TCC (−108, −85) and TAG AGG CAG CAA ACA CAA ATT CTG AGG (1,430, 1,404). PCR-amplified cDNA was purified with the QIAquick PCR Purification Kit (Qiagen) and sequenced with BigDye Terminator v1.1 Cycle Sequencing Kits (Applied Biosystems Inc.) following manufacturer specifications. The reaction products were purified with Performa DTR gel filtration columns (Edge Bio) and analyzed on a 3130xl Genetic Analyzer (Applied Biosystems Inc.). The following primers were used for sequencing: ATC GTG CAC ATC CAG GCT GG (10-29), ATC CAG GCT GGT CAA TG (19-35), CAC CTG TGG CTT CAT TGT AGT ACA C (169-145), CAT GGA CTC TGT TCG CTC AGG (216-236), TCT GGG GCA GGT AAC AAC T (283-301), TGC CTC CTT CCG TAC CAC ATC C (372-351), TCT GCC TCC TTC CGT ACC AC (374-355), CTC TCC GTC CAT CAG TTG GTA GAG (559-582), TTG GTA GAG AAT ACT GAT GAG (572-594), CCT CAG AGT GCG GAA GCA GAT (648-628), CTC ATG GTG GCT GAG ACA AGG (701-681), GAC CTC CGC AAG TTG GCA GTC (745-765), GTT GAC TGC CAA CTT GCG GAG (768-748), CTG CAC GTT AAG CAT CTG CTC (1,002-982), CAG ATG CTT AAC GTG CAG AAC AAG (985-1,002), GGG GAT CCA TTC CAC AAA GT (1,038-1,019), GAT ACT CAG AGA CGA GGT CG (1,267-1,248), GTA CTG CTG ATA CTC AGA GAC GAG G (1,275-1,251), GTA AGA CGG CTA AGG GAA CTG (1,383-1,363), GCA AAC ACA AAT TCT GAG GG (1,423-1,404). All primers were obtained from Invitrogen.
Sequence variation among cells was examined by cloning individual cells from Pel cell cultures, expanding the clonal cultures, and then isolating RNA and sequencing as described above. Cells were cloned by plating in 100 mm dishes at low density (and in drug-free media). Following the appearance of colonies, cloning discs soaked in trypsin-EDTA were used to pick up cells from the colony and transfer them to wells in a 12-well plate for initial expansion.
Modeling and ligand docking.
The modeled binding mode for laulimalide is essentially as presented in Bennett et al.Citation16 The revised binding mode for peloruside is based upon the output of the refined docking study described in Huzil et al.Citation13 Briefly, a search of the binding site in α/β-tubulin identified through H/D exchange methods was conducted in Autodock 3 (http://autodock.scripps.edu), using a conformer for peloruside A built upon that described by Jimenez-Barbero et al.Citation24 and PDB entry 1TVK. This ligand conformer was docked into an area encompassing the binding site and the output of 200 trials clustered using RMSD tolerances of 2 Å. The resulting binding modes and tubulin structures were rendered in all figures using Pymol (pymol.sourceforge.net).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Note added in Proof
While this paper was in final preparation, the paper of Kanakkanthara, et al. appeared [Mol Cancer Ther 10(8):1419], with similar and complementary findings.
Abbreviations
| HDX | = | hydrogen-deuterium exchange |
| MT | = | microtubule |
Figures and Tables
Figure 2 Pel cell lines are resistant to growth inhibition by peloruside but not by paclitaxel or combretastatin A4. Growth and growth inhibition by drugs were determined by the SRB technique as described in methods. Growth is normalized to that observed in the absence of added drug. (A) Relative to the parental cells, Pel A1 and B2 cell lines show greater than 10-fold resistance to growth inhibition by peloruside A. (B) Parental cells and the Pelr lines are equivalently sensitive to growth inhibition by paclitaxel (Taxol). (C) Parental cells and the Pelr lines are equivalently sensitive to growth inhibition by combretastatin A4 (CSA4). Cell lines are identified in the legend in (B). The results shown are from a single experiment. Combined results from multiple experiments are presented in .
