Abstract
Tight regulation of the cell cycle and DNA repair machinery is essential for maintaining genome stability. The APC/CCdh1 ubiquitin ligase complex is a key regulator of protein stability during the G1 phase of the cell cycle. APC/CCdh1 regulates and promotes the degradation of proteins involved in both cell cycle regulation and DNA repair. In a recent study, we identified a novel APC/CCdh1 substrate, the ubiquitin protease USP1. USP1 is a critical regulator of both the Fanconi anemia (FA) and translesion synthesis (TLS) DNA repair pathways. Here, we provide additional mechanistic insights into the regulation of USP1 during the cell cycle. Specifically, we demonstrate that USP1 is phosphorylated in mitosis by cyclin-dependent kinases (Cdks), and that this phosphorylation event may prevent premature degradation of USP1 during normal cell cycle progression. Finally, we provide a unifying hypothesis integrating the role of G1-specific proteolysis of USP1 with the regulation of the transcriptional repressors, Inhibitor of DNA-binding (ID) proteins.
Introduction
USP1 is a deubiquitinating enzyme (DUB) with substrates in both the Fanconi anemia (FA)Citation1 and translesion synthesis (TLS)Citation2 DNA repair pathways. USP1 regulates the FA DNA repair pathway by deubiquitinating the key effector proteins of this pathway, FANCD2 and FANCI.Citation3–Citation6 Disruption of USP1 results in DNA cross-linker hypersensitivity, and USP1-deficient mice possess increased genomic instability and display an FA-like phenotype.Citation7 To date, there have been no reported cases of patients harboring mutations in the USP1 gene, and it is still not well-understood how deubiquitination of FANCD2 and FANCI promotes repair by the FA pathway. It is possible that (1) deubiquitinating FANCD2 and FANCI removes these proteins from chromatin, making them available for additional repair sites (interestingly, it was shown that the cross-link sensitivity in the absence of USP1 could not be rescued by overexpression of FANCD2 alone.Citation7 However, attempts to rescue the sensitivity with both FANCI and FANCD2 have yet to be reported) or (2) removal of FANCI and FANCD2 from chromatin may allow the completion of late DNA repair stages.
In contrast to the FA pathway, USP1 actively inhibits the TLS pathway by deubiquitinating PCNA and preventing the recruitment of error-prone TLS polymerases.Citation8,Citation9 In this setting, USP1 can inhibit mutagenesis and mutagenic repair of plasmids with or without prior exposure to UV damage.Citation8 Despite the identification of critical USP1 substrates, there is still a lot to learn with respect to how USP1 protects against genomic instability. An obvious place to start is whether USP1's role promoting repair by the FA pathway or inhibiting TLS repair is required to suppress the genomic instability seen in USP1-deficient mice and DT40 cells.Citation7,Citation10 Furthermore, understanding how USP1 is regulated with respect to different cell cycle stages and following exposure to different DNA-damaging agents will clarify its role in both of these DNA damage repair pathways.
APC/CCdh1 is one of the major ubiquitin ligase complexes involved in regulating the cell cycle.Citation11 It is typically active during late mitosis and G1 to promote degradation of positive regulators of mitosis and S phase. In the last few years, several studies have shown APC/CCdh1 to have a broader spectrum of substrates.Citation12 Cdh1-knockout mouse embryonic fibroblasts have substantial chromosomal aberrations and a high degree of genomic instability.Citation13 We recently expanded the list of substrates that APC/CCdh1 targets for degradation by showing that USP1 can interact and be poly-ubiquitinated by APC/CCdh1.Citation14 More importantly, failure to degrade USP1 (as we previously showed by the expression of a nondegradable mutant) inhibited PCNA mono-ubiquitition during the G1 UV DNA damage response and failed to recruit TLS polymerases to sites of DNA damage.Citation14
In this review, we will first highlight recent advances made in exploring how the cell cycle regulates USP1 stability and activity. Next, we will include additional data to support the role of Cdk-dependent phosphorylation in the regulation of USP1 protein stability during mitosis. Finally, we will provide new mechanistic insights and propose a working model to explain how and why USP1 needs to be selectively stabilized during mitosis and degraded during the G1 phase of the cell cycle to ensure genomic stability.
USP1 is under Tight Control by Multiple Regulatory Pathways
USP1 activity is tightly regulated by several different mechanisms (). First, the enzymatic activity of USP1 is regulated by binding to a critical catalytic co-factor USP1-associated factor 1 (UAF1), also known as WDR48 or p80.Citation15 Mutating the WD40 repeats on UAF1 disrupts the USP1-UAF1 interaction and leads to USP1 protein destabilization.Citation15 UAF1 was also recently shown to regulate the activity of other deubiquitinating complexes containing USP12 and USP46, suggesting a broader role for UAF1 and possibly other WD40 proteins in the regulation of deubiquitinating enzymes.Citation16 Second, upon UV DNA damage, USP1 is auto-cleaved and degraded by the proteasome.Citation8 Importantly, under the same conditions, the protein level of UAF1 does not change, and the E3 ubiquitin ligase that targets USP1 for degradation upon UV damage still remains elusive.Citation15 Finally, we recently reported that USP1 is also regulated in a cell cycle-dependent manner.Citation14 USP1 is degraded during the G1 phase of the cell cycle by the ubiquitin ligase APC/CCdh1 in order to provide a more permissive environment for mono-ubiquitination of PCNA and TLS recruitment for UV gap repair synthesis.Citation14,Citation17
Regulation of USP1 during the Cell Cycle by APC/CCdh1
APC/C is a multi-subunit E3 ligase complex that is regulated through the binding of one of its two activators: Cdc20 and Cdh1.Citation18 These activators target substrates at specific stages during mitotic exit.Citation19,Citation20 Specifically, Cdc20 is bound to the APC/C prior to Cdh1, which binds during later mitotic stages and throughout G1. Cdh1 is responsible for the degradation of positive regulators of mitosis and S phase and is considered to be a master regulator of the G1 phase.Citation12 In other cell cycle phases, Cdh1 is kept inactive by several mechanisms: (1) Emi1 binding, (2) Cdk-dependent phosphorylation, (3) autodegradation and (4) degradation by SCF complex.Citation21–Citation27 Perturbation by these mechanisms allows accumulation of Cdh1 substrates and aberrant cell cycle progression.
