Abstract
Polyubiquitin-mediated degradation of proteins plays an essential role in various physiological processes including cell cycle progression, transcription and DNA replication and repair. Increasing evidence supports a vital role for the E3 ubiquitin ligase cullin-4, in conjunction with the substrate recognition factor Cdt2 (CRL4Cdt2), for the degradation of multiple cell cycle-regulated proteins to prevent genomic instability. In addition, it is critical for normal cell cycle progression by ensuring the timely destruction of various cell cycle proteins whose deregulated expression impairs cell cycle progression. Here, we summarize our current knowledge about the various roles of the CRL4Cdt2 E3 ubiquitin ligase, and how its activity contributes both to the preservation of genome integrity and to normal cell cycle progression, and how its deregulation may contribute to human cancer.
Introduction
ATP-dependent polyubiquitylation of proteins and their subsequent degradation by the 26S proteasome is involved in all aspects of cell physiology. The highly coordinated process ensures timely downregulation of proteins thereby controlling cellular activity and maintaining cell and tissue homeostasis.Citation1–Citation4 Polyubiquitylation is often triggered by posttranslational modifications of the target substrate. For some substrates, phosphorylation of a single or multiple residues by one or more kinases creates a docking site for ubiquitin-ligases or adaptors of ubiquitin ligases thus signaling its polyubiquitylation prior to their degradation via the proteasome. Such docking sites (also known as degrons) are often conserved in other substrates targeted by the same ubiquitin ligases. While polyubiquitylation of a substrate protein is most frequently associated with its proteolytic degradation via the 26S proteasome, in some cases, addition of ubiquitin is associated with modification of function, enzymatic activity or cellular localization of the substrate protein without affecting its stability.Citation5 The ubiquitylation reaction involves the covalent attachment of the small 76 amino-acid ubiquitin moiety on a specific lysine residue in the target substrate protein. For substrates destined for degradation via the 26S proteasome, polyubiquitin chains are subsequently assembled through the successive attachment of the ubiquitin molecules through the formation of isopeptide bonds between the C-terminus of the incoming ubiquitin to lysine 48 of the attached ubiquitin, thus marking the substrate protein for proteolysis.Citation6 Polyubiquitylation involves three distinct and consecutive enzymatic steps () where ubiquitin is first activated by an E1 ubiquitin-activating enzyme followed by the transfer of the activated (AMP-charged) ubiquitin from the E1 enzyme to an E2 ubiquitin-conjugating enzyme. Finally, the ubiquitin is transferred from the E2 enzyme to the substrate through the activity of an E3 ubiquitin ligase.Citation7 Although the E3 lacks enzymatic activity, it is instrumental for the ubiquitylation reaction as it recognizes, either directly or through adaptor proteins, the specific substrate to promote ubiquitin ligation by the E2 conjugating enzyme. The ability of E3 ubiquitin ligases to recognize, with high specificity, a relatively large number of substrates accounts for the diverse structural complexity of this group of proteins.Citation8
E3 ubiquitin ligases can be categorically classified as members of the HECT (Homologous to E6-AP C-Terminus) or RING (Really Interesting New Gene) family of E3 ubiquitin ligases based on the conservation of sequence motifs and the mechanism of ubiquitin conjugation, and together, they constitute over 1,000 unique E3 ligases. Because these E3 ubiquitin ligases often utilize a large number of different substrate-recognizing proteins, collectively termed substrate recognition factors (SRFs), the number of unique E3 ubiquitin ligase complexes is likely to be several fold higher. Cullin-RING E3 ubiquitin Ligases (CRLs) represent the largest and most complex family of E3 ubiquitin ligases and play significant roles in multiple physiological processes including transcription, differentiation, cell cycle control, proliferation, apoptosis and tumorigenesis.Citation9–Citation11 This class of E3 ubiquitin ligases include cullin 1, 2, 3, 4A, 4B, 5 and cullin 7 as well as the cullin-like proteins PARC and APC2. Several CRLs and many of their target protein substrates are conserved throughout evolution.Citation9
The CRL4 (cullin 4A/B)-based ubiquitin ligase complexes have recently became the subject of intense investigation after publication of the crystal structure of its core components and identification of some of its substrates.Citation12,Citation13 CRL4 E3 complexes adopt an overall architecture similar to the SCF (Skp1-Cullin1-F-Box protein) ubiquitin ligase complex (also known as CRL1) (). The SCF ubiquitin ligase is a prototype of the cullin-family of ubiquitin ligases and the most characterized mammalian cullin-based ubiquitin ligase.Citation14 The core CRL4 ubiquitin ligase complex is composed of one of two scaffold proteins (Cul4A or Cul4B), Ddb1 (damage-specific DNA binding protein-1), an adaptor protein that is analogous to the Skp1 subunit in the SCF ubiquitn ligase complex and functions to bridge one of many substrate recruiting factors (DCAFs; Ddb1 and Cul4 Associated Factors) to the Cul4 E3 subunit, and a small RING finger protein (Rbx1/2) required for the recruitment of a corresponding E2 ubiquitin-conjugating enzyme (UBC) (). DCAFs or WDR (WD repeat-containing proteins) include at least 49 WD motif-rich proteins that function as SRFs to recruit substrates to the CRL4 ubiquitin ligase complex similar to the function of the F-box proteins in the SCF ubiquitin ligase complexes.Citation12,Citation15–Citation17
The CRL4 E3 ligase orchestrates a variety of physiological processes including DNA replication, transcriptional regulation, apoptosis and a number of DNA repair processes (). Recent work demonstrated that CRL4 is critical for preventing genomic instability through its ability to promote the ubiquitin-dependent proteolysis of Cdt1, a replication initiation protein that is essential for pre-RC (pre-replication complex) assembly and the recruitment of the replicative helicase MCM2-7 at replication origins.Citation17–Citation26 In this review, we analyze the various roles of CRL4 in association with the DCAF Cdt2 (also known as DCAF2/RAMP/DTL/L2DTL) (CRL4Cdt2) in preserving the integrity of the genome. We discuss the multiple activities of this E3 ubiquitin ligase in promoting cell cycle progression and various DNA repair processes. Finally, we describe how deregulation of CRL4Cdt2 may contribute to the development of human cancer and how targeting this ubiquitin ligase may be an attractive target for anti-cancer therapeutics.
CRL4Cdt2; a Guardian of the Genome Integrity
Cells that are deficient in Cul4 exhibit re-replication and genomic instability reminiscent to that seen in cells overexpressing the replication initiation factor Cdt1.Citation17,Citation27–Citation29 Consistent with this, inactivation of the Cul4 gene in C. elegans results in massive re-replication with certain cells containing up to 100 C DNA content.Citation30 Similarly, depletion of Ddb1 or Cul4 from mammalian cells is associated with double strand DNA breaks, re-replication and activation of the ATM and ATR-dependent checkpoints which can be partially suppressed by co-depletion of Cdt1.Citation27 CRL4-mediated degradation of Cdt1 is (a) triggered by DNA damage accompanying genotoxic stresses such as gamma or UV irradiation and (b) active constitutively during the S-phase of the cell cycle of unperturbed cells.Citation31 Subsequent studies demonstrated that the DCAF Cdt2 is critical for the ability of CRL4 to promote the degradation of Cdt1, both during the S-phase of the cell cycle and after genotoxic stress. This axis of degradation of Cdt1 is critical for preventing re-licensing of replication origins within the same cell cycle and guards against re-replication and genomic instability.Citation17,Citation32 Significantly, CRL4Cdt2-mediated ubiquitylation and degradation of Cdt1 is dependent on Cdt1 binding to chromatin-bound PCNA ().Citation17,Citation32 DNA re-replication and genomic instability are observed upon depletion of Cdt2 in mammalian cells,Citation17 in C. elegansCitation33 and in zebrafish,Citation32 underscoring the evolutionarily conserved role of Cdt2 in promoting CRL4-mediated destruction of Cdt1 and in maintaining the integrity of the genome. In zebrafish, the loss of Cdt2 results also in the loss of radiation-induced early G2/M checkpoint but this phenotype seems to be independent of deregulated Cdt1, suggesting that additional substrate(s) of CRL4Cdt2 suppress checkpoint activation post-radiation.Citation32 Alternatively, and because this phenotype has not been observed with Cul4 deficiency, Cdt2 may have additional roles that are independent of its role as a component of the CRL4 ubiquitin ligase.