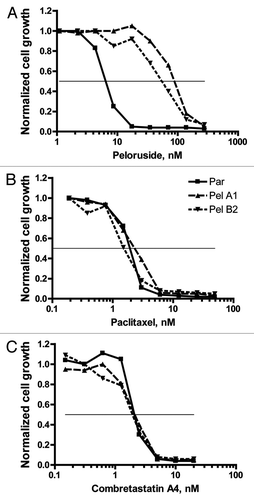
Figure 3 Pelr lines are only resistant to peloruside A/laulimalide binding site drugs. The IC50 for a number of drugs was determined as in for parental and Pelr lines. Fold resistance, calculated as (Pel IC50)/(parental IC50), is presented with error bars representing standard deviation. Resistance was measured for peloruside A (Pel) and five other microtubule stabilizing drugs: laulimalide (Laul), paclitaxel (Ptx), epothilone A (EpoA), dictyostatin (Dictyo) and discodermolide (Disco). In addition four microtubule destabilizing drugs were tested: combretastatin A4 (CSA4), nocodazole (Noc), vinblastine (Vbl) and D24851. Two non-microtubule-targeting drugs were also included: etoposide (VP16), and Romedepsin (or Depsipeptide FK228 = Depsi). Results for all drugs are grouped by Pelr line. The Table below the graph gives the parental cell IC50 and the number or replicates.
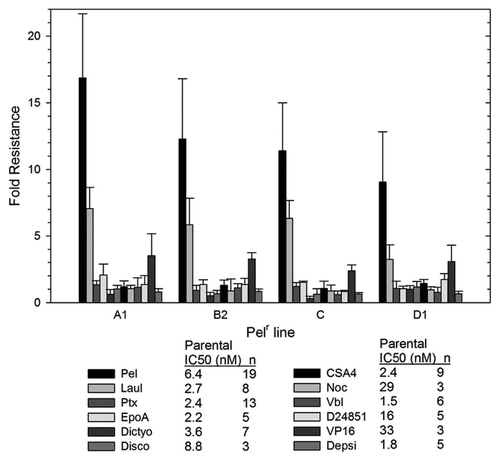
Figure 4 Cell cycle analysis demonstrates that high concentrations of peloruside A induce G2/M accumulation in Pelr lines. Pelr lines and the parental cells were grown in dishes and exposed to varying concentrations of paclitaxel (PTX) or peloruside A (PEL) for 18 h, before trypsinization and staining with propidium iodide to reveal DNA content as described in methods. C-control and drug concentrations were as indicated on the left axis.
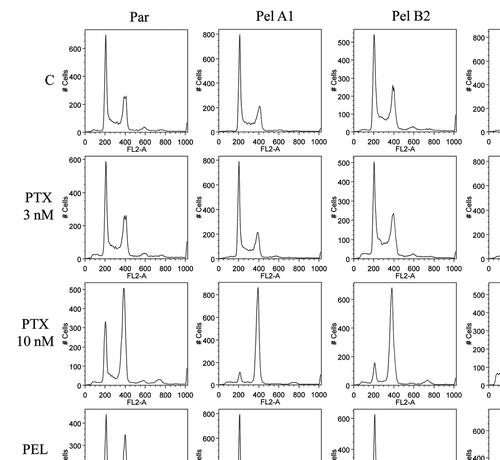
Figure 5 Peloruside A binding site in β-tubulin. (A) Global view of the exterior macrolide binding site on β-tubulin relative to the taxoid binding site on β-tubulin. Protein structure represented as a cartoon model using PBD entry 1JFF, where α-tubulin is colored cyan and β-tubulin pale green. Peloruside A is shown in blue spheres and taxol in red spheres. Exchangeable nucleotide (GDP) is shown as yellow sticks. (B) Closeup view of the binding site showing the position of the Pelr mutations. Binding models showing details of the binding modes for peloruside A (left), based on a reanalysis of docking results and laulimalide (right), adapted from Bennett 2010. Protein structure is rendered in cartoon, highlighting only the binding domain on β-tubulin, and based on PBD file 1JFF as the starting model. Colored secondary structure (orange, yellow, red) represent peptides marking the macrolide binding site by mass shift analysis, from the original determinations of the site (Huzil 2008, Bennett 2010). Ligand structures show only carbon atoms (green) and oxygen atoms (red). Note that both structures orient their long side chains into the cleft defined by in part by A296, R306, N337 and Y340.