APC/CCdh1 also promotes degradation of substrates outside the G1 phase of the cell cycle. In response to DNA damage, APC/CCdh1 can be reactivated in G2 to target Plk1 for degradation in order to control the G2/M checkpoint.Citation28 Cdh1 also degrades Rad17, a cell cycle checkpoint protein required for cell cycle arrest that is part of the DNA damage surveillance complex.Citation29 Additional DNA damage response or repair proteins targeted for degradation by the APC/CCdh1 include Claspin, thymidine kinase 1, the ribonucleotide reductase subunit RRM2 and, more recently, USP1.Citation28,Citation30–Citation32
In our recent study, we reported that USP1 protein levels oscillate throughout the cell cycle. By synchronizing cells at the G0/G1 boundary with serum deprivation or in prophase by incubation with Nocodazole, we were able to show that USP1 protein levels are downregulated during the G1 phase of the cell cycle, begin to accumulate rapidly as cells transition into S phase and remain elevated until the late stages of mitotis.Citation14
During the G1 phase of the cell cycle, APC/CCdh1 also degrades key cell cycle regulators, such as Cyclin A, Cdc6, Cdt1, Aurora A, Aurora B and Plk1.Citation33–Citation38 To facilitate the G1/S transition, the transcription factor E2F induces the expression of several key cell cycle regulators, including Emi1, a central inhibitor of APC/C.Citation22,Citation39 Induction of Emi1 expression leads to inactivation of APC/CCdh1 through the ability of Emi1 zinc finger domain to interact potently with APC/CCdh1 and act as a pseudosubstrate to inhibit its activity and function. Deletion of the zinc domain converts Emi1 into a regular Cdh1 substrates, which can then be degraded by APC/CCdh1.Citation21,Citation23 Thus, inactivation of APC/CCdh1 due to Emi1 expression leads to cyclin A accumulation and further inhibition of APC/C through the phosphorylation of Cdh1. Phosphorylation of Cdh1 by cyclin A-Cdks prevents Cdh1 from binding to the APC/C machinery, resulting in further inactivation of the ligase complex.Citation24,Citation40 Conversely, loss of Emi1 allows activation of APC/CCdh1 prior to S-phase entry and prevents the accumulation of many E2F targets.Citation41
To test whether loss of Emi1 can affect USP1 protein levels, we decided to investigate the effect of silencing Emi1 on USP1 protein stability. Silencing of Emi1 in U2OS cells led to a decrease in USP1 and other Cdh1 substrate protein levels, such as Cdc6 and Plk1 (). However, silencing Emi1 and Cdh1 simultaneously led to the rescue of USP1 levels (not shown). This confirmed previous studies, which showed the importance of Emi1 as a Cdh1 inhibitor in order to allow Cdh1 substrate accumulation.Citation41 Similarly, we also tested the effect of inhibiting Cdks on USP1 protein levels. We treated an asynchronous population of U2OS cells with roscovitine, a purine derivative that can inhibit cyclin-dependent kinase 1 (cdk1), cdk2, cdk5, cdk7 and cdk9.Citation42 Treatment of cells with roscovitine decreased USP1 protein levels, similar to what has been reported previously for Cdc6 ().Citation43 Depletion of Cdh1 in roscotivine-treated cells can rescue the loss of USP1 protein expression (). Together, this data suggests that both Cdk activity and Emi1 are required to stabilize Cdh1 substrates, including USP1.
Cdk-Dependent Phosphorylation of USP1 at Serine 313 during Mitosis
Cdk1 is highly active during mitosis and phosphorylates many substrates, including Cdc6.Citation43 Since we observed a noticeably slower mobility shift in the USP1 protein in U2OS cells arrested in mitosis (not shown), this led us to hypothesize that USP1 might be phosphorylated in M phase, possibly by Cdks. To this end, we searched for putative Cdk phosphorylation sites on USP1 using several publicly available phosphosite databases (Phosida, Phosphosite).Citation44,Citation45 Among the different phosphorylation sites found on USP1, Serine 313 (S313) was a consensus Cdk site and was identified in all the available phospho-site databases searched. Interestingly, this site is located within the degron region, which we previously identified as the Cdh1 recognition site ().Citation14 The Cdh1 recognition site within USP1 was previously mapped to amino acids 295–342. We reported that a USP1 deletion mutant, missing amino acids 295–342, is resistant to the G1 phase APC/CCdh1-mediated degradation.Citation14 Therefore, we decided to analyze the S313 putative Cdk phosphorylation site and determine whether it is indeed phosphorylated in vivo and can regulate the stability of USP1. To study the regulation of S313 phosphorylation, we generated an antiphospho-S313 (P-S313) USP1 antibody. In order to test the antibody specificity, we immunoprecipitated wild-type (WT) Myc-USP1 or Myc-S313A USP1 and immunoblotted with the P-S313 antibody. shows that only the wild-type USP1 was recognized by the phosphospecific antibody, demonstrating that the antibody is specific to the phosphorylated form of S313. Next, we monitored S313 phosphorylation during mitotic exit by synchronizing U2OS cells in prophase with nocodazole and releasing them into the cell cycle. We observed strong S313 phosphorylation during M phase, and the signal progressively decreased as USP1 was degraded during G1 ().
