Abstract
We have recently proposed a new mechanism for explaining energy transfer in cancer metabolism. In this scenario, cancer cells behave as metabolic parasites, by extracting nutrients from normal host cells, such as fibroblasts, via the secretion of hydrogen peroxide as the initial trigger. Oxidative stress in the tumor microenvironment then leads to autophagy-driven catabolism, mitochondrial dys-function, and aerobic glycolysis. This, in turn, produces high-energy nutrients (such as L-lactate, ketones, and glutamine) that drive the anabolic growth of tumor cells, via oxidative mitochondrial metabolism. A logical prediction of this new “parasitic” cancer model is that tumor-associated fibroblasts should show evidence of mitochondrial dys-function (mitophagy and aerobic glycolysis). In contrast, epithelial cancer cells should increase their oxidative mitochondrial capacity. To further test this hypothesis, here we subjected frozen sections from human breast tumors to a staining procedure that only detects functional mitochondria. This method detects the in situ enzymatic activity of cytochrome C oxidase (COX), also known as Complex IV. Remarkably, cancer cells show an over-abundance of COX activity, while adjacent stromal cells remain essentially negative. Adjacent normal ductal epithelial cells also show little or no COX activity, relative to epithelial cancer cells. Thus, oxidative mitochondrial activity is selectively amplified in cancer cells. Although COX activity staining has never been applied to cancer tissues, it could now be used routinely to distinguish cancer cells from normal cells, and to establish negative margins during cancer surgery. Similar results were obtained with NADH activity staining, which measures Complex I activity, and succinate dehydrogenase (SDH) activity staining, which measures Complex II activity. COX and NADH activities were blocked by electron transport inhibitors, such as Metformin. This has mechanistic and clinical implications for using Metformin as an anti-cancer drug, both for cancer therapy and chemo-prevention. We also immuno-stained human breast cancers for a series of well-established protein biomarkers of metabolism. More specifically, we now show that cancer-associated fibroblasts over-express markers of autophagy (cathepsin B), mitophagy (BNIP3L), and aerobic glycolysis (MCT4). Conversely, epithelial cancer cells show the over-expression of a mitochondrial membrane marker (TOMM20), as well as key components of Complex IV (MT-CO1) and Complex II (SDH-B). We also validated our observations using a bioinformatics approach with data from >2,000 breast cancer patients, which showed the transcriptional upregulation of mitochondrial oxidative phosphorylation (OXPHOS) in human breast tumors (p < 10-20), and a specific association with metastasis. Therefore, upregulation of OXPHOS in epithelial tumor cells is a common feature of human breast cancers. In summary, our data provide the first functional in vivo evidence that epithelial cancer cells perform enhanced mitochondrial oxidative phosphorylation, allowing them to produce high amounts of ATP. Thus, we believe that mitochondria are both the “powerhouse” and “Achilles’ heel” of cancer cells.
Introduction
We recently provided experimental evidence that aggressive tumors and skeletal muscle may use similar metabolic strategies, resulting in a form of “symbiotic” metabolic-coupling.Citation1–Citation4 To understand how this applies to human cancer, it is important to first appreciate how skeletal muscle is organized.
Skeletal muscle tissue contains at least two types of muscle fibers: slow-twitch and fast-twitch.Citation5–Citation8 Slow-twitch fibers (type I) have an abundance of mitochondria, undergo oxidative phosphorylation, and produce high amounts of ATP. In contrast, fast-twitch fibers (type II) have few mitochondria, are predominantly glycolytic, produce low amounts of ATP and secrete L-lactate. Secreted L-lactate, generated in fast-twitch fibers, is taken up by slow-twitch muscle fibers, and used as recycled “fuel” for mitochondrial oxidative phosphorylation. This phenomenon is known as the “Lactate Shuttle.”Citation5–Citation8 Thus, fast-twitch and slow-twitch fibers are directly metabolically-coupled.Citation5–Citation8
Over the last 40–50 years, special histo-chemical stains have been utilized to distinguish between glycolytic and oxidative muscle fibers.Citation9–Citation16 These activity-based stains depend on an intact mitochondrial electron transport system (ETC), and are a functional measure of mitochondrial “power” or oxidative capacity. For example, COX (Cytochrome C Oxidase) stainingCitation17 detects Complex IV, the last step in the mitochondrial respiratory chain, also known as Warburg respiratory enzyme. Similarly, NADH staining detects the dehydrogenase activity of Complex I, the first step in the mitochondrial respiratory chain. And, SDH (succinate dehydrogenase) staining detects the activity of Complex II, the second step in the respiratory chain. Thus, slow-twitch muscle fibers are oxidative, and are NADH(+), SDH(+) and COX(+). In contrast, fast-twitch muscle fibers are glycolytic, and are NADH(−), SDH(−) and COX(−). Clinically, these mitochondrial activity stains have been very effective in the diagnosis of mitochondrial-based myopathies, due to genetic defects in the respiratory chain components of either Complex I, Complex II or Complex IV, resulting in defective oxidative phosphorylation.Citation12–Citation16 However, these stains have not been routinely applied to other mitochondrial-based diseases, such as human cancers.
Recently, we proposed that a subset of aggressive tumors use stromal-epithelial metabolic-coupling.Citation1–Citation4 In these cancers, a lactate-shuttle supports the transfer of lactate from glycolytic fibroblasts to oxidative cancer cells, in a pathological process that mirrors the physiological metabolic reciprocity of skeletal muscle fibers.Citation1 We have termed this phenomenon “The Reverse Warburg Effect,” since aerobic glycolysis (lactate production) takes place in cancer-associated fibroblasts, rather than in epithelial cancer cells.Citation18
In this model, autophagy, mitophagy and aerobic glycolysis in the fibroblastic tumor stroma produces high-energy nutrients and chemical building blocks (such as lactate, ketones and glutamine), which can then all be used as “biofuel” for oxidative mitochondrial metabolism in epithelial cancer cells.Citation19–Citation24
A clear prediction of this model is that aggressive tumors are organized on the same or similar metabolic principles as skeletal muscle. For example, the tumor stroma should lack oxidative mitochondrial capacity, and therefore, should be NADH(−), SDH(−) and COX(−). In contrast, aggressive epithelial cancer cells should have increased oxidative mitochondrial capacity, and should be highly NADH(+), SDH(+) and COX(+).
In accordance with this model, here we demonstrate that human breast cancer epithelial cell “nests” show amplified levels of mitochondrial oxidative activity, as seen by intense NADH, SDH and COX functional staining. In contrast, adjacent stromal tissues show little or no mitochondrial oxidative capacity. Cancer cell associated NADH, SDH and COX activity staining was ablated by the addition of well-known Complex I (metformin), Complex II (malonate) or Complex IV (azide) inhibitors, directly demonstrating the mitochondrial-based specificity of our findings. Thus, our data provide the first functional in vivo evidence that epithelial cancer cells perform mitochondrial oxidative phosphorylation, allowing them to produce high amounts of ATP.
As metformin behaves as a “weak” mitochondrial poison, due to its Complex I inhibitor activity,Citation25–Citation28 this could explain its remarkable clinical ability to prevent many different types of human cancers in diabetic patients, and its effectiveness as an anti-cancer agent in pre-clinical animal models.Citation29–Citation36
Results
Detection of mitochondria, autophagy, mitophagy and aerobic glycolysis in human breast cancers, using antibody probes.
We have previously proposed a new paradigm to understand tumor metabolism. In this model, catabolic processes (such as autophagy, mitophagy and aerobic glycolysis) in the tumor stroma “fuel” the anabolic growth of adjacent tumor cells via oxidative mitochondrial metabolism.Citation2–Citation4,Citation18,Citation37–Citation39 Thus, tumor cells and the stroma are metabolically coupled in a type of symbiotic or host-parasite relationship. A logical prediction of this hypothesis is that cancer-associated fibroblasts would undergo autophagy, mitophagy and secrete lactate, while epithelial cancer cells would be rich in functional mitochondria.
To further assess the validity of this hypothesis, we immunostained human breast cancer samples with a well-established panel of markers for autophagy (cathepsin B), mitophagy (BNIP3L), and aerobic glycolysis/lactate secretion (MCT4). Similarly, the same samples were immuno-stained with antibodies directed against TOMM20 (translocase of outer mitochondrial membrane 20), which is a component of the receptor complex responsible for the recognition and translocation of cytosolically-generated mitochondrial precursor proteins.
shows that TOMM20 is specifically localized to the epithelial cancer cell compartment in human breast cancers, and is largely excluded from the adjacent tumor stromal compartment. Thus, it appears that epithelial cancer cells do harbor mitochondria, and show an overall relative increase in mitochondrial mass, as compared with the tumor stroma.
In contrast, cathepsin B (a lysosomal protease; ), BNIP3L (an inducer of mitophagy; ), and MCT4 (a transporter that is specialized in L-lactate efflux; ), are all preferentially localized to the tumor stromal compartment, and are largely absent from epithelial tumor cells. These findings are in accordance with our previous studies showing that the tumor stroma is mainly catabolic.Citation1,Citation22
Thus, autophagy, mitophagy and aerobic glycolysis (lactate production), appear to occur in the tumor stroma, while epithelial cancer cells have the majority of mitochondria. This is consistent with the idea that catabolism in the tumor stroma provides recycled nutrients and chemical building blocks to fuel tumor cell growth and metastasis, via mitochondrial metabolism.
Visualization of functional mitochondria in human breast cancers, using COX staining (complex IV activity).
Cytochrome C oxidase (EC 1.9.3.1; COX; Complex IV; Warburg respiratory enzyme) is the last enzyme complex in the respiratory chain in mitochondria, which participates in driving electron transport, oxidative phosphorylation and ATP synthesis.Citation11 COX normally contains 13 protein subunits in mammalian cells. COX activity staining (which appears brown) has been developed to facilitate the diagnosis of mitochondrial myopathies that are due to defects in respiratory chain subunits, such as those participating in the formation of Complex IV.Citation9
COX staining of frozen sections of normal skeletal muscle tissue allows for the detection of two different types of muscle fibers. Slow-twitch fibers (type I) are oxidative, are mitochondria-rich, and are COX-positive. In contrast, fast-twitch fibers (type II) are glycolytic, are mitochondria-poor, and are COX-negative. Thus, COX staining enables one to distinguish oxidative cells from glycolytic cells, based on functional mitochondrial activity directly related to their capacity for oxidative phosphorylation. shows a typical COX staining pattern in murine skeletal muscle frozen sections, with COX(+) and COX(−) muscle fibers.