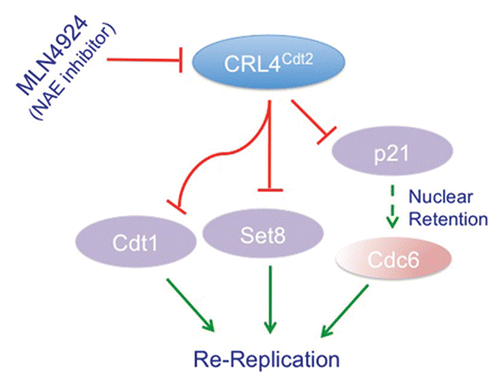
While the re-replication observed upon depletion of Cul4, Ddb1 or Cdt2 from mammalian cells was initially solely attributed to the accumulation of Cdt1 during the S-phase of the cell cycle, we and others have not been able to completely prevent re-replication by co-depleting Cdt1,Citation27 (Abbas T and Dutta A, unpublished results). Furthermore, treatment of various human cancer cell lines with the novel anti-cancer compound, MLN4924, a specific inhibitor of the NEDD8-activating enzyme (NAE), which inhibits protein neddylation, and hence the activity of CRL4 among other cullins, stabilizes Cdt1 and induces significant re-replication.Citation34 Yet, depletion of mammalian cells of Cdt1 by si-RNA inhibited MLN4924-induced re-replication, but only partially.Citation35 These findings suggest that other factor(s), besides Cdt1, are stabilized by the inactivation of CRL4Cdt2 and contribute to the re-replication. Recently, Kim et al. suggested that the stabilization of the CDK2 inhibitor p21, a substrate for CRL4Cdt2,Citation33,Citation36,Citation37 contributes to the re-replication seen in Cdt2-depleted HeLa cells.Citation33 The authors suggested that the stabilized p21 inhibited cyclin-CDK2, thus preventing the cytoplasmic export and inactivation of the CDC6 replication factor.Citation33 This hypothesis would be consistent with the finding that the failure to export Cdc6 during S phase in C. elegans is directly linked to the re-replication phenotype in cul-4 mutants.Citation38 However, a further analysis of CDC6 nuclear retention upon p21 accumulation in Cdt2-depleted cells, led the authors to conclude that Cdc6 nuclear retention was not sufficient to explain how p21 contribute to re-replication.Citation33 Furthermore, the model proposed by Kim et al. does not address the fact that Cdt2 destabilizes p21 only in the context of PCNA bindingCitation33,Citation36,Citation37 and not when bound to cyclin-Cdk complexes. In fact, whether p21 stabilization upon Cdt2 depletion is associated with reduced cyclin E-Cdk2 activity has not been tested. We, on the other hand, found that the depletion of Cdt2 from the human colon cancer cells HCT116 deficient of p21 (HCT116p21-/-) by si-RNA still induced significant re-replication albeit to a lesser extent than wild-type HCT116 (Abbas T, Dutta A, unpublished results), arguing that p21 stabilization was not important for promoting re-replication. Together, these observations leave open the question of whether a yet to be identified factor is stabilized and co-operates with Cdt1 to promote re-replication in cells with inactivated CRL4Cdt2.
Two independent studies have recently shed light on the identity of the second factor promoting re-replication, the histone monomethyl transferase Set8/Pr-Set7.Citation39,Citation40 We and others have demonstrated that CRL4Cdt2 promotes the ubiquitylation and degradation of Set8, both during S-phase of the cell cycle and after UV irradiation in a reaction that is also dependent on Set8-PCNA interaction.Citation39–Citation42 Set8, known to monomethylate histone H4 lysine 20 (H4K20me) in G2 phase of the cell cycle and in mitosis, is a critical enzyme whose inactivation leads to failure of cells to progress through G2,Citation43 global chromosome decondensation,Citation44,Citation45 aberrant centrosome amplification and substantial spontaneous DNA damage.Citation43 Failure to degrade Set8 during S-phase suppressed growth due to activation of the G2/M checkpoint,Citation39,Citation41 and repression of E2F-regulated and histone genes.Citation39 Furthermore, cells expressing PCNA-binding deficient and hence CRL4Cdt2 resistant Set8 exhibited spontaneous DNA damage and induction of the p53 tumor suppressor protein with a concomitant increase of p53-transactivated pro-apoptotic proteins such as Fas and Puma.Citation39 Cells with stable Set8 exhibited large nuclear morphology with roughly 20% of the cells undergoing re-replication even though cells failed to exit mitosis.Citation39 These phenotypes were dependent on Set8 methyltransferase activity and suggest that deregulated Set8 expression in S phase leads to genome instability and may contribute to re-replication observed upon CRL4Cdt2 inactivation (). In fact, depletion of Set8 significantly inhibited re-replication in U2OS depleted of Cdt2 (Abbas T, Dutta A, unpublished results). Similar results were independently reported by Julien and colleagues.Citation40 They further demonstrated that Set8 monomethylates H4K20 at replication origins which coincides with the onset of licensing, and that the expression of PCNA-binding-deficient mutant of Set8 caused the selective maintenance of H4K20me1 at replication origins and re-replication.Citation40 Tethering a catalytically active Set8, but not its catalytically deficient mutant, to a specific genomic locus promoted loading of pre-RC proteins on chromatin.Citation40 Whether other activities of Set8, beside its role in monomethylating H4K20, contribute to the re-replication however, remains to be determined. It also remains unclear as to what is the exact contribution of Cdt1 and Set8 to the re-replication observed upon CRL4Cdt2 inactivation. Nevertheless, these results demonstrate that the ubiquitin-dependent degradation of Set8 via CRL4Cdt2 is critical for preventing re-replication.
CRL4Cdt2 Ubiquitin Ligase and the Regulation of DNA Repair Processes
The CRL4-based ubiquitin ligases regulate genomic stability by participating in a variety of DNA repair processes (). For example, in cooperation with the DCAF Ddb2 (DNA damage-binding protein 2), CRL4Ddb2 promotes nucleotide excision repair (NER) by targeting the DNA damage recognition protein XPC for ubiquitylation. CRL4Ddb2-dependent polyubiquitylation of XPC however, does not promote the proteolysis of XPC and instead targets XPC to damaged DNA.Citation46–Citation49 Mutations in the WD repeat of Ddb2 that prevent its association with Ddb1 and the core CRL4 ligase leads to the radiation-sensitivity disorder Xeroderma Pigmentosa (XP) group E. CRL4 also participates in transcription-coupled repair (TCR) by associating with the DCAF CSA (Cockayne Syndrome A) to promote the polyubiquitylation and degradation of CSB (Cockayne Syndrome B). In the absence of CSA, CSB is not degraded and TCR is impaired in cells irradiated with UV.Citation50,Citation51 Mutations in CSA and CSB are responsible for genetic disorder Cockayne Syndrome due to failure of TCR. It is interesting to speculate that the overexpression of Cdt2, frequently seen in human cancer (see below) may impair NER or TCR by competing with Ddb2 or CSA for binding to CRL4.