Cdk1 is highly active during mitosis, but this activity is rapidly lost as cyclin B becomes degraded during mitotic exit. To test whether S313 phosphorylation was Cdk1-dependent, we arrested cells with nocodazole in the absence or presence of the proteosome inhibitor MG132 and the Cdk1 inhibitor RO-3306.Citation46 Treating prophase-arrested cells with RO-3306 led to the degradation of USP1 as cell exited mitosis (). This degradation was inhibited with MG132 treatment (as expected), and under these conditions, phosphorylation of USP1 S313 was rapidly lost (). To begin to understand the role of S313 phosphorylation, we expressed Myc-USP1 WT, S313A or S313E (phospho-mimic mutant) in U2OS cells and tested for their protein stability in mitosis. Myc-USP1 WT and S313E were both stably expressed during mitosis. In contrast, there was reduced protein expression in the S313A when comparing mitotic cells to the asynchronous population, suggesting that the S313A protein may be unstable during M phase (). Together, these data demonstrate that phosphorylation of USP1 on S313 during M phase is Cdk-dependent and likely important for prolonging USP1 stability during mitosis.
Discussion
Possible roles of Cdk-dependent USP1 phosphorylation.
Protein phosphorylation can lead to different regulatory events. Here, we show that USP1 is phosphorylated in M phase by Cdks on S313. We also provide initial evidence that this phosphorylation event may regulate its protein stability. It has been shown that phosphorylation of APC/C substrates can lead to their stabilization; such is the case for Cdc6.Citation43 Cdc6 is a key regulator for the initiation of DNA replication. Prior to S phase, Cdc6 is kept at low levels by APC/CCdh1-dependent proteolysis.Citation47 Cdk-dependent phosphorylation of Cdc6 allows for its stabilization by preventing its association to Cdh1.Citation43 Perhaps phosphorylation of USP1 might also serve as a protective mechanism against APC/CCdh1-mediated degradation. Since the S313 phosphorylation site is located within the region that we previously characterized as the Cdh1 recognition site, we speculate that USP1 phosphorylation on this site prevents its association to Cdh1. More experiments are necessary to prove this point.
Additionally, phosphorylation of USP1 may have other important regulatory functions besides blocking its degradation. Perhaps phosphorylation of USP1 (either on S313 or other uncharacterized Cdk sites) can serve as an interface for interacting with its co-factor UAF1. As we previously reported, APC/CCdh1 degrades USP1, but it does not target UAF1.Citation14 UAF1 levels do not change throughout the cell cycle. UAF1 serves as an enhancer of USP1 activity and stabilizer of USP1 protein levels. Inability to form the UAF1-USP1 complex can lead to USP1 protein destabilization.Citation15 If phosphorylation of USP1 serves as an interaction site for UAF1, we speculate that once cells exit mitosis, and USP1 becomes dephosphorylated, this might lead to disruption of the USP1-UAF1 complex and subsequent degradation of USP1 by Cdh1 during the G1 phase of the cell cycle. A recent study showed that phosphorylation of USP37 by Cdk2 leads to its full enzymatic activation in order to allow cyclin A accumulation, APC/CCdh1 inhibition and progression into the cell cycle.Citation48 Perhaps outside the G1 phase of the cell cycle, phosphorylation is the first step in the activation of USP1, which is then followed by interaction with UAF1 for full activation of its protease activity and stabilization. Therefore, phosphorylation might act as both a protective and activation mechanism for its enzymatic activity. It will be interesting to see whether USP1 activity changes during different cell cycle phases.
What could be the physiological role of USP1 phosphorylation and G1-specific degradation?
It has been reported that FANCD2 and FANCI can form paired mitotic foci in cells undergoing replication stress after treatment with aphidicolin (an inhibitor of replicative polymerase).Citation49,Citation50 These FANCD2/FANCI mitotic dots co-localize to DNA fragile sites, which are gaps or breaks on chromosomes during mitosis.Citation49,Citation50 Similar to FAND2/FANCI, 53BP1, a component of the DNA damage response, has been reported to form larger nuclear foci (53BP1 nuclear bodies) during the G1 phase of the cell cycle after induced replication stress with low doses of aphidicolin.Citation51,Citation52 It is presumed that replication stress or incomplete DNA synthesis during S-phase forms FANCD2/FANCI foci that persist into mitosis and are probably recognized in the next cell cycle phase as DNA lesion, which then results in the formation of 53BP1 nuclear bodies during the G1 phase. These lesions marked by 53BP1 will be repaired during G1, prior to S-phase entry.Citation51,Citation53 In accordance with this predicted model, the role of USP1 during mitosis may be to deubiquitinate and remove FANCD2/FANCI from their chromatin-bound state. Therefore, USP1 stability (protected against degradation via Cdk-dependent phosphorylation) is critical during mitosis to ensure efficient deubiquitination of FANCD2/FANCI after dealing with replication stress-induced lesions. Then, once the cells enter the G1 phase, USP1 is subsequently degraded to enable efficient PCNA monoubiquitination and TLS polymerase-dependent repair of lingering DNA lesions via gap repair synthesis. Further studies will be necessary to fully address the role of USP1 phosphorylation during mitosis and how it impacts genomic stability in replicating cells.
Possible role of USP1-mediated stabilization of ID proteins in TLS.