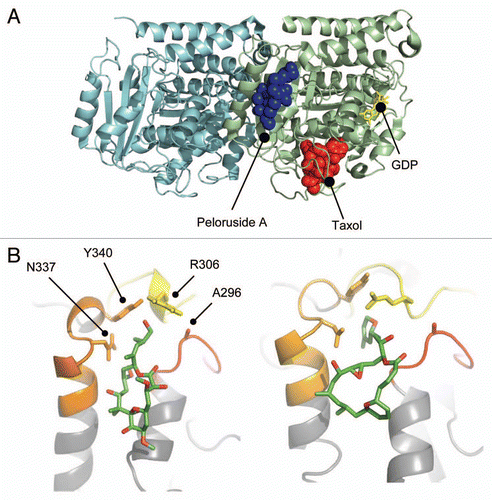
Figure 6 Binding site residues mutated in Pelr mutants are evolutionarily conserved. Residues 296–345 of β-tubulins from a number of sources are aligned to examine sequence conservation. The residues in the binding cavity that are mutated are indicated in bold text and highlighted. (A) Alignment of sequences from human β-tubulin isotypes. (B) Alignment of β sequences from various eukaryotes. Accession numbers: (A) βI NM_178014; βII NP_001060.1; βIII NP_001184110.1: βIVa NP_006078.2; βIVb NP_006079.1; βV NM_032525; βVI NP_110400.1. (B) Human NM_178014; G. gallus AAA49126.1; D. rerio XP_003198209.1; D. melanogaster NP_523795.2; T. thermophila P41352; S. cerevisiae CAA24603; L. tarentolae ABC40567; A. thaliana BAB10059.
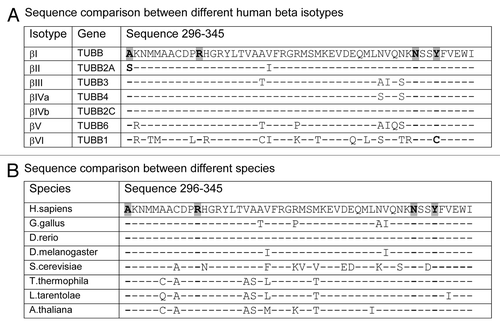
Table 1 Doubling time (hrs) for peloruside A resistant lines
Table 2 Mutations in β-tubulin in Pel lines
Additional material
Download Zip (228.8 KB)Acknowledgements
This work was supported in part by funding from the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH.
References
- Jordan MA, Kamath K. How do microtubule-targeted drugs work?. An overview. Curr Cancer Drug Targets 2007; 7:730 - 742; PMID: 18220533; http://dx.doi.org/10.2174/156800907783220417
- Komlodi-Pasztor E, Sackett D, Wilkerson J, Fojo T. Mitosis is not a key target of microtubule agents in patient tumors. Nat Rev Clin Oncol 2011; 8:244 - 250; PMID: 21283127; http://dx.doi.org/10.1038/nrclinonc.2010.228
- Calligaris D, Verdier-Pinard P, Devred F, Villard C, Braguer D, Lafitte D. Microtubule targeting agents: from biophysics to proteomics. Cell Mol Life Sci 2010; 67:1089 - 1104; PMID: 20107862; http://dx.doi.org/10.1007/s00018-009-0245-6
- Luduena RF, Roach MC. Tubulin sulfhydryls groups as probes for antimitotic and antimicrotubule agents. Pharmacol Ther 1991; 49:133 - 152; PMID: 1852786; http://dx.doi.org/10.1016/0163-7258(91)90027-J
- Sackett DL. Fojo T. Antimicrotubule agents that bind covalently to tubulin. The role of microtubules in cell biology, neurobiology and oncology 2008; Totowa, NJ Humana Press 281 - 306
- West LM, Northcote PT, Battershill CN. peloruside A: a potent cytotoxic macrolide isolated from the new zealand marine sponge Mycale sp. J Org Chem 2000; 65:445 - 449; PMID: 10813954; http://dx.doi.org/10.1021/jo991296y
- Hood KA, West LM, Rouwé B, Northcote PT, Berridge MV, Wakefield SJ, et al. Peloruside A, a novel antimitotic agent with paclitaxel-like microtubule-stabilizing activity. Cancer Res 2002; 62:3356 - 3360; PMID: 12067973
- Gaitanos TN, Buey RM, Díaz JF, Northcote PT, Teesdale-Spittle P, Andreu JM, et al. Peloruside A does not bind to the taxoid site on β-tubulin and retains its activity in multidrug-resistant cell lines. Cancer Res 2004; 64:5063 - 5067; PMID: 15289305; http://dx.doi.org/10.1158/0008-5472.CAN-04-0771
- Gapud EJ, Bai R, Ghosh AK, Hamel E. Laulimalide and paclitaxel: a comparison of their effects on tubulin assembly and their synergistic action when present simultaneously. Mol Pharmacol 2004; 66:113 - 121; PMID: 15213302; http://dx.doi.org/10.1124/mol.66.1.113