Here, for the first time, we applied COX activity staining to frozen sections derived from human breast cancers. shows two low-power images of COX staining. The human epithelial tumor cells are intensely stained, as compared with adjacent stromal cells, which show little or no COX activity, in comparison. Higher power images are also shown in , allowing a better appreciation of the epithelial character of the COX(+) cancer cell nests. The epithelial cancer cells were also more intensely stained than adjacent normal epithelial cells that are part of mammary ductal tissue (). Thus, COX activity staining can be used to distinguish normal epithelial cells from adjacent cancer cells, within the same tumor tissue. Most importantly, our results indicate that the oxidative mitochondrial power of epithelial cancer cells appears to be amplified, relative to adjacent stromal and epithelial tissue.
To assess the specificity of COX activity staining, we used well-known inhibitors for Complex I (metformin) and Complex IV (sodium azide). shows consecutive serial sections stained for COX activity in the presence or absence of mitochondrial inhibitors. Sodium azide (a Complex IV inhibitor) effectively abolished COX activity staining, directly demonstrating high-specificity. Suprisingly, treatment with metformin (a Complex I inhibitor)Citation25 also significantly reduced COX activity staining (Complex IV). How do we explain these results? One possibility is that since the electron transport chain is intact in epithelial cancer cells, Complex I activity helps to boost Complex IV activity as well.
As metformin is now being proposed as an anti-cancer drug for the treatment of a variety of different types of human cancers, and for the chemo-prevention of recurrence,Citation29–Citation36 this assay system may be ideal for assessing the sensitivity of a given patient's tumor to metformin therapy.
Visualization of functional mitochondria in human breast cancers, using NADH staining (complex I activity).
NADH dehydrogenase (EC 1.6.5.3; NADH:ubiquinone reductase; Complex I) is an enzyme complex located within the inner mitochondrial membrane, and is the first step of the mitochondrial electron transport chain (ETC) and oxidative phosphorylation.Citation40,Citation41 More specifically, Complex I transfers electrons from NADH to co-enzyme Q (CoQ).
An activity stain has also been developed for the detection of NADH dehydrogenase (which appears blue). As with the COX activity staining, NADH activity staining was developed for the detection of mitochondrial myopathies. However, this stain will detect defects in Complex I, rather than Complex IV activity.
As for COX staining, NADH staining of normal skeletal muscle frozen sections allows one to distinguish two different types of muscle fibers. Slow-twitch fibers (type I) are oxidative, are mitochondria-rich, and are NADH-positive. In contrast, fast-twitch fibers (type II) are glycolytic, are mitochondria-poor, and are NADH-negative. shows a typical NADH staining pattern in murine skeletal muscle frozen sections, with NADH(+) and NADH(−) fibers.
Next, we applied NADH activity staining to frozen sections derived from human breast cancers. shows that human epithelial tumor cells are intensely stained, as compared with adjacent stromal cells, which show little or no NADH activity, in comparison. Thus, the oxidative mitochondrial activity of epithelial cancer cells appears to be amplified, relative to adjacent stromal tissue.
To further assess the specificity of NADH activity staining, we used a well-known inhibitor for Complex I (metformin).Citation42,Citation43 shows frozen sections stained for NADH activity in the presence or absence of metformin. Note that metformin (a Complex I inhibitor) effectively abolished the NADH activity staining, directly demonstrating high-specificity. As with COX staining, NADH activity staining may be ideal for assessing the sensitivity of a given patient's tumor to metformin therapy, or other mitochondrial inhibitors that could be used for anti-cancer therapy.
Visualization of functional mitochondria in human breast cancers, using SDH staining (complex II activity).
Succinate dehydrogenase (EC 1.3.5.1; succinate-coenzyme Q reductase (SQR) or Complex II) is a multi-subunit enzyme complex (SDH-A, -B, -C) that is attached to the inner mitochondrial membrane. The SDH complex catalyzes the conversion of succinate to fumarate (via oxidation) and ubiquinone to ubiquinol (via reduction). It is the only enzyme that participates in both electron transport and the TCA cycle.
As with COX and NADH, an activity stain has also been developed for the detection of SDH activity (which appears blue). In normal skeletal muscle, SDH staining shows the presence of two different classes of muscle fibers. Slow-twitch fibers (type I) are oxidative, are mitochondria-rich, and are SDH-positive. In contrast, fast-twitch fibers (type II) are glycolytic, are mitochondria-poor, and are SDH-negative. Representative images of murine skeletal muscle frozen sections, with SDH(+) and SDH(−) fibers, are shown in .
As predicted, SDH activity staining of frozen sections derived from human breast cancers demonstrates that human epithelial tumor cells are intensely stained, as compared with adjacent stromal cells (). We also employed a well-known inhibitor for Complex II (malonate), to assess the specificity of SDH activity staining. Importantly, treatment with malonate markedly diminished the SDH activity staining, directly demonstrating high-specificity. shows images of frozen sections stained for SDH activity in the presence or absence of malonate.
Detection of mitochondria in human breast cancers, using complex IV and complex II specific antibody probes.
To further validate our observations with COX activity staining, we also immuno-stained human breast cancers tissues with antibodies directed against a key component of the COX complex, namely mitochondrially-encoded cytochrome c oxidase subunit 1 (MT-CO1). Although Complex IV contains 13 individual components, subunits 1–3 form the active functional core of the enzyme, and subunit 1 is the catalytic element of the complex.
shows that MT-CO1 is specifically localized to the epithelial cancer cells in human breast cancers, and is largely excluded from the adjacent tumor stromal compartment. Similarly, shows that SDH-B, the Fe-S containing component of Complex II, is also largely confined to the epithelial cancer cell compartment.
Thus, these data are consistent with our independent findings that epithelial cancer cells harbor most of the functional COX and SDH mitochondrial activity, and therefore show an overall increase in mitochondrial mass.
Generating an epithelial-specific mitochondrial gene signature using laser-captured compartment-specific profiling data from breast cancer patients.
Next, we used existing genome-wide transcriptional profiling data to further explore the idea that mitochondrial oxidative phosphorylation (OXPHOS) is specifically upregulated in human breast cancer epithelial cells, relative to adjacent stromal tumor tissue. For this purpose, we performed an informatics analysis on the transcriptional profiles of epithelial breast cancer cells and adjacent stromal cells (which were both isolated by laser-capture micro-dissection). Data were generated essentially as we previously described in reference Citation44, using the raw transcriptional profiles of n = 28 breast cancer patients (cancer epithelial vs. stromal cells).Citation45 In this analysis, only the gene transcripts of mitochondrial associated proteins that showed a >4-fold increase in breast cancer epithelial cells (relative to adjacent stromal cells) were selected to generate an epithelial-specific mitochondrial gene signature (). This short signature contains only 38 gene transcripts.
Interestingly, key components of Complex I (NDUF), Complex II (SDH), Complex III (UQCR) and Complex IV (COX) were all transcriptionally upregulated, as well as the mitochondrial ATP synthase (ATP5), specifically in epithelial cancer cells. Conversely, these data also demonstrate that relative to epithelial cancer cells, stromal cells show a transcriptional “down-modulation” of mitochondrial oxidative phosphorylation.
Validation with bioinformatics: an epithelial-specific mitochondrial gene signature predicts metastasis in breast cancer patients.
To further validate our histochemical observations, we next used existing bioinformatics databases to assess the association of an epithelial-specific mitochondrial gene signature (), with the transcriptional profiles of human breast cancer tumor samples and various clinical parameters. As discussed above, this signature consists of mitochondrial associated gene transcripts (mainly OXPHOS-related) that are specifically upregulated only in epithelial breast cancer cells, and not in adjacent stromal tissue.
Importantly, shows that this epithelial-specific mitochondrial gene signature is clearly upregulated in human breast tumors (>2,000 cases examined, with a p value < 10−20), relative to normal healthy breast tissue (102 controls). Similarly, significant associations were obtained with tissue from both ER-positive (>1,600 cases examined, p < 10−20) and ER-negative (nearly 500 cases examined, p < 10−10) breast cancer patients. As such, upregulation of this epithelial MITO/OXPHOS gene signature is a common feature of human breast cancers.
Remarkably, this mitochondrial-based epithelial gene signature also predicts breast cancer cell metastasis, especially in ER-negative patients (). In ER(−) disease, this MITO/OXPHOS signature was associated with a near 30% increase in metastasis, over a 10-year follow-up period (78.6% vs. 50.0% metastasis-free). In support of this observation, injection of L-lactate (a nutrient which stimulates OXPHOS) in an ER(−) xenograft model (MDA-MB-231 cells) is sufficient to increase lung metastasis by >10-fold.Citation44 Furthermore, Berridge and colleagues have shown that specific deletion of mitochondrial DNA from cancer cells prevents both tumor growth and lung metastasis, in mouse animal models.Citation46
It is important to note that for this gene profiling analysis, although the signature we used was epithelial cancer cell-specific, the transcriptional profiling data from breast cancer patients was obtained from whole breast tumors excised during surgery. Thus, our results indicate that the use of this MITO/OXPHOS gene signature for diagnostic and prognostic purposes does not require laser-capture micro-dissection of the primary tumor.
Similar results were obtained using three other mitochondrial OXPHOS-based gene signatures from the KEGG and Molecular Signatures Database (MSigDB) (Fig. S1), providing additional independent support for our hypothesis that mitochondrial oxidative phosphorylation is clearly amplified or hyper-activated in human breast cancers. p-values for these associations between OXPHOS and breast cancers ranged from 10−14 to 10−36. Strong associations were observed with tumors from both ER(+) and ER(−) breast cancer patients.