Additional evidence suggests that CRL4Cdt2 promotes several DNA repair processes by degrading p21. The E3 ubiquitin ligase promotes the ubiquitin-dependent degradation of PCNA-bound p21 following UV irradiation,Citation36,Citation37 freeing PCNA for repair processes. p21, a downstream transcriptional target of the p53 tumor suppressor protein, plays a significant role in mediating not only p53 tumor suppressor activity in response to genotoxic stress, but also functions as a sensor as well as an effector of multiple p53-independent tumor suppressor pathways.Citation52 The main role of p21 under these circumstances is to mediate G1/S growth arrest, both through the direct inhibition of DNA synthesis via its ability to interact with and suppress PCNA-dependent polymerase delta, and indirectly via inhibiting cyclin A and cyclin E-Cdk2 activity necessary for S-phase progression.Citation53 p21 also promotes growth arrest during G2 phase of the cell cycle in response to genotoxic stress and this has been largely attributed to its ability to inhibit the activity of Cdk1 complexes.Citation52,Citation54 The induction of G1/S or G2 growth arrest in response to DNA damage provides a window where DNA damage can be repaired before it is passed to daughter cells.Citation52 An interesting possibility is that excess CRL4Cdt2 (as seen in tumors) leads to excessive downregulation of p21, so that the checkpoint pathways are impaired over the long term and that DNA repair processes do not have time to act.
Whereas p21 is frequently upregulated in response to genotoxic stress, it is downregulated upon DNA damage that is associated with stalled replication forks such as those that accumulate in response to UV irradiation or hydroxyurea treatment.Citation55 This downregulation, mediated via CRL4Cdt2 ubiquitin ligase,Citation36,Citation37 is critical for various DNA repair processes that require PCNA and are inhibited by p21, such as nucleotide excision repair (NER), base excision repair (BER) and DNA translesion synthesis (TLS). The downregulation of p21 in response to UV irradiation or hydroxyurea is required for efficient PCNA monoubiquitylation after UV irradiation.Citation56 PCNA monoubiquitylation at stalled replication forks, both during replication as well as in response to UV irradiation and other genotoxic agents is critical for TLS, an activity where DNA replication can proceed across damaged DNA through the recruitment and utilization of specialized polymerases.Citation57 Besides its ability to inhibit UV-induced PCNA monoubiquitylation, p21, through PCNA binding, also selectively interferes with the recruitment of the TLS polymerase eta (Polη) at DNA lesions.Citation58 Thus, eliminating p21 from PCNA via CRL4Cdt2 is critical for DNA repair by TLS.
In yeast, PCNA monoubiquitylation has been shown to require the Rad6 ubiquitin conjugating enzyme and Rad18 E3 ubiquitin ligase.Citation59 This activity is also conserved in high eukaryotes although residual PCNA monoubiquitylation has been observed in the absence of Rad18.Citation60–Citation62 Recently, we identified a novel role for CRL4Cdt2 in promoting TLS through its ability to directly monoubiquitylate PCNA.Citation63 We showed that CRL4Cdt2 monoubiquitylates PCNA in unperturbed cycling cells and that it synergizes with Rad18 to monoubiquitylate PCNA in response to DNA cross-linking agents such as cisplatin and that this pathway is critical for promoting TLS.Citation63
Just as TLS is important for repairing DNA damage at stalled replication forks, terminating this activity post repair is of equal significance given that TLS is error-prone owing to the lack of proofreading activity of TLS polymerases and hence their low fidelity. One way to ensure the termination of TLS is to induce the rapid degradation of bypass polymerases once they performed their function. Interestingly, in C. elegans, Polη has been shown to undergo such ubiquitin-mediated degradation and surprisingly, this is carried out via the CRL4Cdt2 E3 ligase through a PCNA-coupled reaction.Citation64 Collectively, these observations demonstrate that CRL4Cdt2 is critical, both for promoting TLS via degrading p21 and monoubiquitylating PCNA, as well as controlling the extent of TLS at the damage site via degrading Pol η ().
CRL4Cdt2 plays yet another role in repair of DNA via the repair of double stranded breaks by homologous recombination (HR). Using a genetic screen, Moss et al. demonstrated that deletion of ddb1 or cdt2 significantly increased the sensitivity of fission yeast to agents inducing double-strand breaks (DSB) such as bleomycin and IR.Citation65 This has been attributed to the reduced supply of deoxy-nucleotides required during the HR process. CRL4Cdt2 promotes the degradation of the ribonucleotide reductase (RNR) inhibitor Spd1 so that yeast with cdt2 or ddb1 deleted has reduced RNR activity. Consistent with this, the DNA damage sensitivity and the reduced HR efficiency associated ddb1 or cdt2 deletions are suppressed by spd1 deletion.Citation65 Spd1, the first identified substrate of the CRL4Cdt2 ligase,Citation66 functions as a nuclear anchor of the small RNR subunit. CRL4Cdt2-dependent degradation of Spd1, both during the S-phase and in response to DNA damage in G2, results in the nuclear export of the small RNR subunit and the subsequent assembly of the heterodimeric active RNR complex.Citation66,Citation67 Deletion of cdt2 stabilizes Spd1 and prevents the re-localization of the small RNR subunit from the nucleus to the cytoplasm. Because RNR catalyzes the rate-limiting step for dNTP production and is required for not only for replicative DNA synthesis but also DNA repair,Citation68 Spd1 degradation via CRL4Cdt2 is important to allow for the accumulation of sufficient pool of deoxy-ribonucleotides necessary for high-fidelity DNA repair.