- Wilmes A, Bargh K, Kelly C, Northcote PT, Miller JH. peloruside A synergizes with other microtubule stabilizing agents in cultured cancer cell lines. Mol Pharm 2007; 4:269 - 280; PMID: 17397239; http://dx.doi.org/10.1021/mp060101p
- Wilmes A, O'Sullivan D, Chan A, Chandrahasen C, Paterson I, Northcote PT, et al. Synergistic interactions between peloruside A and other microtubule-stabilizing and destabilizing agents in cultured human ovarian carcinoma cells and murine T cells. Cancer Chemother Pharmacol 2011; In press PMID: 20848285; http://dx.doi.org/10.1007/s00280-010-1461-3
- Pryor DE, O'Brate A, Bilcer G, Díaz JF, Wang Y, Wang Y, et al. The microtubule stabilizing agent laulimalide does not bind in the taxoid site, kills cells resistant to paclitaxel and epothilones, and may not require its epoxide moiety for activity. Biochemistry 2002; 41:9109 - 9115; PMID: 12119025; http://dx.doi.org/10.1021/bi020211b
- Huzil JT, Chik JK, Slysz GW, Freedman H, Tuszynski J, Taylor RE, et al. A unique mode of microtubule stabilization induced by peloruside A. J Mol Biol 2008; 378:1016 - 1030; PMID: 18405918; http://dx.doi.org/10.1016/j.jmb.2008.03.026
- Giannakakou P, Sackett DL, Kang YK, Zhan Z, Buters JT, Fojo T, et al. Paclitaxel-resistant human ovarian cancer cells have mutant β-tubulins that exhibit impaired paclitaxel-driven polymerization. J Biol Chem 1997; 272:17118 - 17125; PMID: 9202030; http://dx.doi.org/10.1074/jbc.272.27.17118
- Poruchynsky MS, Kim JH, Nogales E, Annable T, Loganzo F, Greenberger LM, et al. Tumor cells resistant to a microtubule-depolymerizing hemiasterlin analogue, HTI-286, have mutations in α- or β-tubulin and increased microtubule stability. Biochemistry 2004; 43:13944 - 13954; PMID: 15518543; http://dx.doi.org/10.1021/bi049300
- Bennett MJ, Barakat K, Huzil JT, Tuszynski J, Schriemer DC. Discovery and characterization of the laulimalide-microtubule binding mode by mass shift perturbation mapping. Chem Biol 2010; 17:725 - 734; PMID: 20659685; http://dx.doi.org/10.1016/j.chembiol.2010.05.019
- Sackett DL, Lederberg S. Colcemid-resistant mutants of fission yeast have an altered cell cycle. Exp Cell Res 1986; 163:467 - 476; PMID: 3514248; http://dx.doi.org/10.1016/0014-4827(86)90077-7
- Schibler MJ, Cabral F. Taxol-dependent mutants of Chinese hamster ovary cells with alterations in α- and β-tubulin. J Cell Biol 1986; 102:1522 - 1531; PMID: 2870070; http://dx.doi.org/10.1083/jcb.102.4.1522
- Pera B, Razzak M, Trigili C, Pineda O, Canales A, Buey RM, et al. Molecular recognition of peloruside A by microtubules. The C24 primary alcohol is essential for biological activity. ChemBioChem 2010; 11:1669 - 1678; PMID: 20665616; http://dx.doi.org/10.1002/cbic.201000294
- Paterson I, Menche D, Håkansson AE, Longstaff A, Wong D, Barasoain I, et al. Design, synthesis and biological evaluation of novel, simplified analogues of laulimalide: modification of the side chain. Bioorg Med Chem Lett 2005; 15:2243 - 2247; PMID: 15837302; http://dx.doi.org/10.1016/j.bmcl.2005.03.018
- Nguyen TL, Xu X, Gussio R, Ghosh AK, Hamel E. The assembly-inducing laulimalide/peloruside a binding site on tubulin: molecular modeling and biochemical studies with [3H]peloruside A. J Chem Inf Model 2010; 50:2019 - 2028; PMID: 21028850; http://dx.doi.org/10.1021/ci1002894