Discussion
Over the past 85 years, it has been a hotly debated topic as to whether aggressive cancer cells contain functional mitochondria, and undergo oxidative phosphorylation, or derive their energy from aerobic glycolysis.Citation47 However, glycolysis is an inherently inefficient process for generating energy, resulting in the production of only 2–4 ATPs per glucose molecule.Citation48–Citation50 Nevertheless, it has been argued that cancer cells (with exceedingly high energy demands) use aerobic glycolysis (a.k.a., the Warburg Effect) almost exclusively.Citation48–Citation50
Recently, we proposed an alternative explanation for Warburg's observations that tumors show a shift toward aerobic glycolysis.Citation18 More specifically, we showed that cancer cells induce aerobic glycolysis in adjacent cancer-associated fibroblasts, via oxidative stress.Citation20,Citation21 Oxidative stress in fibroblasts then drives autophagy and mitophagy, resulting in aerobic glycolysis, due to mitochondrial dys-function. These glycolytic fibroblasts, in turn, produce L-lactate to “feed” hungry cancer cells, resulting in a form of “parasitic” stromal-epithelial metabolic coupling ().Citation22,Citation51 In this scenario, epithelial cancer cells then use lactate to perform oxidative mitochondrial respiration, driving the formation of 36–38 ATPs per glucose molecule.Citation52 This model, termed “The Reverse Warburg Effect,” would help resolve the paradox of the conventional “Warburg Effect,” which states that cancer cells with exceedingly high metabolic needs apparently use an inefficient process (aerobic glycolysis) to produce ATP.Citation3,Citation39
In 1931, Otto Warburg was awarded the Nobel Prize in Physiology “for his discovery of the nature and mode of action of the respiratory enzyme” (www.nobelprize.org/nobel_prizes/medicine/laureates/1931). Today, “Warburg respiratory enzyme” is known as Cytochrome C Oxidase (COX) or Complex IV (http://enzyme.expasy.org/EC/1.9.3.1).
Here, we have used a COX-based histochemical activity stain to directly visualize the functional status of oxidative phosphorylation and mitochondria in human breast cancers. We directly demonstrate that epithelial cancer cells have the highest levels of COX activity, higher than normal adjacent epithelial cells, and even higher than stromal cells. The rank order for positive COX staining was: Epithelial Cancer Cells > > > Normal Epithelial Cells > Tumor Stromal Tissue. COX staining allowed us to unambiguously distinguish cancer cells from adjacent normal epithelial cells. This may have important implications for cancer diagnosis and prognosis, especially in demonstrating negative margins and in detecting distant cancer cell metastases.
Similar results were also obtained using NADH and SDH activity staining, as a measure of Complex I and Complex II activity. Positive NADH and SDH staining was also largely confined to epithelial cancer cells, and nearly absent or significantly diminished in the tumor stroma. Thus, the activities of Complex I, II and IV all appear to be amplified in epithelial cancer cells.
The specificity of our COX, NADH and SDH staining in human breast cancer samples was further validated using well-established Complex I (metformin), Complex II (malonate) and Complex IV (azide) inhibitors. Treatment with these respiratory chain inhibitors prevented COX, NADH and SDH staining in human breast cancer cells.
Metformin is reported to be a Complex I inhibitorCitation25 that functions as a “weak” mitochondrial poison, and is currently used extensively for the clinical treatment of diabetes.Citation42,Citation43 By reducing mitochondrial activity, metformin induces cells to take up more glucose, therapeutically resulting in lower blood glucose levels. Metformin toxicity or overdose is associated with lactic acidosis,Citation29,Citation30 consistent with its function as a respiratory chain inhibitor, since it systemically induces aerobic glycolysis, resulting in lactate over-production.
A “side effect” of Metformin therapy in diabetes patients is cancer prevention. In accordance with our data that cancer cells use mitochondrial oxidative phosphorylation for energy production, metformin treatment prevents and/or inhibits tumor formation both in diabetic patients and in mouse animal models.Citation31–Citation36 Metformin also induces apoptosis in “cancer stem cells.”Citation31,Citation34,Citation35 Metabolically, metformin prevents cancer cells from using their mitochondria, induces aerobic glycolysis and lactate production, and shifts cancer cells toward the conventional “Warburg Effect.”Citation30,Citation53 This is all consistent with the idea that cancer cells have functional mitochondria.
Other genetic studies have also suggested that aggressive cancer cells have functional mitochondria.Citation54–Citation56 Mechanistically, in vitro, fibroblasts or mesenchymal stem cells can rescue aerobic glycolysis in adjacent cancer cells, via the direct transfer of live mitochondria or mitochondrial DNA, via intercellular nano-tubes.Citation57 Interestingly, this “mitochondrial transfer” also occurs in murine tumors in vivo, where implanted cancer cells “steal” functional mitochondrial DNA from normal host cells or the tumor microenvironment.Citation58–Citation60 Thus, amplification of oxidative mitochondrial metabolism in human cancer cells in vivo may occur via this mechanism, and could be considered a form of “mitochondrial rejuvenation,” that facilitates the immortality and metastasis of cancer cells.
In direct support of this notion, proteomic analysis of human breast cancer cells with a propensity to undergo brain metastasis, directly showed the upregulation of enzymes involved in the mitochondrial TCA cycle and oxidative phosphorylation.Citation61 Furthermore, the brain is a lactate-rich micro-environment.Citation62 Similarly, MCF7 breast cancer cells treated with L-lactate show a marked increase in mitochondrial massCitation21,Citation22 and a have transcriptional profile that significantly overlaps with neural stem cells.Citation52 These MCF7 cell lactate-induced gene signatures are predictive of tumor recurrence, metastasis and poor clinical outcome, in human breast cancer patients [ER(+)/Luminal A type].Citation52 Our current bioinformatics analysis () also supports an association between mitochondrial oxidative phosphorylation (OXPHOS) in epithelial cancer cells and breast cancer metastasis.
In accordance with our new findings, recent studies on the effectiveness of radiofrequency ablation (RFA) therapy for the surgical removal of human breast cancer tumor tissue used the “NADH-diaphorase reaction” as a measure of cell viability,Citation63 although the authors did not acknowledge that this reaction is mitochondrial-specific. Moreover, their photo-micrographs directly show that the NADH reaction product (blue color) is indeed largely confined to the epithelial tumor cells, and is absent from the adjacent stromal tissue. However, the authors failed to comment on the epithelial-specific compartmentalization of the NADH staining.Citation63 Thus, independent observations already exist to support the specificity of the experiments reported here.
Our results are also consistent with the idea that mitochondrial toxins or poisonsCitation64–Citation66 should be used for cancer chemo-therapy and chemo-prevention, to target cancer cell mitochondria and oxidative phosphorylation. Several groups have recently designed therapies around this principle, by using non-toxic precursor molecules (such as linamarin) that are enzymatically converted to active cyanide (a mitochondrial Complex IV inhibitor) locally at the site of the tumor, thereby inducing mitophagy and cell death in epithelial cancer cells.Citation67–Citation69 In mechanistic support of this notion, recombinant overexpression of mutationally-activated HIF1-α in human breast cancer cells (MDA-MB-231), a known inducer of autophagy/mitophagy and aerobic glycolysis, reduces tumor growth in pre-clinical xenograft models by up to 3-fold.Citation23
Finally, using an independent informatics approach, we were able to directly validate the idea that OXPHOS is selectively upregulated in human breast cancer cells, relative to adjacent stromal cells. For this analysis, we used raw compartment-specific transcriptional profiling data that was obtained by another group, via the laser-capture micro-dissection of epithelial breast cancer cells and adjacent stromal tissue, from n = 28 breast cancer patients.Citation45 Numerous mitochondrial components (including Complexes I–V) were selectively upregulated only in epithelial breast cancer cells, and not in adjacent stromal cells. This allowed us to generate an epithelial cancer cell-specific mitochondrial gene signature that consisted largely of OXPHOS-related genes. Importantly, this OXPHOS signature was upregulated in breast cancer tumor samples (n ≥ 2,000 patients examined, p < 10−20), and was associated with both ER(+) and ER(−) patients. Outcome analysis indicated that this signature was also associated with an increased risk of metastasis, especially in ER(−) breast cancer patients. Three other independent OXPHOS gene signatures were obtained from the MSigDB and KEGG database, and all three OXPHOS signatures were significantly upregulated in human breast cancer tumor tissue (n ≥ 2,000 patients examined), with p values between 10−14 to 10−36. These results directly show that OXPHOS amplification or hyper-activation is clearly an important common characteristic of human breast cancers.
As such, we believe that targeting mitochondrial OXPHOS-related genes has broad implications for both cancer diagnostics and therapeutics, and could be exploited in the pursuit of personalized cancer care.
Materials and Methods
Materials.
Antibodies were obtained as follows: anti-Cav-1 IgG (sc-894, Santa Cruz Biotech); anti-cathepsin B IgG (sc-13985, Santa Cruz Biotech); anti-BNIP3L IgG (ab59908, Abcam); antiMCT4 IgG (sc-50329, Santa Cruz Biotech); anti-TOMM20 IgG (sc-17764, Santa Cruz Biotech); anti-Complex IV mAb (a.k.a., Cytochrome c oxidase subunit 1, Cytochrome c oxidase polypeptide I, MT-CO1, COI, COXI, MTCO1; MS404; MitoSciences, Inc.); anti-Complex II mAb [Succinate dehydrogenase (ubiquinone) iron-sulfur subunit (SDH-B); MS203; MitoSciences, Inc.]. Also, anti-MCT4 isoform-specific antibodies were previously generated and characterized by Dr. Nancy Philip.Citation70 For the NADH, SDH and COX activity staining, validation studies were performed with electron transport inhibitors. More specifically, individual inhibitors were added to the incubation mixture at a final concentration of 1 mM sodium azide (Comple IV inhibitor; BP922-500, Thermo-Fisher), 10 mM sodium malonate (Complex II inhibitor; M1875, Sigma-Aldrich) or 1 mM metformin (Complex I inhibitor; 1,1-dimethylbiguanide hydrochloride, D150959, Sigma-Aldrich).
Immunohistochemistry.
To assess the potential clinical relevance of our findings regarding the cell-type specific compartmentalization of autophagy, mitophagy and aerobic glycolysis, we analyzed human breast cancer tumor samples. For this purpose, we selected cases in which there was a loss of stromal caveolin-1 (Cav-1), an established biomarker for oxidative stress, DNA damage, autophagy and inflammation in the tumor stroma.Citation71 Importantly, loss of stromal Cav-1 is associated with poor clinical outcome in the most common epithelial subtypes of breast cancer [ER(+), PR(+), HER2(+) and triple negative/basal].Citation72–Citation79
Paraffin-embedded sections of human breast cancer samples were immuno-stained as previously described in references Citation1 and Citation22. Briefly, sections were de-paraffinized, rehydrated and washed in PBS. Antigen retrieval was performed in 10 mM sodium citrate, pH 6.0 for 10 min using a pressure cooker. After blocking with 3% hydrogen peroxide for 10 min, sections were incubated with 10% goat serum for 1 hour. Then, sections were incubated with primary antibodies overnight at 4°C. Antibody binding was detected using a biotinylated secondary (Vector Labs) followed by strepavidin-HRP (Dako). Immunoreactivity was revealed using 3,3′-diaminobenzidine.