DUBs have been shown to regulate a wide range of biological processes, and their increasing role in human pathogenesis, such as cancer, underscores the need to target DUBs for therapeutic intervention.Citation54 A recent study has linked overexpression of USP1 to osteogenic sarcoma and suggested USP1 as a potential therapeutic target.Citation55 Elegant work by Dixit and colleagues identified three new USP1 substrates called inhibitor of DNA-binding (ID) proteins, ID1, ID2 and ID3. While ID4 is also associated with cancer, its protein stability is likely not regulated by USP1.Citation55,Citation56 ID proteins antagonize basic-helix-loop-helix (bHLH) transcription factors, such as the Cdk inhibitor p21. They showed that USP1 can interact with ID proteins and prevent their proteasomal degradation.Citation55 Interestingly, a subset of primary osterosarcomas presented high levels of USP1 and ID proteins concomitantly. Knockdown of USP1 led to the degradation of ID1/2/3, cell cycle arrest and induction of p21. Conversely, overexpression of USP1 led to an increase in ID1/2/3 abundance and cell proliferation.Citation55 A previous study showed that in neurons, APC/CCdh1 interacts with ID2 and promotes its degradation in cells undergoing quiescence to prevent axonal growth.Citation57 Therefore, we speculate that during the G1 phase of the cell cycle, APC/CCdh1 targets USP1 for degradation, so it can no longer deubiquitinate the ID proteins, making them susceptible for degradation by APC/CCdh1. This ensures upregulation of p21 and proper G1 timing to prevent unscheduled DNA synthesis (see model, ).
A unifying hypothesis for USP1 regulation during cell cycle progression and DNA repair.
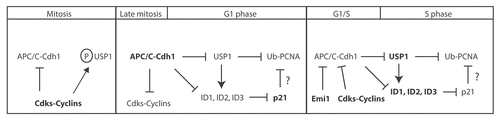
Here, we present our working model to highlight how different cell cycle phases (namely, M, G1 and S phase) can regulate both USP1 and ID proteins stability and speculate how these events can ultimately affect PCNA-mediated DNA repair (). (1) In the early mitotic stages, Cdh1 is inhibited by Cdk phosphorylation, which prevents Cdh1 binding to APC/C.Citation24,Citation25 During mitosis, Cdks can also phosphorylate USP1 on at least S313 ( and ). We speculate that phosphorylation of USP1 serves as (a) an interaction site for UAF1 and/or (b) a protective mechanism against Cdh1 interaction and subsequent degradation. (2) In the later mitotic stages, Cdh1 is de-phosphorylated, Emi1 gets degraded by SCFBTRCP and Cdh1 becomes active and continues the degradation of cyclins.Citation25,Citation27,Citation58 (3) Once cells enter the G1 phase of the cell cycle, Cdk activity is very low or absent due to degradation of their cyclin partners by APC/C and therefore are unable to maintain USP1 phosphorylation (and/or active phosphatase).Citation33,Citation59 Dephosphorylated USP1 is then susceptible to degradation by APC/CCdh1 in the G1 phase. (4) Low levels of USP1 during the G1 phase leads to two possible outcomes: first, it allows for a permissive environment for PCNA mono-ubiquitination. Upon DNA damage during the G1 phase, cells are arrested in G1 and USP1 degradation by APC/CCdh1 is prolonged, allowing PCNA mono-ubiquitination, recruitment of TLS polymerase and DNA repair;Citation14 second, low levels of USP1 during the G1 phase of the cell cycle might be unable to inhibit the poly-ubiquitination and degradation of ID proteins by APC/CCdh1. Therefore, degradation of USP1 in G1 will lead to subsequent degradation of ID proteins. Loss of ID proteins will relieve the transcriptional repression of p21, leading to the accumulation of p21 during G1.Citation55 In the absence of DNA damage, p21 might inhibit PCNA-mediated function during the G1 phase of the cell cycle by controlling PCNA-interacting TLS polymerases.Citation60 Several studies have shown that p21 can interact with PCNA and control the loading of TLS polymerases onto PCNA by displacing them.Citation61–Citation65 (5) At the G1/S transition, induction of Emi1 expression inhibits APC/CCdh1 activity and leads to the accumulation of APC/C substrates including cyclins, USP1 and, perhaps, ID proteins.Citation14,Citation22,Citation23,Citation57 Accumulation of cyclins leads to an increase in Cdk-cyclin activity that can further inhibit APC/CCdh1 activity by phosphorylation and allow progression into S phase. (6) In the absence of DNA damage during S phase, USP1 can stabilize ID proteins and transcriptionally repress p21. Since p21 can no longer suppress TLS activity at the replication fork, USP1 becomes more critical in preventing aberrant error-prone TLS polymerase usage in the absence or present of DNA damage.
Abbreviations
| APC/C | = | anaphase promoting complex/cyclosome |
| USP1 | = | ubiquitin-specific protease 1 |
| Cdk | = | cyclin-dependent kinases |
| UAF1 | = | USP1 associated factor 1 |
| DUBs | = | deubiquitinating enzymes |
| FA | = | fanconi anemia |
| PCNA | = | proliferating cell nuclear antigen |
| SCF complex | = | Skp1 cullin F-box-containing complex |
| ID | = | inhibitor of DNA-binding protein |
Figures and Tables
Figure 1 Multiple mechanisms regulate USP1 activity. (A) USP1 requires the association of UAF1 for its full enzymatic activity and protein stability. (B) Upon UV DNA damage, USP1 is auto-cleaved and degraded by the proteasome by an unknown ubiquitin E3 ligase. (C) APC/CCdh1 binds to and degrades USP1 during the G1 phase of the cell cycle.
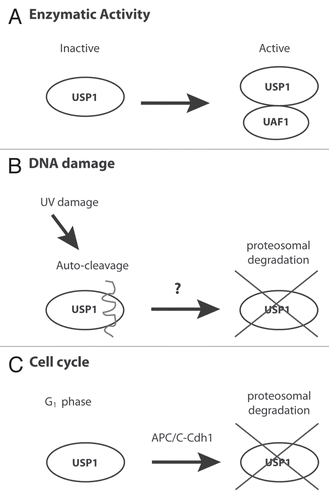
Figure 2 Inhibition of Emi1 or Cdks can destabilize USP1. (A) U2OS cells were transfected for 48 h with a control (Ctrl) siRNA oligo (Qiagen AllStar Neg siRNA) or a siRNA oligo directed against Emi1 (5′-CAT GTT CAT TCC GGA CTT AAA-3′). Samples were collected, lysed and analyzed by protein gel blot with the indicated antibodies: Plk1 (Abcam), Cdc6 (Santa Cruz), Emi1 (Invitrogen), Cdk2 (Bethyl), Cdh1 (CalBiochem), Tubulin (Abcam). (B) U2OS cells were left untreated (UN) or treated with Roscovitine (CalBiochem) (5 uM) for the indicated time points, analyzed by protein gel blot and probed with the indicated antibodies. (C) U2OS cells were transfected with the indicated siRNAs as in (A) or Cdh1 (AAT GAG AA G TCT CCC AGT CAG) and treated for 6 h with Roscovitine (5 uM). Samples were analyzed by protein gel blot and probe with indicated antibodies.