- Khrapunovich-Baine M, Menon V, Yang CP, Northcote PT, Miller JH, Angeletti RH, et al. Hallmarks of molecular action of microtubule stabilizing agents: effects of epothilone B, ixabepilone, peloruside A and laulimalide on microtubule conformation. J Biol Chem 2011; 286:11765 - 11778; PMID: 21245138; http://dx.doi.org/10.1074/jbc.M110.162214
- Hamel E, Day BW, Miller JH, Jung MK, Northcote PT, Ghosh AK, et al. Synergistic effects of peloruside A and laulimalide with taxoid site drugs, but not with each other, on tubulin assembly. Mol Pharmacol 2006; 70:1555 - 1564; PMID: 16887932; http://dx.doi.org/10.1124/mol.106.027847
- Jiménez-Barbero J, Canales A, Northcote PT, Buey RM, Andreu JM, Díaz JF. NMR determination of the bioactive conformation of peloruside A bound to microtubules. J Am Chem Soc 2006; 128:8757 - 8765; PMID: 16819869; http://dx.doi.org/10.1021/ja0580237
- Pineda O, Farrès J, Maccari L, Manetti F, Botta M, Vilarrasa J. Computational comparison of microtubule-stabilising agents laulimalide and peloruside with taxol and colchicine. Bioorg Med Chem Lett 2004; 14:4825 - 4829; PMID: 15341932; http://dx.doi.org/10.1016/j.bmcl.2004.07.053
- Wang Y, O'Brate A, Zhou W, Giannakakou P. Resistance to microtubule-stabilizing drugs involves two events: β-tubulin mutation in one allele followed by loss of the second allele. Cell Cycle 2005; 4:1847 - 1853; PMID: 16294009; http://dx.doi.org/10.4161/cc.4.12.2264
- Cabral F. Fojo T. Resistance to microtubule-targeting drugs. The role of microtubules in cell biology, neurobiology and oncology 2008; Totowa, NJ Humana Press 357 - 394
- Yin S, Bhattacharya R, Cabral F. Human mutations that confer paclitaxel resistance. Mol Cancer Ther 2010; 9:327 - 335; PMID: 20103599; http://dx.doi.org/10.1158/1535-7163.MCT-09-0674
- Kavallaris M. Microtubules and resistance to tubulin-binding agents. Nat Rev Cancer 2010; 10:194 - 204; PMID: 20147901; http://dx.doi.org/10.1038/nrc2803
- Yang CP, Liu L, Ikui AE, Horwitz SB. The interaction between mitotic checkpoint proteins, CENP-E and BubR1, is diminished in epothilone B-resistant A549 cells. Cell Cycle 2010; 9:1207 - 1213; PMID: 20237434; http://dx.doi.org/10.4161/cc.9.6.11122
- Sharma S, Poliks B, Chiauzzi C, Ravindra R, Blanden AR, Bane S. Characterization of the colchicine binding site on avian tubulin isotype βVI. Biochemistry 2010; 49:2932 - 2942; PMID: 20178367; http://dx.doi.org/10.1021/bi100159p
- Suffness M. Is taxol a surrogate for a universal regulator of mitosis?. In Vivo 1994; 8:867 - 878; PMID: 7727737
- Park H, Kim M, Fygenson DK. Tau-isoform dependent enhancement of taxol mobility through microtubules. Arch Biochem Biophys 2008; 478:119 - 126; PMID: 18691553; http://dx.doi.org/10.1016/j.abb.2008.07.020
- Kar S, Fan J, Smith MJ, Goedert M, Amos LA. Repeat motifs of tau bind to the insides of microtubules in the absence of taxol. EMBO J 2003; 22:70 - 77; PMID: 12505985; http://dx.doi.org/10.1093/emboj/cdg001
- Jin M, Taylor RE. Total synthesis of (+)-peloruside A. Org Lett 2005; 7:1303 - 1305; PMID: 15787492; http://dx.doi.org/10.1021/ol050070g