Cytochrome C oxidase (COX)/complex IV activity staining.
Cryostat sections (7 or 10 µm) were prepared from human breast carcinoma samples and stored at −80°C until use. For the COX activity staining, frozen sections were brought to room temperature, washed for 5 min with 25 mM sodium phosphate buffer, pH 7.4 and then incubated for 0.5, 1 or 2 hours at 37°C with the COX incubation mixture.Citation9,Citation80,Citation81 The COX solution consisted of 10 mg Cytochrome C (C7752, Sigma-Aldrich), 10 mg 3,3′-diaminobenzidine tetrahydrochloride hydrate (D5637, Sigma-Aldrich) and 2 mg catalase (C1345, Sigma-Aldrich) dissolved in 10 ml of 25 mM sodium phosphate buffer. The solution was filtered after preparation and the pH was adjusted to 7.2–7.4 with 1 N NaOH. Cryostat sections obtained from normal mouse skeletal muscle (hind-limb/gastrocnemius) were used as a positive control.
NADH dehydrogenase/complex I activity staining.
For the NADH mitochondrial assay, frozen sections were brought to room temperature, washed for 5 min with 50 mM TRIS-HCl buffer, pH 7.4 and then incubated for 90 min at 37°C with the NADH incubation mixture.Citation9 The NADH solution consisted of 2 mg β-nicotinamide adenine dinucelotide (N7410, Sigma-Aldrich) dissolved in 1 ml of 77 mM TRIS-HCl buffer, pH 7.4 containing 38 mM cobalt (II) chloride (C8661, Sigma-Aldrich) and 380 µg nitrotetrazolium blue chloride (N6876, Sigma-Aldrich).
Succinate dehydrogenase (SDH)/complex II activity staining.
For the SDH mitochondrial assay, frozen sections were brought to room temperature, washed for 5 min with 50 mM sodium phosphate buffer, pH 7.4 and then incubated for 30, 60 or 90 min at 37°C with the SDH incubation mixture.Citation9,Citation80,Citation82 The SDH solution consisted of 50 mM succinic acid (S3674, Sigma-Aldrich), 0.2 mM phenazine methosulfate (P9625, Sigma-Aldrich), 5 mM EDTA, 1 mM potassium cyanide (60178, Sigma-Aldrich) and 1.5 mM nitrotetrazolium blue chloride (N6876, Sigma-Aldrich) in 50 mM sodium phosphate buffer pH 7.4–7.6. The competitive inhibitor sodium malonate (M1875, Sigma-Aldrich) was used at a 10 mM final concentration to verify the specificity of the reaction.
Bioinformatics analysis of >2,000 human breast cancer samples.
A microarray data set that was previously compiled from the public repositories Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo)Citation83 and ArrayExpress (www.ebi.ac.uk/arrayexpress)Citation84 was used to evaluate the combined expression signature of 38 mitochondria- and oxidative phosphorylation-associated genes in the context of clinical samples.Citation85 This clinical data set includes 102 healthy breast tissue samples and 2,152 breast cancer cases, of which 1,674 were identified as ER-positive and 478 were identified as ER-negative, based on ESR1 mRNA expression.Citation85 Samples were analyzed in separate groups based on ER status. To analyze the combined 38-gene MITO/OXPHOS signature in the micro-array data set, a signature magnitude was computed by median-centering each gene and averaging their expression in each sample. Differential expression of the averaged gene signature magnitude between sample groups was evaluated using a two-tailed t-test. Kaplan-Meier analysis was used to evaluate survival trends among 831 samples annotated with metastasis-free survival time (684 ER-pos., 147 ER-neg.). X-Tile software was employed to identify subpopulation cut-points to observe maximum survival differences between the high expression and low expression subpopulations.Citation86 The Log-rank test was used to evaluate the significance of differences in survival curves for high vs. low signature-expressing populations.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Figures and Tables
Figure 1 TOMM20, a mitochondrial marker protein, is preferentially expressed in human epithelial cancer cells, in breast cancer patients. Paraffin-embedded sections of human breast cancer samples lacking stromal Cav-1 were immuno-stained with antibodies directed against TOMM20 (brown color). Slides were then counter-stained with hematoxylin (blue color). Note that TOMM20 is highly expressed in the epithelial compartment of human breast cancers that lack stromal Cav-1. Two representative images are shown. Original magnification, 40x and 60x, as indicated.
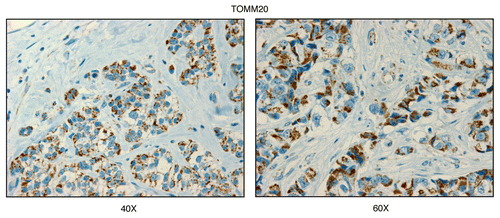
Figure 2 Cathepsin B, a lysosomal protease and autophagy marker, is selectively increased in the stroma of human breast cancers. Paraffin-embedded sections of human breast cancer samples lacking stromal Cav-1 were immuno-stained with antibodies directed against Cathepsin B. Slides were then counter-stained with hematoxylin. Note that Cathepsin B is highly expressed in the stromal compartment of human breast cancers that lack stromal Cav-1. Two representative images are shown. Original magnification, 40x and 60x, as indicated.
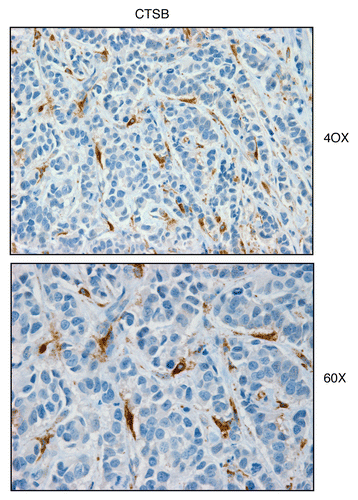
Figure 3 BNIP3L, a mitophagy marker, is elevated in the stroma of human breast cancers. Paraffin-embedded sections of human breast cancer samples lacking stromal Cav-1 were immuno-stained with antibodies directed against BNIP3L. Slides were then counter-stained with hematoxylin. Note that BNIP3L is highly expressed in the stromal compartment of human breast cancers that lack stromal Cav-1. Two representative images are shown. Original magnification, 40x and 60x, as indicated.
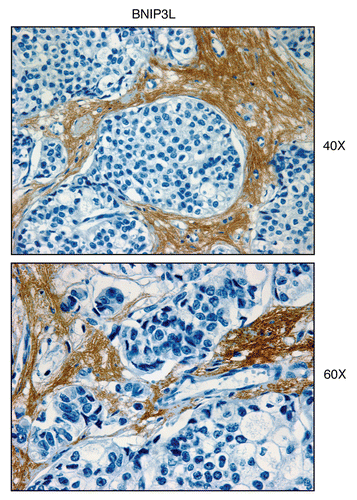
Figure 4 MCT4, a marker of aerobic glycolysis and lactate efflux, is elevated in the stroma of human breast cancers. Paraffin-embedded sections of human breast cancer samples lacking stromal Cav-1 were immuno-stained with antibodies directed against MCT4. Slides were then counter-stained with hematoxylin. Note that MCT4 is highly expressed in the stromal compartment of human breast cancers that lack stromal Cav-1. Three representative images are shown. Original magnification, 40x and 60x, as indicated.
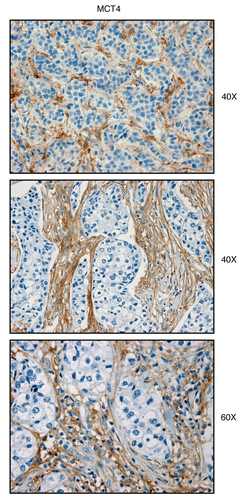
Figure 5 Visualizing mitochondrial complex IV (COX) activity in murine skeletal muscle tissue. Frozen sections of murine skeletal muscle (hind-limb/gastronemius) were subjected to COX activity staining (brown color). Slides were then counter-stained with hematoxylin (blue color). Note that slow-twitch fibers (type I) are oxidative, are mitochondria-rich, and are COX-positive (see red arrows). In contrast, fast-twitch fibers (type II) are glycolytic, are mitochondria-poor, and are COX-negative. Two representative images are shown. Original magnification, 20x and 40x, as indicated.
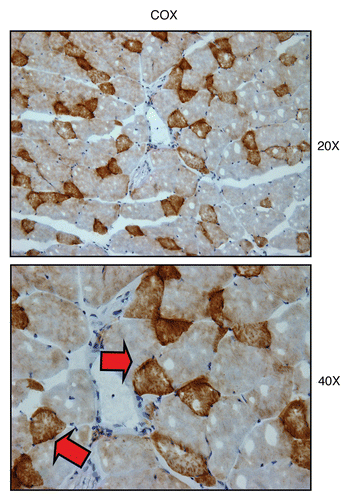
Figure 6 Mitochondrial complex IV (COX) activity is amplified in human epithelial cancer cells, in breast cancer patients (low-power images). Frozen sections of human breast cancer samples were subjected to COX activity staining (brown color). Slides were then counter-stained with hematoxylin (blue color). Note that human epithelial tumor cells are intensely stained, as compared with adjacent stromal cells, which show little or no COX activity, in comparison. Original magnification, 20x, as indicated.
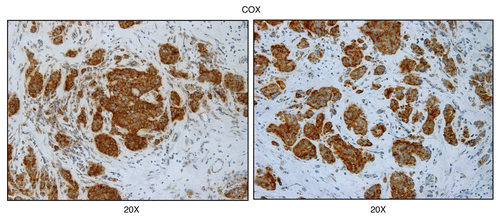
Figure 7 Mitochondrial complex IV (COX) activity is amplified in human epithelial cancer cells, in breast cancer patients (higher-power images). As in , except higher power images are shown. Note that human epithelial cancer cell “nests” are intensely stained, as compared with adjacent stromal cells. Original magnification, 40x, as indicated.