CRL4Cdt2, a Major Coordinator of Normal Cell Cycle Progression
CRL4Cdt2 promotes the ubiquitin-dependent degradation of several of its substrates not only in response to genotoxic stress, but also in unperturbed proliferating cells, specifically during the S phase of the cell cycle. Several studies demonstrate that CRL4Cdt2 is critical for the proper progression through the cell cycle. As discussed above, CRL4Cdt2-mediated ubiquitylation and degradation of Spd1 is important to allow for the full activation of RNR and the accumulation of sufficient pool of deoxy-ribonucleotides necessary for normal S-phase progression. Furthermore, CRL4Cdt2 promotes the degradation of the D. melanogaster E2F1 transcription factor during the S phase of the cell cycle also in a reaction that is dependent on E2F1-PCNA interaction.Citation69 Inhibiting CRL4Cdt2-mediated degradation of E2F1 accelerates S-phase progression and induces apoptosis.Citation69
In yeast, metazoans and mammalian cells, CRL4Cdt2 promotes the ubiquitin-dependent degradation of chromatin and PCNA-bound Cdt1 not only in response to DNA damage but also during the S phase of the cell cycle. Along with the aforementioned CRL4Cdt2-mediated degradation of PCNA-bound Set8 in mammalian cells, this activity ensures the timely destruction of Cdt1 and Set8 to prevent the loading of Pre-RC required for licensing of replication origins until the end of mitosis.Citation31,Citation39,Citation40 Failure to degrade Cdt1 initiates re-replication that is accompanied by spontaneous DNA damage, activation of ATM and ATR-dependent G2/M checkpoint and the induction of apoptosis. Failure to degrade Set8 via CRL4Cdt2 also triggers re-replication, induces spontaneous DNA damage, activates G2/M checkpoint and significantly delays progression through G2 phase of the cell cycle without significantly impacting S-phase progression.Citation39,Citation41 In addition, the persistent expression of CRL4Cdt2-resistant Set8 represses transcription from E2F-regulated genes as well as transcription from histone genes leading to significant depletion of all four canonical histone proteins and global chromatin decompaction, contributing to the potent growth inhibition.Citation39 Transcriptional repression from the histone genes was associated with an increase in the repressive trimethylation mark (H4K20me3) at the promoters of these genes that results from unrestricted Set8 activity.Citation39
CRL4Cdt2 plays another important role for cell cycle progression through the targeted destruction of the cyclin-dependent kinase (Cdk) and cell cycle inhibitor p21.Citation36,Citation37 The small Cdk inhibitor exhibits growth suppressive activities that are primarily dependent on its ability to inhibit Cdk2 and PCNA.Citation52 Cdk2 is freed from inhibitory p21 through the activity of the SCFSkp2 ubiquitin ligase, both during G1 and in S phases of the cell cycle, but how PCNA-bound p21 is degraded to allow reactivation of PCNA during S phase has remained unknown until recently. We and others have shown that CRL4Cdt2 promote the ubiquitin-dependent destruction of p21 specifically during S-phase in a reaction that is strictly dependent on PCNA.Citation37,Citation39 This activity is conserved in C. elegansCitation33 and a similar mechanism has recently been uncovered for the X. laevis Cip/Kip-type Cdk inhibitor Xic1.Citation70 The discovery of CRL4Cdt2 as the mediator of such activity, not only underscores the significance of this ubiquitin ligase for S-phase progression, but also provides a keen example where a given protein, in this case p21, is selectively and differentially targeted for proteolysis via different mechanisms based on changes in its interaction partners and spatio-temporal distribution.
PCNA, a Universal Co-Factor for CRL4Cdt2-Mediated Ubiquitylation?
A unique feature of ubiquitylation of various substrates via the CRL4Cdt2 E3 ubiquitin ligase complex is the requirement of their physical association with the DNA polymerase delta processivity factor PCNA. Initial studies have demonstrated that CRL4Cdt2-dependent ubiquitylation of Cdt1 is dependent on Cdt1 binding to PCNA through a conserved PIP (PCNA-Interacting Peptide) box located in the extreme N-terminus of Cdt1.Citation17,Citation21–Citation25 Cdt2 is the substrate specificity factor that recruits chromatin-associated and PCNA-bound Cdt1 for ubiquitylation via the CRL4 core E3 ubiquitin ligase and its subsequent degradation by the 26S proteasome.Citation31 Subsequent studies demonstrated that most other substrates of CRL4Cdt2 also bind to PCNA through similar PIP boxes and that their physical association with PCNA was required for their ubiquitylation via CRL4Cdt2. The only exception to this rule is the S-phase and DNA damage-dependent degradation of Spd1 mentioned above. However, whether Spd1 interacts with PCNA has not been determined and it remains possible that CRL4Cdt2-dependent degradation of Spd1 follows the same rule.
The exact role PCNA plays in CRL4Cdt2-mediated ubiquitylation and degradation of various substrates remains largely unclear. Insights into the mechanism by which various substrates are targeted for ubiquitin-dependent degradation via the CRL4Cdt2 complex came from the finding that unlike the SCFSkp2 mediated ubiquitylation of Cdt1, CRL4Cdt2-mediated ubiquitylation of Cdt1 occurs exclusively on chromatin and requires the physical association of Cdt1 with PCNA.Citation71 Cdt1 selectively binds chromatin-bound PCNA and the formation of chromatin-bound PCNA-Cdt1 complex is essential for the recruitment of Cdt2 and hence the rest of the CRL4 complex.Citation22,Citation71 While many proteins interact with PCNA through similar PIP boxes as those common to Cdt1 and other CRL4Cdt2 substrates and follows the consensus [QXX(I/L/M)XX(F/Y)(F/Y)],Citation72 only a small subset of these are subject to CRL4Cdt2-mediated ubiquitylation and degradation. Bioinformatic analysis of the PCNA interacting peptide of Cdt1, and that of other substrates of CRL4Cdt2, revealed that these select proteins contain unique sequences not present in other PCNA-interacting proteins.Citation71 Two specific residues within Cdt1 PIP box, and conserved among other CRL4Cdt2 substrates, were particularly important for Cdt1 ubiquitylation via CRL4Cdt2; a threonine at position 5 of the Cdt1 PIP box conferred high affinity binding with chromatin-bound PCNA and a positively charged (lysine or arginine) residue at position +4 from the PIP box is important for the recruitment of Cdt2 to chromatin-bound PCNA. The significance of these critical residues for CRL4Cdt2 and PCNA-mediated ubiquitylation and degradation of various substrates was elegantly demonstrated by the replacement of the PIP box of FEN1, a stable binding partner of PCNA, with a PIP box containing the conserved threonine and arginine of the p21 PIP box. This substitution triggered FEN 1 degradation via CRL4Cdt2.Citation71 These results imply that a specialized PIP box in some PCNA-interacting proteins dock onto PCNA to create a degron essential for CRL4Cdt2-dependent ubiquitylation.Citation71
As is the case for Cdt1, CRL4Cdt2-mediated ubiquitylation and degradation of p21 and Set8 requires their physical association with PCNA through a similar specialized PIP box.Citation33,Citation36,Citation37,Citation39,Citation41,Citation42 Although this activity removes chromatin-bound p21 and Set8,Citation36,Citation39 it is not clear whether CRL4Cdt2 is capable of ubiquitylating p21 and Set8 that bind PCNA off-chromatin. Although we were able to demonstrate that CRL4Cdt2 can directly ubiquitylate immuno-purified p21 from mammalian cells in vitro, our attempts at reconstituting this reaction with recombinant p21, even in the presence of PCNA, have been unsuccessful. Thus, we speculate that like Cdt1, p21 and Set8 can be ubiquitylated via the CRL4Cdt2 E3 ligase, only when bound to PCNA on chromatin. At least for p21, this conclusion is supported by the finding that human p21 transcribed and translated in vitro can be polyubiquitylated by recombinant Cdt2 in a reaction that is supplemented with Xenopus interphase extract only in the presence of single-stranded DNA.Citation70
Same Substrates, Different E3 Ligases: Redundancy vs. Specificity
Several of the CRL4Cdt2 substrates are also targeted for ubiquitylation and degradation via the SCFSkp2 ubiquitin ligase. During the S phase of the cell cycle, Cdt1 is phosphorylated by cyclin A-Cdk2 and this phosphorylation triggers its ubiquitin-dependent degradation via the SCFSkp2 E3 ligase.Citation73 Similarly, Cdk2-phosphorylated p21 is also degraded via the SCFSkp2 ubiquitin ligase. Furthermore, Set8 has also been suggested to be targeted for degradation via the SCFSkp2 pathway,Citation42,Citation74 although it is not clear whether such activity occurs specifically during S phase or is dependent on a prior phosphorylation of Set8. An obvious question is whether CRL4Cdt2 and SCFSkp2 function redundantly to target the degradation of various substrates during the S phase of the cell cycle?