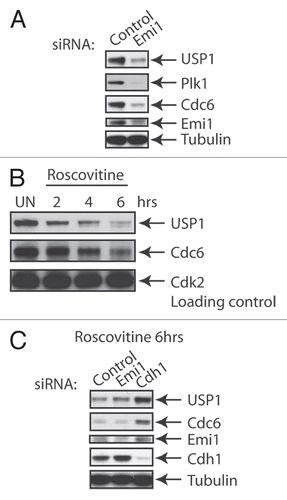
Figure 3 Cdk-dependent phosphorylation of USP1 on S313. (A) Schematic diagram of USP1 catalytic domains, Cdh1 recognition region and putative Cdk phosphorylation site S313 on human USP1. (B) U2OS cells were transfected with Myc-USP1 WT or Myc-USP1 S313A using Fugene6 transfection reagent (Roche Applied Science). Samples were lysed and immunoprecipitated with anti-Myc antibody (Santa Cruz) and protein gel blot was performed using anti-Myc and P-S313 USP1 antibodies (Thermo Scientific). Lysing, immunoprecipitation and washing buffers were done using the low IPB buffer [25 mM Tris, pH 7.5, 150 mM NaCl, 2 mM EDTA and 0.5% NP-40 with protease inhibitor cocktail (Roche)]. (C) U2OS cells were synchronized in M phase by incubating cells with Nocodazole (0.1 µg/ml) for 12–16 h. After 12-16 h, mitotic cells were shaken off and collected and washed twice with 1x PBS, then plated with fresh media and collected for the indicated time points. Samples were analyzed by protein gel blot and probe with the indicated antibodies: P-S313 USP1 antibody (Thermo Scientific), H3 Ser10 (Millipore) and others described above. (D) U2OS cells synchronized in M phase, as in (C), and were treated (or left untreated) with Cdk1 inhibitor RO-3306 (Calbiochem) (10 uM) in the presence or absence of MG132 (10 uM). Samples were analyzed by protein gel blot using the indicated antibodies. (E) U2OS cells were transfected as in (C), with Myc-USP1 WT, S313A or S313E, then cells were split, left unsynchronized (AS) or synchronized in mitosis (M). Samples were analyzed by protein gel blot and probe with the indicated antibodies.
![Figure 3 Cdk-dependent phosphorylation of USP1 on S313. (A) Schematic diagram of USP1 catalytic domains, Cdh1 recognition region and putative Cdk phosphorylation site S313 on human USP1. (B) U2OS cells were transfected with Myc-USP1 WT or Myc-USP1 S313A using Fugene6 transfection reagent (Roche Applied Science). Samples were lysed and immunoprecipitated with anti-Myc antibody (Santa Cruz) and protein gel blot was performed using anti-Myc and P-S313 USP1 antibodies (Thermo Scientific). Lysing, immunoprecipitation and washing buffers were done using the low IPB buffer [25 mM Tris, pH 7.5, 150 mM NaCl, 2 mM EDTA and 0.5% NP-40 with protease inhibitor cocktail (Roche)]. (C) U2OS cells were synchronized in M phase by incubating cells with Nocodazole (0.1 µg/ml) for 12–16 h. After 12-16 h, mitotic cells were shaken off and collected and washed twice with 1x PBS, then plated with fresh media and collected for the indicated time points. Samples were analyzed by protein gel blot and probe with the indicated antibodies: P-S313 USP1 antibody (Thermo Scientific), H3 Ser10 (Millipore) and others described above. (D) U2OS cells synchronized in M phase, as in (C), and were treated (or left untreated) with Cdk1 inhibitor RO-3306 (Calbiochem) (10 uM) in the presence or absence of MG132 (10 uM). Samples were analyzed by protein gel blot using the indicated antibodies. (E) U2OS cells were transfected as in (C), with Myc-USP1 WT, S313A or S313E, then cells were split, left unsynchronized (AS) or synchronized in mitosis (M). Samples were analyzed by protein gel blot and probe with the indicated antibodies.](/cms/asset/d1ff67f0-f5fe-4db9-b10f-c456a413726f/kccy_a_10918501_f0003.gif)
Figure 4 Model showing how USP1-mediated events are regulated during normal cell cycle progression. Schematic diagram of the proposed model of how different cell cycle stages can affect USP1 protein stability, regulation of ID proteins and PCNA-directed DNA repair. Briefly, in M phase, both USP1 and APC/CCdh1 are phosphorylated by Cdks, which prevents USP1 from being prematurely targeted for degradation by the APC/CCdh1. In late M and early G1, USP1 and APC/CCdh1 become dephosphorylated, which leads to the degradation of both USP1 and cyclins. USP1 normally protects ID proteins from ubiquitin-mediated degradation. However, without USP1, ID proteins become subsequently degraded. Loss of ID proteins prevents transcriptional repression of p21, leading to p21 protein accumulation and possible inhibition of TLS activity on PCNA. During the G1-S or S-phase entry, levels of cyclins rise to inhibit APC/CCdh1, which lead to the accumulation of USP1 and presumed stabilization of ID proteins.