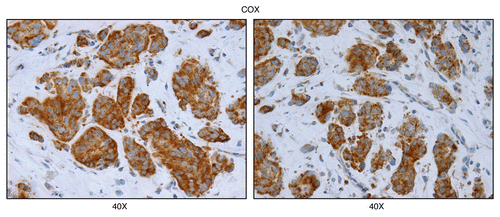
Figure 8 COX activity is higher in human epithelial cancer cells, relative to normal adjacent epithelial ductal cells. Frozen sections of human breast cancer samples were subjected to COX activity staining (brown color). Slides were then counter-stained with hematoxylin (blue color). Note that epithelial cancer cells were also more intensely stained than adjacent normal epithelial cells that were part of mammary ductal tissue. Thus, COX activity staining can be used to distinguish normal epithelial cells from adjacent cancer cells, within the same tumor tissue. Original magnification, 20x and 40x, as indicated.
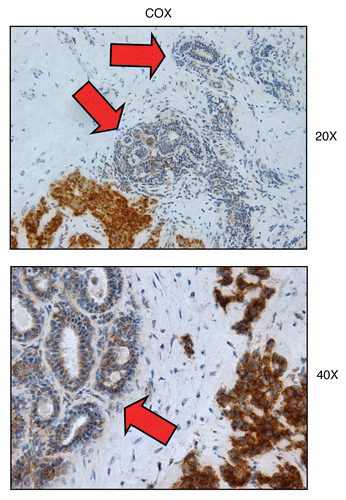
Figure 9 Mitochondrial poisons (azide and metformin) validate the specificity of COX activity staining in human epithelial cancer cells. Serial frozen sections of human breast cancer samples were subjected to COX activity staining (brown color). Slides were then counter-stained with hematoxylin (blue color). Note that sodium azide (1 mM; a Complex IV inhibitor) effectively abolished the COX activity staining, directly demonstrating high-specificity. Treatment with metformin (1 mM; a Complex I inhibitor) also significantly reduced the COX activity staining (Complex IV).
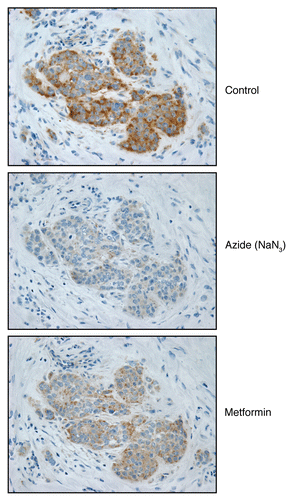
Figure 10 Visualizing mitochondrial complex I (NADH) activity in murine skeletal muscle tissue. Frozen sections of murine skeletal muscle (hind-limb/gastronemius) were subjected to NADH activity staining (blue color). Slides were then counter-stained with nuclear fast-red (pink color). Note that slow-twitch fibers (type I) are oxidative, are mitochondria-rich, and are NADH-positive (see red arrows). In contrast, fast-twitch fibers (type II) are glycolytic, are mitochondria-poor, and are NADH-negative. Two representative images are shown. Original magnification, 20x and 40x, as indicated.
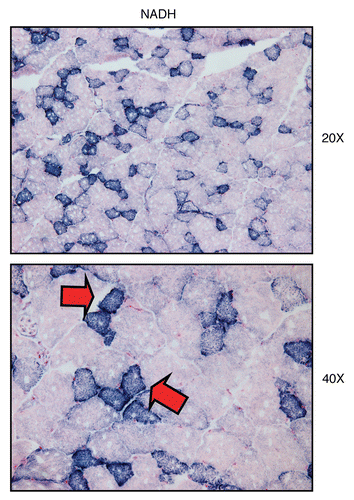
Figure 11 Mitochondrial complex I (NADH) activity is amplified in human epithelial cancer cells, in breast cancer patients. Frozen sections of human breast cancer samples were subjected to NADH activity staining (blue color). However, slides were not counter-stained with nuclear fast-red (pink color), and were mounted without counter-staining, to better visualize the blue reaction product. Note that human epithelial tumor cells are intensely stained dark blue (see red arrows), as compared with adjacent stromal cells, which show little or no NADH activity, in comparison. Original magnification, 40x, as indicated.
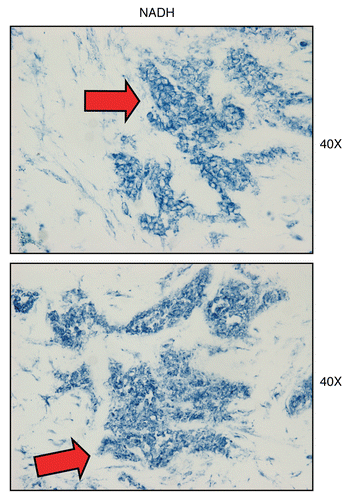
Figure 12 Treatment with metformin (a complex I inhibitor) validates the specificity of NADH activity staining in human epithelial cancer cells. Frozen sections of human breast cancer samples were subjected to NADH activity staining (blue color) in the presence or absence of metformin. However, slides were not counter-stained with nuclear fast-red (pink color), and were mounted without counter-staining, to better visualize the blue reaction product. Note that meformin (1 mM; a Complex I inhibitor) effectively abolished the NADH activity staining, directly demonstrating high-specificity. Original magnification, 40x.
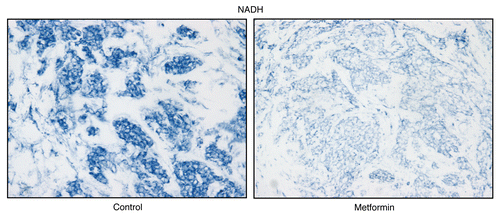
Figure 13 Visualizing mitochondrial complex II (SDH) activity in murine skeletal muscle tissue. Frozen sections of murine skeletal muscle (hind-limb/gastronemius) were subjected to SDH activity staining (blue color). Note that slow-twitch fibers (type I) are oxidative, are mitochondria-rich, and are SDH-positive (see red arrows). In contrast, fast-twitch fibers (type II) are glycolytic, are mitochondria-poor, and are SDH-negative. Two representative images are shown. Original magnification, 20x and 40x, as indicated.
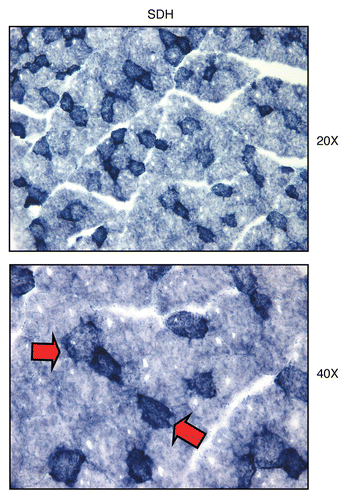
Figure 14 Mitochondrial complex II (SDH) activity is amplified in human epithelial cancer cells, in breast cancer patients. Frozen sections of human breast cancer samples were subjected to SDH activity staining (blue color). However, slides were not counter-stained. Note that human epithelial tumor cells are intensely stained dark blue, as compared with adjacent stromal cells, which show little or no SDH activity, in comparison. Original magnification, 40x, as indicated.
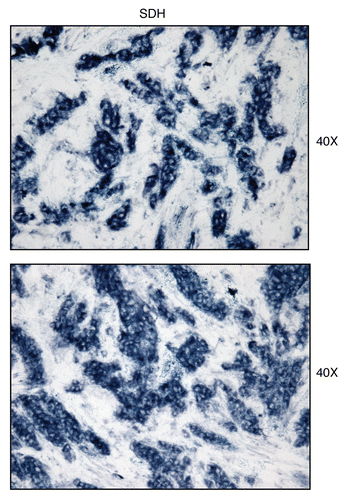
Figure 15 Treatment with malonate (a complex II inhibitor) validates the specificity of SDH activity staining in human epithelial cancer cells. Frozen sections of human breast cancer samples were subjected to SDH activity staining (blue color) in the presence or absence of malonate. However, slides were not counter-stained. Note that sodium malonate (10 mM; a Complex II inhibitor) effectively abolished the SDH activity staining, directly demonstrating high-specificity. Original magnification, 40x.
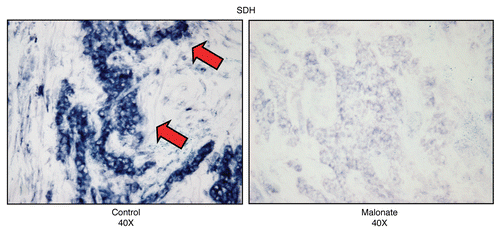
Figure 16 Complex IV (MT-CO1), a component of the respiratory chain, is preferentially expressed in human epithelial cancer cells, in breast cancer patients. Paraffin-embedded sections of human breast cancer samples lacking stromal Cav-1 were immuno-stained with antibodies directed against Complex IV (MT-CO1) (brown color). Slides were then counter-stained with hematoxylin (blue color). Note that Complex IV is highly expressed in the epithelial compartment of human breast cancers that lack stromal Cav-1. Two representative images are shown. Original magnification, 40x, as indicated.
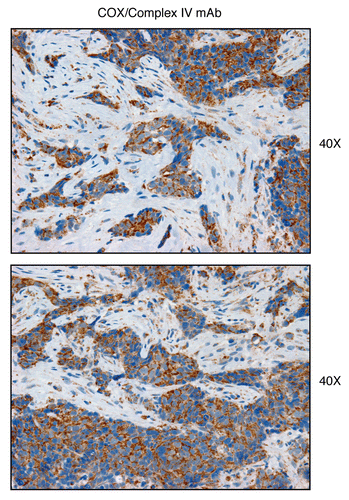
Figure 17 Complex II (SDH-B), a component of the respiratory chain, is preferentially expressed in human epithelial cancer cells, in breast cancer patients. Paraffin-embedded sections of human breast cancer samples lacking stromal Cav-1 were immuno-stained with antibodies directed against Complex II (SDH-B) (brown color). Slides were then counter-stained with hematoxylin (blue color). Note that Complex II is highly expressed in the epithelial compartment of human breast cancers that lack stromal Cav-1. Two representative images are shown. Original magnification, 40x, as indicated.