The current evidence suggests that the S-phase-specific degradation of Cdt1 via the CRL4Cdt2 and SCFSkp2 pathways are not entirely redundant. For example, mice with targeted disruption of the Skp2 gene are viable although cells from these animals exhibit markedly enlarged nuclei with polyploidy and multiple centrosomes, and show a reduced growth rate and increased apoptosis.Citation75 These phenotypes have been attributed to the failure to degrade cyclin E and the cyclin-dependent kinase inhibitor p27, both during S and G2 phase of the cell cycle.Citation75 Whether failure to degrade Cdt1 via the SCFSkp2 ubiquitin ligase in these animals contribute to some of these cellular phenotypes remain to be determined. However, acute depletion of Skp2 by si-RNA does not lead to re-replication.Citation33 On the other hand, Cdt2 deletion in the mouse results in early embryonic lethality with embryos failing to develop beyond the 4–8 cell stage.Citation76 Furthermore, acute depletion of Cdt2 from human cells by siRNA is sufficient to induce significant re-replication that is not affected by co-depleting Skp2.Citation33 These findings raise the interesting question of why is Cdt1 resistant to degradation via the cyclin A-Cdk2-SCFSkp2 pathway in cells with inactivated CRL4Cdt2? One possibility is that the SCFSkp2 pathway can only degrade soluble Cdt1, while chromatin and/or PCNA-bound Cdt1 is maintained in a confirmation that is resistant to phosphorylation by cyclin A-Cdk2 or is not recognized by the SCFSkp2 ubiquitin ligase. A similar argument can be made for Set8 where only soluble, but not chromatin-bound Set8 is sensitive to SCFSkp2-mediated degradation in a manner that is analogous to Cdt1 degradation.Citation42
Deregulation of CRL4Cdt2 in Human Cancer
The central role CRL4Cdt2 in regulating the expression of various cell cycle regulatory proteins as well as its role in affecting the integrity of DNA replication and repair suggest that various components of this ubiquitin machinery could be involved in tumorigenesis. In fact, the CUL4A gene was amplified in a significant number of human malignancies including squamous cell carcinomas, primary breast cancer, adrenocortical carcinomas and hepatocellular carcinomas and to associate with shorter overall disease-free survival.Citation77 Furthermore, conditional Cul4a knockout in the skin, render mice markedly resistant to UV-induced skin carcinogenesis.Citation78 Although the oncogenic function of Cul4A can be attributed to activities associated with other DCAFs, it is tempting to speculate that Cdt2 may promote some of these Cul4A-driven tumors. This is supported by the finding that the expression of Cdt2 itself is frequently elevated in aggressive hepatocellular carcinomas (HCC) and that its level correlates positively with tumor grade and poor patient survival.Citation79 Cdt2 expression is also elevated in human breast and gastric cancers and in various cell lines derived from these primary tumors.Citation80,Citation81 In vitro, Cdt2 can promote the growth of mammary epithelial and gastric cells and silencing Cdt2 by siRNA significantly impairs the growth of these cells with defects in chromosomal segregation and cytokinesis and induction of apoptosis.Citation80,Citation81
How Cdt2 overexpression may contribute to tumorigenesis is not entirely clear but may depend on the ability of CRL4Cdt2 to promote S-phase progression by reducing the steady state basal levels of the cell cycle inhibitor p21, thus eliminating a primary mechanism by which p53-proficient cells respond to DNA damage. In fact, we found that overexpression of Cdt2 in the human osteosarcoma-derived U2OS cells is sufficient to decrease p21 protein levels specifically during the S phase of the cell cycle.Citation36 Importantly, the increased resistance to UV-induced skin carcinogenesis in mice with conditional knockout of Cul4a was dependent on failure to degrade p21 and Ddb2 or ubiquitylate the NER protein XPC.Citation78 While these observations underscore the significance of CRL4Cdt2-mediated degradation of p21 in promoting cancer, the ubiquitin-dependent degradation of other CRL4Cdt2 substrates are likely to play an equally important role in Cul4A and Cdt2-driven tumors.
The importance of CRL4Cdt2 in promoting the degradation of Cdt1 during the S phase explains the rather surprising finding that expression of constitutively nuclear cyclin D1 protein in human cancer-derived cells, as a model for deregulated cyclin D1 expression commonly occurring in human malignancies, initiates Cdt1-induced re-replication.Citation82 The stabilized Cdt1 in cyclin D1-expressing cells resulted from cyclin D1-Cdk4-dependent phosphorylation of MEP50, a cofactor of the protein arginine methyltransferase PRMT5, leading to enhanced arginine methylation of histones at the Cul4A/B promoters with consequent transcriptional repression from these genes.Citation82,Citation83 Importantly, inactivation of p53 in transgenic mice expressing constitutively nuclear cyclin D1 resulted in genomic instability and neoplastic growth in a lymphoma mouse model.Citation82 These results demonstrate that the CRL4Cdt2 may interact with other tumor-suppressor or oncogenic pathways to derive tumorigenesis.
The role CRL4Cdt2 plays in tumorigenesis is likely to introduce new avenues that can potentially be exploited for therapeutic purposes. Because failure of CRL4Cdt2 to promote the degradation of Cdt1 leads to DNA damage with the induction of apoptosis, inactivating the CRL4Cdt2 ubiquitin ligase may be an attractive target for therapeutic intervention. This conclusion gains support from the recent finding that the NAE and cullin inhibitor MLN4924 is effective at killing a large number of cancer cell lines by inducing the stabilization of Cdt1 with concurrent re-replication.Citation34,Citation35 Interestingly, MLN4924 is equally toxic in wild-type and p53 mutant cells, perhaps because p53 mutation allows more extensive re-replication.Citation28,Citation35 While inducing re-replication and consequent senescence and apoptosis are attractive therapeutically, the challenge lies in preventing potential deleterious effects that may arise in treating tumors with certain genetic makeup, as in the case for cyclin-D1-induced mouse lymphomas in the absence of wild-type p53. Hence, a clear understanding of the role CRL4Cdt2 may play in tumorigenesis and how this role is played in various genetic backgrounds is required.
Conclusions and Perspective
In the last few decades, significant progress has been made in understanding the molecular mechanisms that govern protein synthesis and turnover and how this contributes to cellular and organismal homeostasis. At the heart of this tightly regulated and finely tuned process is the regulation of proteins whose synthesis and degradation must be coordinated to allow the timely and successful transition from one stage of the cell cycle to the next while faithfully passing on genetic material to daughter cells. Several master regulators of these processes, such as cyclin-Cdk kinases, Cdc25 phosphatases, and multiple transcription factors, have already been described and characterized. The CRL4Cdt2 ubiquitin ligase is emerging as a new regulator of various cell cycle-regulated proteins and plays significant roles in promoting progression through S- and G2 phases of the cell cycle. It is also a major player for preventing aberrant DNA replication and is required for multiple DNA repair pathways.
The identification of several CRL4Cdt2 substrates has already provided valuable information on the role of this ubiquitin ligase but several questions remained unanswered. First, the signaling cascade leading to CRL4Cdt2-mediated ubiquitylation of various substrates is yet to be defined. We know that for most substrates, binding of the substrate with a “specialized PIP box” to PCNA on chromatin is essential but we do not know whether it is sufficient to trigger the ubiquitylation of the substrate. For p21, phosphorylation of serine 114 by GSK3β is required for its ubiquitylation and degradation in response to UV irradiation,Citation84 and phosphomimetic mutant p21 is better ubiquitylated via CRL4Cdt2 than wild type p21 in vitro.Citation36 While it is possible that such phosphorylation may modulate PCNA-p21 interaction, these results demonstrate that, at least for some substrates, posttranslational modification of the substrate may affect CRL4Cdt2-mediated ubiquitylation.
