Abstract
Comment on: Peña-Llopis S, et al. EMBO J 2011; 30:3242-58.
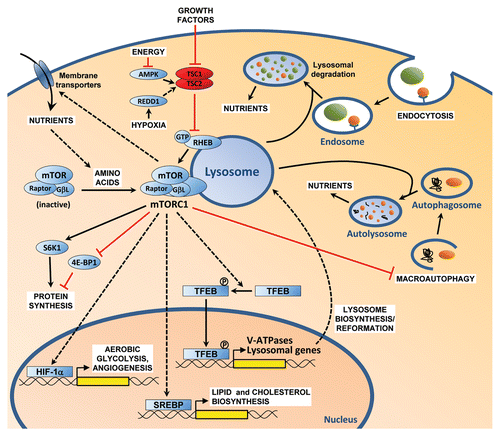
Cell growth involves the coordinated biosynthesis of macromolecules when conditions are favorable. Mammalian target of rapamycin (mTOR) complex 1 (mTORC1) regulates cell growth and is activated in response to nutrients and growth factors and inhibited by low energy and hypoxia. Nutrients promote the translocation of mTORC1 to the surface of the late endosome/lysosome, where it becomes receptive to activation by the GTP-bound form of the Ras homolog enriched in brain (Rheb).Citation1 Rheb is, in turn, regulated by the GTPase-activating tumor suppressor complex, composed of the tuberous sclerosis complex 1 (TSC1) and TSC2 proteins, which relays signals from growth factors, energy stores and oxygen. mTORC1 increases cap-dependent translation, promotes ribosome biogenesis and suppresses autophagy, thereby stimulating cell growth.
We recently discovered that the transcription factor EB (TFEB) is regulated by mTORC1.Citation2 To identify mTORC1-regulated genes, we manipulated mTORC1 activity using an orthogonal approach: mTORC1 was activated via disruption of TSC2 and inactivated using the allosteric and selective inhibitor rapamycin. Under serum-starved conditions, mTORC1 is inhibited, and rapamycin has little added effect. However, in Tsc2-deficient cells, mTORC1 remains active and becomes inactivated by rapamycin. Using microarrays, we searched for transcripts that were low [in wild-type (wt) untreated], low (in wt rapamycin treated), high (in Tsc2−/−) and low (in Tsc2−/− rapamycin treated), conforming to a LLHL pattern. By comparing to alternate patterns (HLLL, LHLL and LLLH), we determined that the probability that the association of genes with the LLHL pattern was random was below 1.6 × 10−28. Among the genes identified, there was a statistically significant over-representation of lysosomal genes and, in particular, of genes encoding components of the vacuolar H+-ATPase (V-ATPase), a complex essential for the acidification of late endosomes/lysosomes.
Lysosome biogenesis is regulated by TFEB,Citation3 and we asked whether TFEB was implicated downstream of mTORC1. TFEB depletion abrogated V-ATPase upregulation in TSC2-deficient cells, indicating that TFEB was necessary. Subcellular fractionation and immunofluorescence experiments showed that the subcellular localization of TFEB was controlled by mTORC1. Whereas in serum-starved cells, TFEB was largely cytoplasmic, in TSC2deficient cells, TFEB localized to the nucleus. Furthermore, mTORC1 inactivation in TSC2-deficient cells by rapamycin (or by raptor knockdown) excluded TFEB from the nucleus. By introducing deletions, a serine-rich region was identified in the C terminus of TFEB (462SSRRSSFS469) that was necessary for mTORC1-depedendent nuclear localization. Nuclear TFEB was found to be phosphorylated and alanine substitution of potential phospho-acceptor sites in the serine-rich motif abrogated mTORC1-depedendent nuclear localization. Conversely, phosphomimetic amino acid substitution of serine residues phenocopied the effects of active mTORC1. These data led us to propose that mTORC1 regulates the nuclear localization of TFEB by promoting phosphorylation in the serine-rich motif.