Acknowledgments
We especially would like to thank Luca Colnaghi and Miklos Bekes for helpful discussions of the manuscript. We also thank members of the T. Huang, M. Pagano, D. Bar-Sagi and D. Reinberg laboratories for their valuable tools, reagents, equipment and technical assistance. This work was supported by a National Institute of Health grant to T.T. Huang (R01-GM084244).
References
- Moldovan GL, D'Andrea AD. How the fanconi anemia pathway guards the genome. Annu Rev Genet 2009; 43:223 - 249; PMID: 19686080; http://dx.doi.org/10.1146/annurev-genet-102108-134222
- Lehmann AR. Translesion synthesis in mammalian cells. Exp Cell Res 2006; 312:2673 - 2676; PMID: 16854411; http://dx.doi.org/10.1016/j.yexcr.2006.06.010
- Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D'Andrea AD, et al. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell 2005; 17:331 - 339; PMID: 15694335; http://dx.doi.org/10.1016/j.molcel.2005.01.008
- Sims AE, Spiteri E, Sims RJ, Arita AG, Lach FP, Landers T, et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol 2007; 14:564 - 567; PMID: 17460694; http://dx.doi.org/10.1038/nsmb1252
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, Hurov KE, Luo J, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 2007; 129:289 - 301; PMID: 17412408; http://dx.doi.org/10.1016/j.cell.2007.03.009
- Dorsman JC, Levitus M, Rockx D, Rooimans MA, Oostra AB, Haitjema A, et al. Identification of the Fanconi anemia complementation group I gene, FANCI. Cell Oncol 2007; 29:211 - 218; PMID: 17452773
- Kim JM, Parmar K, Huang M, Weinstock DM, Ruit CA, Kutok JL, et al. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev Cell 2009; 16:314 - 320; PMID: 19217432; http://dx.doi.org/10.1016/j.devcel.2009.01.001
- Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol 2006; 8:339 - 347; PMID: 16531995; http://dx.doi.org/10.1038/ncb1378
- Brown S, Niimi A, Lehmann AR. Ubiquitination and deubiquitination of PCNA in response to stalling of the replication fork. Cell Cycle 2009; 8:689 - 692; PMID: 19221475; http://dx.doi.org/10.4161/cc.8.5.7707
- Oestergaard VH, Langevin F, Kuiken H, Pace P, Niedzwiedz W, Simpson L, et al. Deubiquitination of FANCD2 is required for DNA cross-link repair. Mol Cell 2007; 28:798 - 809; PMID: 18082605; http://dx.doi.org/10.1016/j.molcel.2007.09.020
- Qiao X, Zhang L, Gamper AM, Fujita T, Wan Y. APC/C-Cdh1: from cell cycle to cellular differentiation and genomic integrity. Cell Cycle 2010; 9:3904 - 3912; PMID: 20935501; http://dx.doi.org/10.4161/cc.9.19.13585
- Skaar JR, Pagano M. Cdh1: a master G0/G1 regulator. Nat Cell Biol 2008; 10:755 - 757; PMID: 18591966; http://dx.doi.org/10.1038/ncb0708-755
- García-Higuera I, Manchado E, Dubus P, Cañamero M, Méndez J, Moreno S, et al. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat Cell Biol 2008; 10:802 - 811; PMID: 18552834; http://dx.doi.org/10.1038/ncb1742
- Cotto-Rios XM, Jones MJ, Busino L, Pagano M, Huang TT. APC/CCdh1-dependent proteolysis of USP1 regulates the response to UV-mediated DNA damage. J Cell Biol 2011; 194:177 - 186; PMID: 21768287; http://dx.doi.org/10.1083/jcb.201101062
- Cohn MA, Kowal P, Yang K, Haas W, Huang TT, Gygi SP, et al. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol Cell 2007; 28:786 - 797; PMID: 18082604; http://dx.doi.org/10.1016/j.molcel.2007.09.031
- Cohn MA, Kee Y, Haas W, Gygi SP, D'Andrea AD. UAF1 is a subunit of multiple deubiquitinating enzyme complexes. J Biol Chem 2009; 284:5343 - 5351; PMID: 19075014; http://dx.doi.org/10.1074/jbc.M808430200
- Ogi T, Limsirichaikul S, Overmeer RM, Volker M, Takenaka K, Cloney R, et al. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol Cell 2010; 37:714 - 727; PMID: 20227374; http://dx.doi.org/10.1016/j.molcel.2010.02.009
- McLean JR, Chaix D, Ohi MD, Gould KL. State of the APC/C: organization, function and structure. Crit Rev Biochem Mol Biol 2011; 46:118 - 136; PMID: 21261459; http://dx.doi.org/10.3109/10409238.2010.541420
- Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science 1997; 278:460 - 463; PMID: 9334304; http://dx.doi.org/10.1126/science.278.5337.460
- Fang G, Yu H, Kirschner MW. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol Cell 1998; 2:163 - 171; PMID: 9734353; http://dx.doi.org/10.1016/S1097-2765(00)80126-4
- Reimann JD, Gardner BE, Margottin-Goguet F, Jackson PK. Emi1 regulates the anaphase-promoting complex by a different mechanism than Mad2 proteins. Genes Dev 2001; 15:3278 - 3285; PMID: 11751633; http://dx.doi.org/10.1101/gad.945701
- Hsu JY, Reimann JD, Sørensen CS, Lukas J, Jackson PK. E2F-dependent accumulation of hEmi1 regulates S phase entry by inhibiting APC(Cdh1). Nat Cell Biol 2002; 4:358 - 366; PMID: 11988738; http://dx.doi.org/10.1038/ncb785