Other unanswered questions pertain to the regulation of the CRL4Cdt2 activity. For example, we do not know the identity of the E2 ubiquitin-conjugating enzyme(s) that cooperates with CRL4Cdt2. We also do not know the factors that trigger the assembly and disassembly of the CRL4Cdt2 E3 complex or whether the complex is assembled prior to or after its recruitment to chromatin-bound PCNA. Given that all currently identified CRL4Cdt2 substrates are targeted for degradation in S-phase or after DNA damage or both, and that the ubiquitylation requires chromatin-bound PCNA, it is likely that CRL4Cdt2 functions exclusively as an S-phase-specific ligase that can be activated by DNA damage. However, the exact role of PCNA in CRL4Cdt2-mediated activity also remains to be clearly defined. Does it function solely by creating a docking site for these substrates or does it have additional roles, for example by activating the ubiquitin ligase? Finally, significant future work is necessary to determine the exact role Cdt2 plays in the development of human tumors, both as a component of the CRL4Cdt2 ubiquitin ligase and potentially via a CRL4-independent activity.
Abbreviations
| Cdk | = | cyclin-dependent kinase |
| CRL | = | cullin-ring ligase |
| dNTP | = | deoxy-neocleotide triphosphate |
| DDB1 | = | damage-specific DNA binding protein-1 |
| DDB2 | = | damage-specific DNA binding protein-2 |
| NER | = | nucleotide excision repair |
| BER | = | base excision repair |
| CPD | = | cyclobutane pyrimidine dimers |
| TCR | = | transcription-coupled repair |
| PCNA | = | proliferating cell nuclear antigen |
| ATM | = | ataxia talengectasia-mutated |
| ATR | = | ataxia talengectasia-mutated and Rad3-related |
| XP | = | xeroderma pigmentosa |
| CSA | = | Cockayne syndrome A |
| CSB | = | Cockayne syndrome B |
| DCAF | = | DDB1 and Cul4-associated factors |
| SRFs | = | substrate recognition factors |
| pre-RC | = | pre-replication complex |
| NAE | = | NEDD8-activating enzyme |
| TLS | = | translesion DNA synthesis |
| AMP | = | adenosine monophosphate |
| HECT | = | homologous to E6-AP C-terminus |
| RING | = | really interesting new gene |
| GSK3β | = | glycogen synthase kinase-3beta |
Figures and Tables
Figure 1 Schematic illustration of the various steps involved in the ubiquitin proteasome pathway and general architecture of the cullin 1 (CRL1) and cullin 4 (CRL4)-based E3 ubiquitin ligases. (A) Ubiquitin molecules (red circles) are covalently attached to the substrate in three consecutive steps. The last step of the reaction is mediated by the specific recognition of the substrate (light magenta), often tagged with a degradation signal, by an E3 ubiquitin ligase (blue) and the transfer of ubiquitin from the E2 ubiquitin-conjugating enzyme (blue) to the substrate. In many cases the degradation signal results in the phosphorylation of the substrate at a specific residues(s). Also shown in blue, is the E1 ubiquitin-activating enzyme involved in charging ubiquitin in the first step of the reaction. The polyubiquitylated protein substrate is directed to the 26S proteasome (light green) where it is degraded proteolytically. (B and C) The scaffold cullin 1 and cullin 4 (cullin 4A or cullin 4B) proteins (light blue) in complex with Rbx1 or Rbx2 small ring finger proteins (purple) form the catalytic core of CRL1 (also known as SCF) and CRL4 respectively. Rbx1/2 proteins recruit the ubiquitin-conjugating enzyme E2 (beige) to mediate the covalent attachment of ubiquitin (red) to the substrate (light green). Skp1 and Ddb1 (damage-specific DNA binding protein 1) (orange) are adaptor proteins for CRL1 and CRL4 respectively and function to bridge the cullin proteins with a number of substrate recognition factors (SRFs) (brown). For Ddb1, one β-propeller domain (BPB) interacts with cullin 4, whereas the two other β-propeller domains (BPA and BPC) make contacts with the SRFs. For CRL1, the SRF are collectively termed F-box proteins because they invariably contain the conserved F-box motif, whereas for CRL4 ubiquitin ligases, SRFs are collectively termed DCAFs (Ddb1 and cullin 4-associated factors). (C) A schematic of the CRL4Cdt2 E3 ligase showing the DCAF Cdt2 (brown) and its ability to recognize substrates only when they are bound to the trimeric PCNA ring (red) encircling DNA (blue helix).
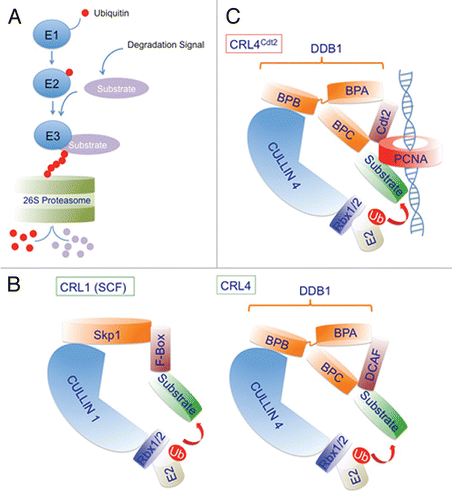
Figure 2 CRL4 ubiquitin ligase regulates several cellular activities via associating with multiple DCAFs. Several DCAFs (Ddb1 and Cul4 associating factors; shown in blue) promote the degradation of various substrates via the CRL4 E3 ubiquitin ligase. The DCAFs, DET1-COP1, Cdt2, CSA and Ddb2 (damage-specific DNA binding protein 2) (shown in blue) recognize the indicated substrates and promote their ubiquitin-dependent proteolysis to regulate various physiological activities such as transcription, chromatin remodeling and DNA repair. The targeted degradation of several of these substrates via CRL4 complexes regulates more than one physiological process.
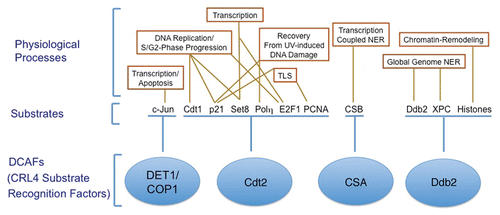
Figure 3 CRL4Cdt2 role in preventing re-replication and genomic instability. A schematic of the various pathways regulated by the CRL4Cdt2 E3 ubiquitin ligase to prevent re-initiation of DNA replication within the same cell cycle (re-replication). By promoting the degradation of the replication-licensing factor Cdt1 in S-phase, CRL4Cdt2 prevents relicensing of replication origins during the S and G2 phases of the cell cycle.Citation31 CRL4Cdt2 also promotes the degradation of the histone H4 lysine 20 methyltransferase Set8 during S-phase thus preventing the premature accumulation of momomethylated histone 4 lysine 20 (H4K20me1) at replication origins, thus inhibiting licensing of these origins.Citation39,Citation41 Additionally, the accumulation of the Cdk inhibitor p21 in cells with inactivated CRL4Cdt2,Citation33,Citation36,Citation37 is hypothesized to inhibit the cytoplasmic export of the licensing factor Cdc6 thereby contributing to re-replication and genomic instability.Citation33 These multiple activities of CRL4Cdt2 are thought to contribute to the re-replication and subsequent apoptosis induced by the novel anti-cancer drug MLN4924, which leads to the inhibition of cullin 4 activities among other cullins.Citation34
Acknowledgements
Work in authors' laboratory is supported by grants from the NIH R01 CA089406 and GM084465. T. Abbas is supported by cancer training grant from the NIH (T32CA009109) and by NCI grant (KCA140774A).