To assess the importance of TFEB in mTORC1-dependent gene expression, microarrays were performed in Tsc2-deficient cells stably depleted of Tfeb. Interestingly, TFEB was required for the upregulation of as many as 25% of mTORC1-induced genes. While it remains to be determined how many mTORC1-regulated genes are directly regulated by TFEB, a ChIP-seq study using cells stably expressing epitope-tagged TFEB showed that many V-ATPase genes were bound by TFEB.Citation4
TFEB and V-ATPases were also found to be implicated in a poorly characterized function of mTORC1, endocytosis.Citation2 TFEB and V-ATPase knockdown, like rapamycin treatment, diminished caveolin-dependent endocytosis. Given the role of mTORC1 in promoting anabolic processes, endocytosis may provide a means to increase nutrient uptake.Citation5 Consistent with this notion, we found that mTORC1 increased the uptake of albumin, which can serve as a source of amino acids. Since lysosomes are essential for endosome fusion, by increasing lysosome biogenesis, mTORC1 may indirectly promote endocytosis. Interestingly, lysosome trafficking may also be regulated by TFEB.Citation6
Lysosome biogenesis may be important under conditions of persistent starvation and autophagy (macroautophagy), which may deplete the lysosomal pool.Citation7 During prolonged autophagy, the release of nutrients from autophagosomes into the cytosol may reactivate mTORC1,Citation7 which, based on our findings, could lead to the phosphorylation and translocation of TFEB into the nucleus, lysosome biosynthesis and replenishment of the lysosomal pool.
Since mTORC1 activation requires its translocation to the surface of the late endosome/lysosome, the synthesis of lysosomes may also serve as a feedforward mechanism and reinforce mTORC1 activation.
Using a similar experimental approach to the one that led us to TFEB identification, we previously reported that hypoxiainducible factor (HIF)-1α was regulated by mTORC1.Citation8 Further studies showed that HIF-1 is implicated in coordinately regulating the expression of glycolytic enzymes downstream of mTORC1.Citation9,Citation10 HIF-1 promotes a switch from oxidative phosphorylation to aerobic glycolysis, also known as the Warburg effect. This facilitates the shunting of glucose toward the pentose phosphate pathway, resulting in the production of ribose and NADPH, which are necessary for nucleotide and lipid synthesis. Lipid and cholesterol biosynthesis are also increased by activation of sterol regulatory element binding transcription factor (SREBP), whose nuclear localization is regulated by mTORC1.Citation10,Citation11 Our results add TFEB to the list of transcriptional mTORC1 effectors implicated in cell growth and metabolism control ().
While our study focused on mTORC1-induced genes and TFEB was identified as an important mediator, there are likely to be other transcriptional effectors. Genes were identified whose expression was repressed by mTORC1, and other genes emerged whose expression was regulated by TSC1/TSC2 independently of mTORC1.Citation2
Comment on: Peña-Llopis S, et al. EMBO J 2011; 30:3242 - 3258; PMID: 21804531; http://dx.doi.org/10.1038/emboj.2011.257
References
- Zoncu R, et al. Nat Rev Mol Cell Biol 2011; 12:21 - 35; PMID: 21157483; http://dx.doi.org/10.1038/nrm3025
- Peña-Llopis S, et al. EMBO J 2011; 30:3242 - 3258; PMID: 21804531; http://dx.doi.org/10.1038/emboj.2011.257
- Sardiello M, et al. Science 2009; 325:473 - 477; PMID: 19556463
- Palmieri M, et al. Hum Mol Genet 2011; 20:3852 - 3866; PMID: 21752829; http://dx.doi.org/10.1093/hmg/ddr306
- Edinger AL, et al. Mol Biol Cell 2002; 13:2276 - 2288; PMID: 12134068; http://dx.doi.org/10.1091/mbc.01-12-0584
- Medina DL, et al. Dev Cell 2011; 21:421 - 430; PMID: 21889421; http://dx.doi.org/10.1016/j.devcel.2011.07.016
- Yu L, et al. Nature 2010; 465:942 - 946; PMID: 20526321; http://dx.doi.org/10.1038/nature09076
- Brugarolas JB, et al. Cancer Cell 2003; 4:147 - 158; PMID: 12957289; http://dx.doi.org/10.1016/S1535-6108(03)00187-9
- Majumder PK, et al. Nat Med 2004; 10:594 - 601; PMID: 15156201; http://dx.doi.org/10.1038/nm1052
- Düvel K, et al. Mol Cell 2010; 39:171 - 183; PMID: 20670887; http://dx.doi.org/10.1016/j.molcel.2010.06.022
- Porstmann T, et al. Cell Metab 2008; 8:224 - 236; PMID: 18762023; http://dx.doi.org/10.1016/j.cmet.2008.07.007