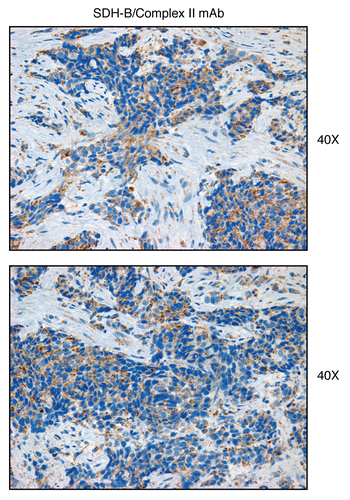
Figure 18 An epithelial-specific mitochondrial gene signature predicts metastasis in breast cancer patients. We examined whether a 38-member MITO/OXPHOS gene signature (see for the complete list) is associated with the transcriptional profiles of human breast cancer tumor tissue. This signature contains mitochondrial associated gene transcripts specifically upregulated only in epithelial breast cancer cells, and not in adjacent stromal tissue. (A) Box Plot. Note that this epithelial-specific mitochondrial gene signature is upregulated in human breast tumors (>2,000 cases examined, with a p value < 10−20), relative to normal healthy breast tissue (102 controls). In addition, significant associations were obtained with tumor tissue from both ER-positive (>1,600 cases examined, p < 10−20) and ER-negative (nearly 500 cases examined, p < 10−10) breast cancer patients. Thus, upregulation of this MITO/OXPHOS gene signature is a general feature of human breast cancers. (B) Kaplan-Meier Analysis. Note that this mitochondrial-based epithelial gene signature also predicts breast cancer cell metastasis, and especially in ER-negative patients. In this patient group, the MITO/OXPHOS signature was associated with a near 30% increase is metastasis, over a 10-year follow-up period. Numbers of cases with annotation are shown.
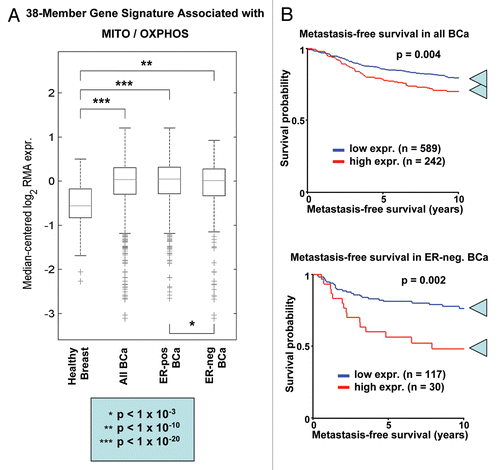
Figure 19 Energy transfer in cancer metabolism. We have recently proposed a new paradigm for understanding energy transfer in cancer metabolism. In this model, cancer cells behave as metabolic parasites, by extracting nutrients from normal host cells, such as fibroblasts, by secreting hydrogen peroxide. Oxidative stress in the tumor microenvironment leads to autophagy, mitophagy and aerobic glycolysis. This, in turn, produces high-energy nutrients (such as L-lactate, ketones and glutamine) that fuel the anabolic growth of tumor cells, via oxidative mitochondrial metabolism (OXPHOS ). Thus, cancer cells and the tumor stroma are metabolically coupled in a type of symbiotic or host-parasite relationship. In accordance with this model, here we provide evidence that cancer-associated fibroblasts undergo autophagy, mitophagy and secrete lactate, while epithelial cancer cells are rich in functional mitochondria.
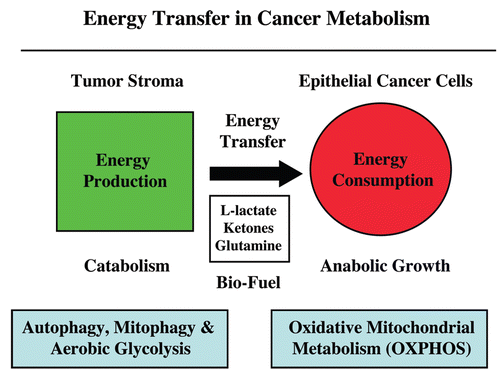
Table 1 An epithelial-specific mitochondrial gene signature in breast cancer
Acknowledgments
F.S. and M.P.L. wish to thank Dr. Salvatore (Billi) DiMauro (Columbia University, NY) for his friendship, valuable mentorship and inspiration to venture into the mitochondrial field. We also thank Dr. Nancy J. Philip (Thomas Jefferson University, PA) for her generous gift of MCT4-specific antibodies. F.S. and her laboratory were supported by grants from the Breast Cancer Alliance (BCA) and the American Cancer Society (ACS). U.E.M. was supported by a Young Investigator Award from the Margaret Q. Landenberger Research Foundation. M.P.L. was supported by grants from the NIH/NCI (R01-CA-080250; R01-CA-098779; R01-CA-120876; R01-AR-055660) and the Susan G. Komen Breast Cancer Foundation. A.K.W. was supported by a Susan G. Komen Career Catalyst Grant. R.G.P. was supported by grants from the NIH/NCI (R01-CA-70896, R01-CA-75503, R01-CA-86072 and R01-CA-107382) and the Dr. Ralph and Marian C. Falk Medical Research Trust. The Kimmel Cancer Center was supported by the NIH/NCI Cancer Center Core grant P30-CA-56036 (to R.G.P.). Funds were also contributed by the Margaret Q. Landenberger Research Foundation (to M.P.L.). This project is funded, in part, under a grant with the Pennsylvania Department of Health (to M.P.L. and F.S.). The Department specifically disclaims responsibility for any analyses, interpretations or conclusions. This work was also supported, in part, by a Centre grant in Manchester from Breakthrough Breast Cancer in the UK (to A.H.) and an Advanced ERC Grant from the European Research Council.
References
- Whitaker-Menezes D, Martinez-Outschoorn UE, Lin Z, Ertel A, Flomenberg N, Witkiewicz AK, et al. Evidence for a stromal-epithelial “lactate shuttle” in human tumors: MCT4 is a marker of oxidative stress in cancer-associated fibroblasts. Cell Cycle 2011; 10:1772 - 1783; PMID: 21558814; http://dx.doi.org/10.4161/cc.10.11.15659
- Sotgia F, Martinez-Outschoorn UE, Pavlides S, Howell A, Pestell RG, Lisanti MP. Understanding the Warburg effect and the prognostic value of stromal caveolin-1 as a marker of a lethal tumor microenvironment. Breast Cancer Res 2011; 13:213; PMID: 21867571; http://dx.doi.org/10.1186/bcr2892
- Martinez-Outschoorn UE, Pavlides S, Howell A, Pestell RG, Tanowitz HB, Sotgia F, et al. Stromalepithelial metabolic coupling in cancer: integrating autophagy and metabolism in the tumor microenvironment. Int J Biochem Cell Biol 2011; 43:1045 - 1051; PMID: 21300172; http://dx.doi.org/10.1016/j.biocel.2011.01.023
- Pavlides S, Vera I, Gandara R, Sneddon S, Pestell R, Mercier I, et al. Warburg Meets Autophagy: Cancer Associated Fibroblasts Accelerate Tumor Growth and Metastasis Via Oxidative Stress, Mitophagy and Aerobic Glycolysis. Antioxid Redox Signal 2011; In press PMID: 21883043; http://dx.doi.org/10.1089/ars.2011.4243
- Brooks GA. Current concepts in lactate exchange. Med Sci Sports Exerc 1991; 23:895 - 906; PMID: 1956262; http://dx.doi.org/10.1249/00005768-199108000-00003
- Brooks GA. Lactate shuttles in nature. Biochem Soc Trans 2002; 30:258 - 264; PMID: 12023861; http://dx.doi.org/10.1042/BST0300258
- Brooks GA. Lactate: link between glycolytic and oxidative metabolism. Sports Med 2007; 37:341 - 343; PMID: 17465603; http://dx.doi.org/10.2165/00007256200737040-00017
- Gladden LB. A lactatic perspective on metabolism. Med Sci Sports Exerc 2008; 40:477 - 485; PMID: 18379210; http://dx.doi.org/10.1249/MSS.0b013e31815fa580
- Tritschler HJ, Bonilla E, Lombes A, Andreetta F, Servidei S, Schneyder B, et al. Differential diagnosis of fatal and benign cytochrome c oxidase-deficient myopathies of infancy: an immunohistochemical approach. Neurology 1991; 41:300 - 305; PMID: 1846953
- Lombes A, Nakase H, Tritschler HJ, Kadenbach B, Bonilla E, DeVivo DC, et al. Biochemical and molecular analysis of cytochrome c oxidase deficiency in Leigh's syndrome. Neurology 1991; 41:491 - 498; PMID: 1849240
- DiMauro S, Lombes A, Nakase H, Mita S, Fabrizi GM, Tritschler HJ, et al. Cytochrome c oxidase deficiency. Pediatr Res 1990; 28:536 - 541; PMID: 2175026; http://dx.doi.org/10.1203/00006450-199011000-00025
- DiMauro S. Pathogenesis and treatment of mitochondrial myopathies: recent advances. Acta Myol 2010; 29:333 - 338; PMID: 21314015
- DiMauro S, Garone C. Historical perspective on mitochondrial medicine. Dev Disabil Res Rev 2010; 16:106 - 113; PMID: 20818724; http://dx.doi.org/10.1002/ddrr.102
- DiMauro S, Garone C, Naini A. Metabolic myopathies. Curr Rheumatol Rep 2010; 12:386 - 393; PMID: 20676808; http://dx.doi.org/10.1007/s11926-010-0119-9
- DiMauro S, Hirano M. Pathogenesis and treatment of mitochondrial disorders. Adv Exp Med Biol 2009; 652:139 - 170; PMID: 20225024; http://dx.doi.org/10.1007/978-90481-2813-6_10
- Dimauro S, Rustin P. A critical approach to the therapy of mitochondrial respiratory chain and oxidative phosphorylation diseases. Biochim Biophys Acta 2009; 1792:1159 - 1167; PMID: 19026744
- Seligman AM, Karnovsky MJ, Wasserkrug HL, Hanker JS. Nondroplet ultrastructural demonstration of cytochrome oxidase activity with a polymerizing osmiophilic reagent, diaminobenzidine (DAB). J Cell Biol 1968; 38:1 - 14; PMID: 4300067; http://dx.doi.org/10.1083/jcb.38.1.1
- Pavlides S, Whitaker-Menezes D, Castello-Cros R, Flomenberg N, Witkiewicz AK, Frank PG, et al. The reverse Warburg effect: Aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle 2009; 8:3984 - 4001; PMID: 19923890; http://dx.doi.org/10.4161/cc.8.23.10238
- Pavlides S, Tsirigos A, Migneco G, Whitaker-Menezes D, Chiavarina B, Flomenberg N, et al. The Autophagic Tumor Stroma Model of Cancer: Role of Oxidative Stress and Ketone Production in Fueling Tumor Cell Metabolism. Cell Cycle 2010; 9:3485 - 3505; PMID: 20861672; http://dx.doi.org/10.4161/cc.9.17.12721