References
- Amir R, Ciechanover A, Cohen S. The ubiquitin-proteasome system: the relationship between protein degradation and human diseases. Harefuah 2001; 140:1172 - 1176
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 2002; 82:373 - 428
- Kornitzer D, Ciechanover A. Modes of regulation of ubiquitin-mediated protein degradation. J Cell Physiol 2000; 182:1 - 11
- Ciechanover A, Schwartz AL. Ubiquitin-mediated degradation of cellular proteins in health and disease. Hepatology 2002; 35:3 - 6
- Grillari J, Grillari-Voglauer R, Jansen-Durr P. Post-translational modification of cellular proteins by ubiquitin and ubiquitin-like molecules: role in cellular senescence and aging. Adv Exp Med Biol 694:172 - 196
- Groll M, Huber R. Substrate access and processing by the 20S proteasome core particle. Int J Biochem Cell Biol 2003; 35:606 - 616
- Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem 2001; 70:503 - 533
- Zimmerman ES, Schulman BA, Zheng N. Structural assembly of cullin-RING ubiquitin ligase complexes. Curr Opin Struct Biol 2010; 20:714 - 721
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 2005; 6:9 - 20
- Hotton SK, Callis J. Regulation of cullin RING ligases. Annu Rev Plant Biol 2008; 59:467 - 489
- Bosu DR, Kipreos ET. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div 2008; 3:7
- Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 2006; 443:590 - 593
- Higa LA, Zhang H. Stealing the spotlight: CUL4-DDB1 ubiquitin ligase docks WD40-repeat proteins to destroy. Cell Div 2007; 2:5
- Nakayama KI, Nakayama K. Regulation of the cell cycle by SCF-type ubiquitin ligases. Semin Cell Dev Biol 2005; 16:323 - 333
- He YJ, McCall CM, Hu J, Zeng Y, Xiong Y. DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev 2006; 20:2949 - 2954
- Higa LA, Wu M, Ye T, Kobayashi R, Sun H, Zhang H. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat Cell Biol 2006; 8:1277 - 1283
- Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell 2006; 23:709 - 721
- Higa LA, Mihaylov IS, Banks DP, Zheng J, Zhang H. Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint. Nat Cell Biol 2003; 5:1008 - 1015
- Hu J, McCall CM, Ohta T, Xiong Y. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat Cell Biol 2004; 6:1003 - 1009
- Arias EE, Walter JC. Replication-dependent destruction of Cdt1 limits DNA replication to a single round per cell cycle in Xenopus egg extracts. Genes Dev 2005; 19:114 - 126
- Hu J, Xiong Y. An evolutionarily conserved function of proliferating cell nuclear antigen for Cdt1 degradation by the Cul4-Ddb1 ubiquitin ligase in response to DNA damage. J Biol Chem 2006; 281:3753 - 3756
- Arias EE, Walter JC. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat Cell Biol 2006; 8:84 - 90
- Senga T, Sivaprasad U, Zhu W, Park JH, Arias EE, Walter JC, et al. PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J Biol Chem 2006; 281:6246 - 6252
- Higa LA, Banks D, Wu M, Kobayashi R, Sun H, Zhang H. L2DTL/CDT2 interacts with the CUL4/DDB1 complex and PCNA and regulates CDT1 proteolysis in response to DNA damage. Cell Cycle 2006; 5:1675 - 1680
- Nishitani H, Sugimoto N, Roukos V, Nakanishi Y, Saijo M, Obuse C, et al. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J 2006; 25:1126 - 1136
- Ralph E, Boye E, Kearsey SE. DNA damage induces Cdt1 proteolysis in fission yeast through a pathway dependent on Cdt2 and Ddb1. EMBO Rep 2006; 7:1134 - 1139
- Lovejoy CA, Lock K, Yenamandra A, Cortez D. DDB1 maintains genome integrity through regulation of Cdt1. Mol Cell Biol 2006; 26:7977 - 7990
- Vaziri C, Saxena S, Jeon Y, Lee C, Murata K, Machida Y, et al. A p53-dependent checkpoint pathway prevents rereplication. Mol Cell 2003; 11:997 - 1008
- Tatsumi Y, Sugimoto N, Yugawa T, Narisawa-Saito M, Kiyono T, Fujita M. Deregulation of Cdt1 induces chromosomal damage without rereplication and leads to chromosomal instability. J Cell Sci 2006; 119:3128 - 3140
- Zhong W, Feng H, Santiago FE, Kipreos ET. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature 2003; 423:885 - 889
- O'Connell BC, Harper JW. Ubiquitin proteasome system (UPS): what can chromatin do for you?. Curr Opin Cell Biol 2007; 19:206 - 214
- Sansam CL, Shepard JL, Lai K, Ianari A, Danielian PS, Amsterdam A, et al. DTL/CDT2 is essential for both CDT1 regulation and the early G2/M checkpoint. Genes Dev 2006; 20:3117 - 3129
- Kim Y, Starostina NG, Kipreos ET. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev 2008; 22:2507 - 2519
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 2009; 458:732 - 736
- Lin JJ, Milhollen MA, Smith PG, Narayanan U, Dutta A. NEDD8 targeting drug MLN4924 elicits DNA re-replication by stabilizing Cdt1 in S Phase, triggering checkpoint activation, apoptosis and senescence in cancer cells. Cancer Res In press
- Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev 2008; 22:2496 - 2506
- Nishitani H, Shiomi Y, Iida H, Michishita M, Takami T, Tsurimoto T. CDK inhibitor p21 is degraded by a PCNA coupled Cul4-DDB1Cdt2 pathway during S phase and after UV irradiation. J Biol Chem 2008;
- Kim J, Feng H, Kipreos ET. C. elegans CUL-4 prevents rereplication by promoting the nuclear export of CDC-6 via a CKI-1-dependent pathway. Curr Biol 2007; 17:966 - 972
- Abbas T, Shibata E, Park J, Jha S, Karnani N, Dutta A. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol Cell 40:9 - 21
- Tardat M, Brustel J, Kirsh O, Lefevbre C, Callanan M, Sardet C, et al. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat Cell Biol 12:1086 - 1093
- Centore RC, Havens CG, Manning AL, Li JM, Flynn RL, Tse A, et al. CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol Cell 40:22 - 33
- Oda H, Hubner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, et al. Regulation of the histone H4 monomethylase PR-Set7 by CRL4Cdt2-mediated PCNA-dependent degradation during DNA damage. Mol Cell 40:364 - 376
- Houston SI, McManus KJ, Adams MM, Sims JK, Carpenter PB, Hendzel MJ, et al. Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J Biol Chem 2008; 283:19478 - 19488
- Karachentsev D, Sarma K, Reinberg D, Steward R. PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes Dev 2005; 19:431 - 435
- Trojer P, Li G, Sims RJ 3rd, Vaquero A, Kalakonda N, Boccuni P, et al. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell 2007; 129:915 - 928
- Fitch ME, Nakajima S, Yasui A, Ford JM. In vivo recruitment of XPC to UV-induced cyclobutane pyrimidine dimers by the DDB2 gene product. J Biol Chem 2003; 278:46906 - 46910