- Miller JJ, Summers MK, Hansen DV, Nachury MV, Lehman NL, Loktev A, et al. Emi1 stably binds and inhibits the anaphase-promoting complex/cyclosome as a pseudosubstrate inhibitor. Genes Dev 2006; 20:2410 - 2420; PMID: 16921029; http://dx.doi.org/10.1101/gad.1454006
- Zachariae W, Schwab M, Nasmyth K, Seufert W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science 1998; 282:1721 - 1724; PMID: 9831566; http://dx.doi.org/10.1126/science.282.5394.1721
- Jaspersen SL, Charles JF, Morgan DO. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr Biol 1999; 9:227 - 236; PMID: 10074450; http://dx.doi.org/10.1016/S0960-9822(99)80111-0
- Rape M, Kirschner MW. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature 2004; 432:588 - 595; PMID: 15558010; http://dx.doi.org/10.1038/nature03023
- Margottin-Goguet F, Hsu JY, Loktev A, Hsieh HM, Reimann JD, Jackson PK. Prophase destruction of Emi1 by the SCF(betaTrCP/Slimb) ubiquitin ligase activates the anaphase promoting complex to allow progression beyond prometaphase. Dev Cell 2003; 4:813 - 826; PMID: 12791267; http://dx.doi.org/10.1016/S1534-5807(03)00153-9
- Bassermann F, Frescas D, Guardavaccaro D, Busino L, Peschiaroli A, Pagano M. The Cdc14B-Cdh1-Plk1 axis controls the G2 DNA-damage-response checkpoint. Cell 2008; 134:256 - 267; PMID: 18662541; http://dx.doi.org/10.1016/j.cell.2008.05.043
- Zhang L, Park CH, Wu J, Kim H, Liu W, Fujita T, et al. Proteolysis of Rad17 by Cdh1/APC regulates checkpoint termination and recovery from genotoxic stress. EMBO J 2010; 29:1726 - 1737; PMID: 20424596; http://dx.doi.org/10.1038/emboj.2010.55
- Ke PY, Chang ZF. Mitotic degradation of human thymidine kinase 1 is dependent on the anaphase-promoting complex/cyclosome-CDH1-mediated pathway. Mol Cell Biol 2004; 24:514 - 526; PMID: 14701726; http://dx.doi.org/10.1128/MCB.24.2.514-26.2004
- Ke PY, Hu CM, Chang YC, Chang ZF. Hiding human thymidine kinase 1 from APC/C-mediated destruction by thymidine binding. FASEB J 2007; 21:1276 - 1284; PMID: 17227951; http://dx.doi.org/10.1096/fj.06-7272com
- Simpson-Lavy KJ, Sajman J, Zenvirth D, Brandeis M. APC/CCdh1 specific degradation of Hsl1 and Clb2 is required for proper stress responses of S. cerevisiae. Cell Cycle 2009; 8:3003 - 3009; PMID: 19713762; http://dx.doi.org/10.4161/cc.8.18.9616
- Geley S, Kramer E, Gieffers C, Gannon J, Peters JM, Hunt T. Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J Cell Biol 2001; 153:137 - 148; PMID: 11285280; http://dx.doi.org/10.1083/jcb.153.1.137
- Sugimoto N, Kitabayashi I, Osano S, Tatsumi Y, Yugawa T, Narisawa-Saito M, et al. Identification of novel human Cdt1-binding proteins by a proteomics approach: proteolytic regulation by APC/CCdh1. Mol Biol Cell 2008; 19:1007 - 1021; PMID: 18162579; http://dx.doi.org/10.1091/mbc.E07-09-0859
- Littlepage LE, Ruderman JV. Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Genes Dev 2002; 16:2274 - 2285; PMID: 12208850; http://dx.doi.org/10.1101/gad.1007302
- Taguchi S, Honda K, Sugiura K, Yamaguchi A, Furukawa K, Urano T. Degradation of human Aurora-A protein kinase is mediated by hCdh1. FEBS Lett 2002; 519:59 - 65; PMID: 12023018; http://dx.doi.org/10.1016/S0014-5793(02)02711-4
- Nguyen HG, Chinnappan D, Urano T, Ravid K. Mechanism of Aurora-B degradation and its dependency on intact KEN and A-boxes: identification of an aneuploidy-promoting property. Mol Cell Biol 2005; 25:4977 - 4992; PMID: 15923616; http://dx.doi.org/10.1128/MCB.25.12.4977-92.2005
- Stewart S, Fang G. Destruction box-dependent degradation of aurora B is mediated by the anaphase-promoting complex/cyclosome and Cdh1. Cancer Res 2005; 65:8730 - 8735; PMID: 16204042; http://dx.doi.org/10.1158/0008-5472.CAN-05-1500
- Guardavaccaro D, Pagano M. Stabilizers and destabilizers controlling cell cycle oscillators. Mol Cell 2006; 22:1 - 4; PMID: 16600864; http://dx.doi.org/10.1016/j.molcel.2006.03.017
- Mitra J, Enders GH, Azizkhan-Clifford J, Lengel KL. Dual regulation of the anaphase promoting complex in human cells by cyclin A-Cdk2 and cyclin A-Cdk1 complexes. Cell Cycle 2006; 5:661 - 666; PMID: 16582612; http://dx.doi.org/10.4161/cc.5.6.2604
- Verschuren EW, Ban KH, Masek MA, Lehman NL, Jackson PK. Loss of Emi1-dependent anaphase-promoting complex/cyclosome inhibition deregulates E2F target expression and elicits DNA damage-induced senescence. Mol Cell Biol 2007; 27:7955 - 7965; PMID: 17875940; http://dx.doi.org/10.1128/MCB.00908-07
- Taylor SL, Kinchington PR, Brooks A, Moffat JF. Roscovitine, a cyclin-dependent kinase inhibitor, prevents replication of varicella-zoster virus. J Virol 2004; 78:2853 - 2862; PMID: 14990704; http://dx.doi.org/10.1128/JVI.78.6.2853-62.2004