- Martinez-Outschoorn UE, Pavlides S, Whitaker-Menezes D, Daumer KM, Milliman JN, Chiavarina B, et al. Tumor Cells Induce the Cancer Associated Fibroblast Phenotype Via Caveolin-1 Degradation: Implications for Breast Cancer and DCIS Therapy with Autophagy Inhibitors. Cell Cycle 2010; 9:2423 - 2433; PMID: 20562526; http://dx.doi.org/10.4161/cc.9.12.12048
- Martinez-Outschoorn UE, Balliet RM, Rivadeneira DB, Chiavarina B, Pavlides S, Wang C, et al. Oxidative Stress in Cancer Associated Fibroblasts Drives Tumor-Stroma Co-Evolution: A New Paradigm for Understanding Tumor Metabolism, the Field Effect and Genomic Instability in Cancer Cells. Cell Cycle 2010; 9:3256 - 3276; PMID: 20814239; http://dx.doi.org/10.4161/cc.9.16.12553
- Martinez-Outschoorn UE, Trimmer C, Lin Z, Whitaker-Menezes D, Chiavarina B, Zhou J, et al. Autophagy in Cancer Associated Fibroblasts Promotes Tumor Cell Survival: Role of Hypoxia, HIF1 Induction and NFκB Activation in the Tumor Stromal Microenvironment. Cell Cycle 2010; 9:3515 - 3533; PMID: 20855962; http://dx.doi.org/10.4161/cc.9.17.12928
- Chiavarina B, Whitaker-Menezes D, Migneco G, Martinez-Outschoorn UE, Pavlides S, Howell A, et al. HIF-1alpha Functions as a Tumor Promoter in Cancer Associated Fibroblasts, and as a Tumor Suppressor in Breast Cancer Cells: Autophagy Drives Compartment-Specific Oncogenesis. Cell Cycle 2010; 9:3534 - 3551; PMID: 20864819; http://dx.doi.org/10.4161/cc.9.17.12908
- Trimmer C, Sotgia F, Whitaker-Menezes D, Balliet RM, Eaton G, Martinez-Outschoorn UE, et al. Caveolin-1 and mitochondrial SOD2 (MnSOD) function as tumor suppressors in the stromal microenvironment: a new genetically tractable model for human cancer associated fibroblasts. Cancer Biol Ther 2011; 11:383 - 394; PMID: 21150282; http://dx.doi.org/10.4161/cbt.11.4.14101
- Brunmair B, Staniek K, Gras F, Scharf N, Althaym A, Clara R, et al. Thiazolidinediones, like metformin, inhibit respiratory complex I: a common mechanism contributing to their antidiabetic actions?. Diabetes 2004; 53:1052 - 1059; PMID: 15047621; http://dx.doi.org/10.2337/diabetes.53.4.1052
- Detaille D, Guigas B, Leverve X, Wiernsperger N, Devos P. Obligatory role of membrane events in the regulatory effect of metformin on the respiratory chain function. Biochem Pharmacol 2002; 63:1259 - 1272; PMID: 11960602; http://dx.doi.org/10.1016/S00062952(02)00858-4
- El-Mir MY, Nogueira V, Fontaine E, Averet N, Rigoulet M, Leverve X. Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. J Biol Chem 2000; 275:223 - 228; PMID: 10617608; http://dx.doi.org/10.1074/jbc.275.1.223
- Owen MR, Doran E, Halestrap AP. Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem J 2000; 348:607 - 614; PMID: 10839993; http://dx.doi.org/10.1042/0264-6021:3480607
- Biradar V, Moran JL, Peake SL, Peter JV. Metforminassociated lactic acidosis (MALA): clinical profile and outcomes in patients admitted to the intensive care unit. Crit Care Resusc 2010; 12:191 - 195; PMID: 21261578
- Dykens JA, Jamieson J, Marroquin L, Nadanaciva S, Billis PA, Will Y. Biguanide-induced mitochondrial dysfunction yields increased lactate production and cytotoxicity of aerobically-poised HepG2 cells and human hepatocytes in vitro. Toxicol Appl Pharmacol 2008; 233:203 - 210; PMID: 18817800; http://dx.doi.org/10.1016/j.taap.2008.08.013
- Hirsch HA, Iliopoulos D, Tsichlis PN, Struhl K. Metformin selectively targets cancer stem cells, and acts together with chemotherapy to block tumor growth and prolong remission. Cancer Res 2009; 69:7507 - 7511; PMID: 19752085; http://dx.doi.org/10.1158/0008-5472.CAN09-2994
- Memmott RM, Mercado JR, Maier CR, Kawabata S, Fox SD, Dennis PA. Metformi prevents tobacco carcinogen-induced lung tumorigenesis. Cancer Prev Res (Phila) 2010; 3:1066 - 1076; PMID: 20810672; http://dx.doi.org/10.1158/1940-6207.CAPR-10-0055
- Pollak M. Metformin and other biguanides in oncology: advancing the research agenda. Cancer Prev Res (Phila) 2010; 3:1060 - 1065; PMID: 20810670; http://dx.doi.org/10.1158/1940-6207.CAPR-10-0175
- Vazquez-Martin A, Oliveras-Ferraros C, Barco SD, Martin-Castillo B, Menendez JA. The anti-diabetic drug metformin suppresses self-renewal and proliferation of trastuzumab-resistant tumor-initiating breast cancer stem cells. Breast Cancer Res Treat 2011; 126:355 - 364; PMID: 20458531; http://dx.doi.org/10.1007/s10549010-0924-x
- Vazquez-Martin A, Oliveras-Ferraros C, Cufi S, Del Barco S, Martin-Castillo B, Menendez JA. Metformin regulates breast cancer stem cell ontogeny by transcriptional regulation of the epithelial-mesenchymal transition (EMT) status. Cell Cycle 2010; 9:3807 - 3814; PMID: 20890129; http://dx.doi.org/10.4161/cc.9.18.13131
- Vazquez-Martin A, Oliveras-Ferraros C, Cufi S, Martin-Castillo B, Menendez JA. Metformin and energy metabolism in breast cancer: from insulin physiology to tumour-initiating stem cells. Curr Mol Med 2010; 10:674 - 691; PMID: 20712585; http://dx.doi.org/10.2174/156652410792630625
- Pavlides S, Tsirigos A, Vera I, Flomenberg N, Frank PG, Casimiro MC, et al. Transcriptional evidence for the “Reverse Warburg Effect” in human breast cancer tumor stroma and metastasis: similarities with oxidative stress, inflammation, Alzheimer's disease and “Neuron-Glia Metabolic Coupling”. Aging (Albany NY) 2010; 2:185 - 199; PMID: 20442453
- Pavlides S, Tsirigos A, Vera I, Flomenberg N, Frank PG, Casimiro MC, et al. Loss of Stromal Caveolin-1 Leads to Oxidative Stress, Mimics Hypoxia and Drives Inflammation in the Tumor Microenvironment, Conferring the “Reverse Warburg Effect”: A Transcriptional Informatics Analysis with Validation. Cell Cycle 2010; 9:2201 - 2219; PMID: 20519932; http://dx.doi.org/10.4161/cc.9.11.11848
- Martinez-Outschoorn UE, Whitaker-Menezes D, Pavlides S, Chiavarina B, Bonuccelli G, Casey T, et al. The Autophagic Tumor Stroma Model of Cancer or “Battery-Operated Tumor Growth”: A Simple Solution to the Autophagy Paradox. Cell Cycle 2010; 9:4297 - 4306; PMID: 21051947; http://dx.doi.org/10.4161/cc.9.21.13817
- Clark JB, Hayes DJ, Morgan-Hughes JA, Byrne E. Mitochondrial myopathies: disorders of the respiratory chain and oxidative phosphorylation. J Inherit Metab Dis 1984; 7:62 - 68; PMID: 6434847; http://dx.doi.org/10.1007/BF03047377
- Sengers RC, Stadhouders AM, Trijbels JM. Mitochondrial myopathies. Clinical, morphological and biochemical aspects. Eur J Pediatr 1984; 141:192 - 207; PMID: 6329761; http://dx.doi.org/10.1007/BF00572761
- Leverve XM, Guigas B, Detaille D, Batandier C, Koceir EA, Chauvin C, et al. Mitochondrial metabolism and type-2 diabetes: a specific target of metformin. Diabetes Metab 2003; 29:88 - 94; PMID: 14502105; http://dx.doi.org/10.1016/S1262-3636(03)72792-X
- Batandier C, Guigas B, Detaille D, El-Mir MY, Fontaine E, Rigoulet M, et al. The ROS production induced by a reverse-electron flux at respiratory-chain complex 1 is hampered by metformin. J Bioenerg Biomembr 2006; 38:33 - 42; PMID: 16732470; http://dx.doi.org/10.1007/s10863-006-9003-8
- Bonuccelli G, Tsirigos A, Whitaker-Menezes D, Pavlides S, Pestell RG, Chiavarina B, et al. Ketones and Lactate “Fuel” Tumor Growth and Metastasis: Evidence that Epithelial Cancer Cells Use Oxidative Mitochondrial Metabolism. Cell Cycle 2010; 9:3506 - 3514; PMID: 20818174; http://dx.doi.org/10.4161/cc.9.17.12731
- Casey T, Bond J, Tighe S, Hunter T, Lintault L, Patel O, et al. Molecular signatures suggest a major role for stromal cells in development of invasive breast cancer. Breast Cancer Res Treat 2009; 114:47 - 62; PMID: 18373191; http://dx.doi.org/10.1007/s10549-008-9982-8
- Berridge MV, Tan AS. Effects of mitochondrial gene deletion on tumorigenicity of metastatic melanoma: reassessing the Warburg effect. Rejuvenation Res 2010; 13:139 - 141; PMID: 20370492; http://dx.doi.org/10.1089/rej.2009.0948
- Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer 2011; 11:325 - 337; PMID: 21508971; http://dx.doi.org/10.1038/nrc3038
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009; 324:1029 - 1033; PMID: 19460998; http://dx.doi.org/10.1126/science.1160809
- Kaelin W Jr, Thompson CB. Q&A: Cancer: clues from cell metabolism. Nature 2010; 465:562 - 564; PMID: 20520704; http://dx.doi.org/10.1038/465562a
- Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science 2010; 330:1340 - 1344; PMID: 21127244; http://dx.doi.org/10.1126/science.1193494