- Wang QE, Zhu Q, Wani G, Chen J, Wani AA. UV radiation-induced XPC translocation within chromatin is mediated by damaged-DNA binding protein, DDB2. Carcinogenesis 2004; 25:1033 - 1043
- Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G, et al. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 2005; 121:387 - 400
- Li J, Wang QE, Zhu Q, El-Mahdy MA, Wani G, Praetorius-Ibba M, et al. DNA damage binding protein component DDB1 participates in nucleotide excision repair through DDB2 DNA-binding and cullin 4A ubiquitin ligase activity. Cancer Res 2006; 66:8590 - 8597
- Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, et al. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 2003; 113:357 - 367
- Groisman R, Kuraoka I, Chevallier O, Gaye N, Magnaldo T, Tanaka K, et al. CSA-dependent degradation of CSB by the ubiquitin-proteasome pathway establishes a link between complementation factors of the Cockayne syndrome. Genes Dev 2006; 20:1429 - 1434
- Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 2009; 9:400 - 414
- Zhu W, Abbas T, Dutta A. DNA replication and genomic instability. Adv Exp Med Biol 2005; 570:249 - 279
- Cazzalini O, Scovassi AI, Savio M, Stivala LA, Prosperi E. Multiple roles of the cell cycle inhibitor p21(CDKN1A) in the DNA damage response. Mutat Res 704:12 - 20
- Soria G, Gottifredi V. PCNA-coupled p21 degradation after DNA damage: The exception that confirms the rule?. DNA Repair (Amst) 9:358 - 364
- Soria G, Podhajcer O, Prives C, Gottifredi V. P21Cip1/WAF1 downregulation is required for efficient PCNA ubiquitination after UV irradiation. Oncogene 2006; 25:2829 - 2838
- Waters LS, Minesinger BK, Wiltrout ME, D'Souza S, Woodruff RV, Walker GC. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev 2009; 73:134 - 154
- Soria G, Speroni J, Podhajcer OL, Prives C, Gottifredi V. p21 differentially regulates DNA replication and DNA-repair-associated processes after UV irradiation. J Cell Sci 2008; 121:3271 - 3282
- Lee KY, Myung K. PCNA modifications for regulation of post-replication repair pathways. Mol Cells 2008; 26:5 - 11
- Brun J, Chiu R, Lockhart K, Xiao W, Wouters BG, Gray DA. hMMS2 serves a redundant role in human PCNA polyubiquitination. BMC Mol Biol 2008; 9:24
- Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol 2006; 8:339 - 347
- Simpson LJ, Ross AL, Szuts D, Alviani CA, Oestergaard VH, Patel KJ, et al. RAD18-independent ubiquitination of proliferating-cell nuclear antigen in the avian cell line DT40. EMBO Rep 2006; 7:927 - 932
- Terai K, Abbas T, Jazaeri AA, Dutta A. CRL4(Cdt2) E3 ubiquitin ligase monoubiquitinates PCNA to promote translesion DNA synthesis. Mol Cell 37:143 - 149
- Kim SH, Michael WM. Regulated proteolysis of DNA polymerase eta during the DNA-damage response in C. elegans. Mol Cell 2008; 32:757 - 766
- Moss J, Tinline-Purvis H, Walker CA, Folkes LK, Stratford MR, Hayles J, et al. Break-induced ATR and Ddb1-Cul4Cdt2 ubiquitin ligase-dependent nucleotide synthesis promotes homologous recombination repair in fission yeast. Genes Dev 24:2705 - 2716
- Liu C, Poitelea M, Watson A, Yoshida SH, Shimoda C, Holmberg C, et al. Transactivation of Schizosaccharomyces pombe cdt2+ stimulates a Pcu4-Ddb1-CSN ubiquitin ligase. EMBO J 2005; 24:3940 - 3951
- Liu C, Powell KA, Mundt K, Wu L, Carr AM, Caspari T. Cop9/signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint-dependent and -independent mechanisms. Genes Dev 2003; 17:1130 - 1140
- Elledge SJ, Zhou Z, Allen JB. Ribonucleotide reductase: regulation, regulation, regulation. Trends Biochem Sci 1992; 17:119 - 123
- Shibutani ST, de la Cruz AF, Tran V, Turbyfill WJ 3rd, Reis T, Edgar BA, et al. Intrinsic negative cell cycle regulation provided by PIP box- and Cul4Cdt2-mediated destruction of E2f1 during S phase. Dev Cell 2008; 15:890 - 900
- Kim DH, Budhavarapu VN, Herrera CR, Nam HW, Kim YS, Yew PR. The CRL4Cdt2 ubiquitin ligase mediates the proteolysis of cyclin-dependent kinase inhibitor Xic1 through a direct association with PCNA. Mol Cell Biol 30:4120 - 4133
- Havens CG, Walter JC. Docking of a specialized PIP Box onto chromatin-bound PCNA creates a degron for the ubiquitin ligase CRL4Cdt2. Mol Cell 2009; 35:93 - 104
- Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell 2007; 129:665 - 679
- Takeda DY, Parvin JD, Dutta A. Degradation of Cdt1 during S phase is Skp2-independent and is required for efficient progression of mammalian cells through S phase. J Biol Chem 2005; 280:23416 - 23423
- Yin Y, Yu VC, Zhu G, Chang DC. SET8 plays a role in controlling G1/S transition by blocking lysine acetylation in histone through binding to H4 N-terminal tail. Cell Cycle 2008; 7:1423 - 1432
- Nakayama K, Nagahama H, Minamishima YA, Matsumoto M, Nakamichi I, Kitagawa K, et al. Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. EMBO J 2000; 19:2069 - 2081
- Liu CL, Yu IS, Pan HW, Lin SW, Hsu HC. L2dtl is essential for cell survival and nuclear division in early mouse embryonic development. J Biol Chem 2007; 282:1109 - 1118
- Lee J, Zhou P. Cullins and Cancer. Genes Cancer 1:690 - 699
- Liu L, Lee S, Zhang J, Peters SB, Hannah J, Zhang Y, et al. CUL4A abrogation augments DNA damage response and protection against skin carcinogenesis. Mol Cell 2009; 34:451 - 460
- Pan HW, Chou HY, Liu SH, Peng SY, Liu CL, Hsu HC. Role of L2DTL, cell cycle-regulated nuclear and centrosome protein, in aggressive hepatocellular carcinoma. Cell Cycle 2006; 5:2676 - 2687
- Ueki T, Nishidate T, Park JH, Lin ML, Shimo A, Hirata K, et al. Involvement of elevated expression of multiple cell cycle regulator, DTL/RAMP (denticleless/RA-regulated nuclear matrix associated protein), in the growth of breast cancer cells. Oncogene 2008; 27:5672 - 5683
- Li J, Ng EK, Ng YP, Wong CY, Yu J, Jin H, et al. Identification of retinoic acid-regulated nuclear matrix-associated protein as a novel regulator of gastric cancer. Br J Cancer 2009; 101:691 - 698
- Aggarwal P, Lessie MD, Lin DI, Pontano L, Gladden AB, Nuskey B, et al. Nuclear accumulation of cyclin D1 during S phase inhibits Cul4-dependent Cdt1 proteolysis and triggers p53-dependent DNA rereplication. Genes Dev 2007; 21:2908 - 2922
- Aggarwal P, Vaites LP, Kim JK, Mellert H, Gurung B, Nakagawa H, et al. Nuclear cyclin D1/CDK4 kinase regulates CUL4 expression and triggers neoplastic growth via activation of the PRMT5 methyltransferase. Cancer Cell 18:329 - 340
- Jeng YM, Cai-Ng S, Li A, Furuta S, Chew H, Chen PL, et al. Brca1 heterozygous mice have shortened life span and are prone to ovarian tumorigenesis with haploinsufficiency upon ionizing irradiation. Oncogene 2007; 26:6160 - 6166