- Mailand N, Diffley JF. CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell 2005; 122:915 - 926; PMID: 16153703; http://dx.doi.org/10.1016/j.cell.2005.08.013
- Gnad F, Gunawardena J, Mann M. PHOSIDA 2011: the posttranslational modification database. Nucleic Acids Res 2011; 39:253 - 260; PMID: 21081558; http://dx.doi.org/10.1093/nar/gkq1159
- Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics 2004; 4:1551 - 1561; PMID: 15174125; http://dx.doi.org/10.1002/pmic.200300772
- Ma HT, Tsang YH, Marxer M, Poon RY. Cyclin A2-cyclin-dependent kinase 2 cooperates with the PLK1-SCFbeta-TrCP1-EMI1-anaphase-promoting complex/cyclosome axis to promote genome reduplication in the absence of mitosis. Mol Cell Biol 2009; 29:6500 - 6514; PMID: 19822658; http://dx.doi.org/10.1128/MCB.00669-09
- Petersen BO, Wagener C, Marinoni F, Kramer ER, Melixetian M, Lazzerini Denchi E, et al. Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes Dev 2000; 14:2330 - 2343; PMID: 10995389; http://dx.doi.org/10.1101/gad.832500
- Huang X, Summers MK, Pham V, Lill JR, Liu J, Lee G, et al. Deubiquitinase USP37 is activated by CDK2 to antagonize APC(CDH1) and promote S phase entry. Mol Cell 2011; 42:511 - 523; PMID: 21596315; http://dx.doi.org/10.1016/j.molcel.2011.03.027
- Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol 2009; 11:753 - 760; PMID: 19465922; http://dx.doi.org/10.1038/ncb1882
- Naim V, Rosselli F. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat Cell Biol 2009; 11:761 - 768; PMID: 19465921; http://dx.doi.org/10.1038/ncb1883
- Harrigan JA, Belotserkovskaya R, Coates J, Dimitrova DS, Polo SE, Bradshaw CR, et al. Replication stress induces 53BP1-containing OPT domains in G1 cells. J Cell Biol 2011; 193:97 - 108; PMID: 21444690; http://dx.doi.org/10.1083/jcb.201011083
- Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol 2011; 13:243 - 253; PMID: 21317883; http://dx.doi.org/10.1038/ncb2201
- Chan KL, Hickson ID. New insights into the formation and resolution of ultra-fine anaphase bridges. Semin Cell Dev Biol 2011; 22:906 - 912; PMID: 21782962
- Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem 2009; 78:363 - 397; PMID: 19489724; http://dx.doi.org/10.1146/annurev.biochem.78.082307.091526
- Williams SA, Maecker HL, French DM, Liu J, Gregg A, Silverstein LB, et al. USP1 Deubiquitinates ID Proteins to Preserve a Mesenchymal Stem Cell Program in Osteosarcoma. Cell 2011; 146:918 - 930; PMID: 21925315; http://dx.doi.org/10.1016/j.cell.2011.07.040
- Dell'Orso S, Ganci F, Strano S, Blandino G, Fontemaggi G. ID4: a new player in the cancer arena. Oncotarget 2010; 1:48 - 58; PMID: 21293053
- Lasorella A, Stegmüller J, Guardavaccaro D, Liu G, Carro MS, Rothschild G, et al. Degradation of Id2 by the anaphase-promoting complex couples cell cycle exit and axonal growth. Nature 2006; 442:471 - 474; PMID: 16810178; http://dx.doi.org/10.1038/nature04895
- Peters JM. Emi1 proteolysis: how SCF(beta-Trcp1) helps to activate the anaphase-promoting complex. Mol Cell 2003; 11:1420 - 1421; PMID: 12820955; http://dx.doi.org/10.1016/S1097-2765(03)00233-8
- Raff JW, Jeffers K, Huang JY. The roles of Fzy/Cdc20 and Fzr/Cdh1 in regulating the destruction of cyclin B in space and time. J Cell Biol 2002; 157:1139 - 1149; PMID: 12082076; http://dx.doi.org/10.1083/jcb.200203035
- Prives C, Gottifredi V. The p21 and PCNA partnership: a new twist for an old plot. Cell Cycle 2008; 7:3840 - 3846; PMID: 19066467; http://dx.doi.org/10.4161/cc.7.24.7243
- Cazzalini O, Scovassi AI, Savio M, Stivala LA, Prosperi E. Multiple roles of the cell cycle inhibitor p21(CDKN1A) in the DNA damage response. Mutat Res 2010; 704:12 - 20; PMID: 20096807; http://dx.doi.org/10.1016/j.mrrev.2010.01.009
- Waga S, Hannon GJ, Beach D, Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature 1994; 369:574 - 578; PMID: 7911228; http://dx.doi.org/10.1038/369574a0
- Flores-Rozas H, Kelman Z, Dean FB, Pan ZQ, Harper JW, Elledge SJ, et al. Cdk-interacting protein 1 directly binds with proliferating cell nuclear antigen and inhibits DNA replication catalyzed by the DNA polymerase delta holoenzyme. Proc Natl Acad Sci USA 1994; 91:8655 - 8659; PMID: 7915843; http://dx.doi.org/10.1073/pnas.91.18.8655
- Chen J, Peters R, Saha P, Lee P, Theodoras A, Pagano M, et al. A 39 amino acid fragment of the cell cycle regulator p21 is sufficient to bind PCNA and partially inhibit DNA replication in vivo. Nucleic Acids Res 1996; 24:1727 - 1733; PMID: 8649992; http://dx.doi.org/10.1093/nar/24.9.1727
- Oku T, Ikeda S, Sasaki H, Fukuda K, Morioka H, Ohtsuka E, et al. Functional sites of human PCNA which interact with p21 (Cip1/Waf1), DNA polymerase delta and replication factor C. Genes Cells 1998; 3:357 - 369; PMID: 9734782; http://dx.doi.org/10.1046/j.1365-2443.1998.00199.x