- Migneco G, Whitaker-Menezes D, Chiavarina B, Castello-Cros R, Pavlides S, Pestell RG, et al. Glycolytic cancer associated fibroblasts promote breast cancer tumor growth, without a measurable increase in angiogenesis: Evidence for stromal-epithelial metabolic coupling. Cell Cycle 2010; 9:2412 - 2422; PMID: 20562527; http://dx.doi.org/10.4161/cc.9.12.11989
- Martinez-Outschoorn UE, Prisco M, Ertel A, Tsirigos A, Lin Z, Pavlides S, et al. Ketones and lactate increase cancer cell “stemness,” driving recurrence, metastasis and poor clinical outcome in breast cancer: achieving personalized medicine via Metabolo-Genomics. Cell Cycle 2011; 10:1271 - 1286; PMID: 21512313; http://dx.doi.org/10.4161/cc.10.8.15330
- Martinez-Outschoorn UE, Goldberg AF, Lin Z, Ko YH, Flomenberg N, Wang C, et al. Anti-estrogen resistance in breast cancer is induced by the tumor microenvironment and can be overcome by inhibiting mitochondrial function in epithelial cancer cells. Cancer Biol Ther 2011; 12:924 - 938
- Reich NC. STAT3 revs up the powerhouse. Sci Signal 2009; 2:61; PMID: 19797267; http://dx.doi.org/10.1126/scisignal.290pe61
- Gough DJ, Corlett A, Schlessinger K, Wegrzyn J, Larner AC, Levy DE. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science 2009; 324:1713 - 1716; PMID: 19556508; http://dx.doi.org/10.1126/science.1171721
- Wegrzyn J, Potla R, Chwae YJ, Sepuri NB, Zhang Q, Koeck T, et al. Function of mitochondrial Stat3 in cellular respiration. Science 2009; 323:793 - 797; PMID: 19131594; http://dx.doi.org/10.1126/science.1164551
- Spees JL, Olson SD, Whitney MJ, Prockop DJ. Mitochondrial transfer between cells can rescue aerobic respiration. Proc Natl Acad Sci USA 2006; 103:1283 - 1288; PMID: 16432190; http://dx.doi.org/10.1073/pnas.0510511103
- Rebbeck CA, Leroi AM, Burt A. Mitochondrial capture by a transmissible cancer. Science 2011; 331:303; PMID: 21252340; http://dx.doi.org/10.1126/science.1197696
- Villanueva T. Metabolism: the mitochondria that wag the dog. Nat Rev Cancer 2011; 11:155; PMID: 21451550; http://dx.doi.org/10.1038/nrc3026
- Berridge MV, Tan AS. Mitochondrial Gene Transfer to Transplantable Tumours Lacking a Mitochondrial Genome. Rejuvenation Res 2011; 14:13 - 14
- Chen EI, Hewel J, Krueger JS, Tiraby C, Weber MR, Kralli A, et al. Adaptation of energy metabolism in breast cancer brain metastases. Cancer Res 2007; 67:1472 - 1486; PMID: 17308085; http://dx.doi.org/10.1158/00085472.CAN-06-3137
- Wyss MT, Jolivet R, Buck A, Magistretti PJ, Weber B. In vivo evidence for lactate as a neuronal energy source. J Neurosci 2011; 31:7477 - 7485; PMID: 21593331; http://dx.doi.org/10.1523/JNEUROSCI.0415-11.2011
- Tsuda H, Seki K, Hasebe T, Sasajima Y, Shibata T, Iwamoto E, et al. A histopathological study for evaluation of therapeutic effects of radiofrequency ablation in patients with breast cancer. Breast Cancer 2011; 18:24 - 32; PMID: 20862572; http://dx.doi.org/10.1007/s12282010-0222-9
- Rohlena J, Dong LF, Ralph SJ, Neuzil J. Anticancer Drugs Targeting the Mitochondrial Electron Transport Chain. Antioxid Redox Signal 2011; 15:2951 - 2974; PMID: 21777145; http://dx.doi.org/10.1089/ars.2011.3990
- Siegelin MD, Dohi T, Raskett CM, Orlowski GM, Powers CM, Gilbert CA, et al. Exploiting the mitochondrial unfolded protein response for cancer therapy in mice and human cells. J Clin Invest 2011; 121:1349 - 1360; PMID: 21364280; http://dx.doi.org/10.1172/JCI44855
- Fulda S, Galluzzi L, Kroemer G. Targeting mitochondria for cancer therapy. Nat Rev Drug Discov 2010; 9:447 - 464; PMID: 20467424; http://dx.doi.org/10.1038/nrd3137
- Kousparou CA, Epenetos AA, Deonarain MP. Antibody-guided enzyme therapy of cancer producing cyanide results in necrosis of targeted cells. Int J Cancer 2002; 99:138 - 148; PMID: 11948505; http://dx.doi.org/10.1002/ijc.10266
- Gargini R, Garcia-Escudero V, Izquierdo M. Therapy mediated by mitophagy abrogates tumor progression. Autophagy 2011; 7:466 - 476; PMID: 21270513; http://dx.doi.org/10.4161/auto.7.5.14731
- García-Escudero V, Gargini R. Autophagy induction as an efficient strategy to eradicate tumors. Autophagy 2008; 4:923 - 925; PMID: 18716458
- Gallagher SM, Castorino JJ, Wang D, Philp NJ. Monocarboxylate transporter 4 regulates maturation and trafficking of CD147 to the plasma membrane in the metastatic breast cancer cell line MDA-MB-231. Cancer Res 2007; 67:4182 - 4189; PMID: 17483329; http://dx.doi.org/10.1158/0008-5472.CAN-06-3184
- Witkiewicz AK, Kline J, Queenan M, Brody JR, Tsirigos A, Bilal E, et al. Molecular profiling of a lethal tumor microenvironment, as defined by stromal caveolin-1 status in breast cancers. Cell Cycle 2011; 10:1794 - 1809; PMID: 21521946; http://dx.doi.org/10.4161/cc.10.11.15675
- Witkiewicz AK, Dasgupta A, Sotgia F, Mercier I, Pestell RG, Sabel M, et al. An Absence of Stromal Caveolin-1 Expression Predicts Early Tumor Recurrence and Poor Clinical Outcome in Human Breast Cancers. Am J Pathol 2009; 174:2023 - 2034; PMID: 19411448; http://dx.doi.org/10.2353/ajpath.2009.080873
- Witkiewicz AK, Dasgupta A, Nguyen KH, Liu C, Kovatich AJ, Schwartz GF, et al. Stromal caveolin-1 levels predict early DCIS progression to invasive breast cancer. Cancer Biol Ther 2009; 8:1071 - 1079; PMID: 19502809; http://dx.doi.org/10.4161/cbt.8.11.8874
- Witkiewicz AK, Dasgupta A, Sammons S, Er O, Potoczek MB, Guiles F, et al. Loss of Stromal Caveolin-1 Expression Predicts Poor Clinical Outcome in Triple Negative and Basal-like Breast Cancers. Cancer Biol Ther 2010; 10:135 - 143; PMID: 20431349; http://dx.doi.org/10.4161/cbt.10.2.11983
- Witkiewicz AK, Casimiro MC, Dasgupta A, Mercier I, Wang C, Bonuccelli G, et al. Towards a new “stromalbased” classification system for human breast cancer prognosis and therapy. Cell Cycle 2009; 8:1654 - 1658; PMID: 19448435; http://dx.doi.org/10.4161/cc.8.11.8544
- Ghajar CM, Meier R, Bissell MJ. Quis custodiet ipsos custodies: who watches the watchmen?. Am J Pathol 2009; 174:1996 - 1999; PMID: 19465642; http://dx.doi.org/10.2353/ajpath.2009.090363
- Sloan EK, Ciocca D, Pouliot N, Natoli A, Restall C, Henderson M, et al. Stromal Cell Expression of Caveolin-1 Predicts Outcome in Breast Cancer. Am J Pathol 2009; 174:2035 - 2043; PMID: 19411449; http://dx.doi.org/10.2353/ajpath.2009.080924
- Koo JS, Park S, Kim SI, Lee S, Park BW. The impact of caveolin protein expression in tumor stroma on prognosis of breast cancer. Tumour Biol 2011; 32:787 - 799; PMID: 21584795; http://dx.doi.org/10.1007/s13277-011-0181-6
- Qian N, Ueno T, Kawaguchi-Sakita N, Kawashima M, Yoshida N, Mikami Y, et al. Prognostic significance of tumor/stromal caveolin-1 expression in breast cancer patients. Cancer Sci 2011; 102:1590 - 1596; PMID: 21585620; http://dx.doi.org/10.1111/j.13497006.2011.01985.x
- Tanji K, Bonilla E. Light microscopic methods to visualize mitochondria on tissue sections. Methods 2008; 46:274 - 280; PMID: 18929660; http://dx.doi.org/10.1016/j.ymeth.2008.09.027
- Mahad DJ, Ziabreva I, Campbell G, Laulund F, Murphy JL, Reeve AK, et al. Detection of cytochrome c oxidase activity and mitochondrial proteins in single cells. J Neurosci Methods 2009; 184:310 - 319; PMID: 19723540; http://dx.doi.org/10.1016/j.jneumeth.2009.08.020
- Nachlas MM, Tsou KC, De Souza E, Cheng CS, Seligman AM. Cytochemical demonstration of succinic dehydrogenase by the use of a new p-nitrophenyl substituted ditetrazole. J Histochem Cytochem 1957; 5:420 - 436; PMID: 13463314; http://dx.doi.org/10.1177/5.4.420
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, et al. NCBI GEO: mining tens of millions of expression profiles-database and tools update. Nucleic Acids Res 2007; 35:760 - 765; PMID: 17099226; http://dx.doi.org/10.1093/nar/gkl887
- Brazma A, Parkinson H, Sarkans U, Shojatalab M, Vilo J, Abeygunawardena N, et al. ArrayExpress—a public repository for microarray gene expression data at the EBI. Nucleic Acids Res 2003; 31:68 - 71; PMID: 12519949; http://dx.doi.org/10.1093/nar/gkg091
- Ertel A, Dean JL, Rui H, Liu C, Witkiewicz A, Knudsen KE, et al. RB-pathway disruption in breast cancer: Differential association with disease subtypes, disease-specific prognosis and therapeutic response. Cell Cycle 2010; 9:4153 - 4163; PMID: 20948315; http://dx.doi.org/10.4161/cc.9.20.13454
- Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res 2004; 10:7252 - 7259; PMID: 15534099; http://dx.doi.org/10.1158/1078-0432.CCR-04-0713