Abstract
Different molecular pathways involved in maintaining mitochondrial function are of fundamental importance to control cellular homeostasis. Mitochondrial i-AAA protease is part of such a surveillance system and PaIAP is the putative ortholog in the fungal aging model Podospora anserina. Here we investigated the role of PaIAP in aging and development. Deletion of the gene encoding PaIAP resulted in a specific phenotype. When incubated at 27°C, spore germination and fruiting body formation are not different from that of the corresponding wild-type strain. Unexpectedly, the lifespan of the deletion strain is strongly increased. In contrast, cultivation at an elevated temperature of 37°C leads to impairments in spore germination and fruiting body formation, and to a reduced lifespan. The higher PaIAP abundance in wild-type strains of the fungus grown at elevated temperature and the phenotype of the deletion strain unmasks a temperaturerelated role of the protein. The protease appears to be part of a molecular system that has evolved to allow survival under changing temperatures as they characteristically occur in nature.
Introduction
Mitochondria, best known as the power plants of eukaryotic cells, generate most cellular adenosine triphosphate (ATP) that is required to drive the various energy-consuming processes in biological systems. In addition, they are involved in a number of other important processes, like lipid and amino acid biosynthesis, the assembly of iron-sulfur clusters, calcium storage, signaling and programmed cell death. As a side reaction of electron transfer through the respiratory chain that drives ATP synthesis at the inner mitochondrial membrane, reactive oxygen species (ROS) are generated,Citation1 which have the potential to damage all kinds of biomolecules, including lipids, nucleic acids and proteins. The accumulation of such damage ultimately leads to physiological impairments and mitochondrial dysfunction. Fortunately, different pathways have evolved to cope with this hazardous situation and to maintain a functional population of mitochondria.Citation2–Citation4
Important components of the mitochondrial surveillance system are pathways controlling the quality of mitochondrial proteins. These pathways rely on enzymes able to repair damaged proteins, chaperones involved in refolding of misfolded polypeptides, as well as ATP-dependent proteases that selectively degrade irreversibly damaged proteins.Citation2 The inner mitochondrial membrane harbors two AAA proteases, the i-AAA protease and the m-AAA protease. Both belong to the highly conserved family of AAA proteins found in all eukaryotes.Citation5 The two proteases expose their catalytic sites to opposite surfaces of the membrane. While the i-AAA protease is oriented toward the intermembrane space, the m-AAA protease is active at the matrix side.Citation6 Apart from their metal-dependent proteolytic function, both enzymes contain a chaperone-like activity suppressing the aggregation of unfolded polypeptides. This chaperone activity is independent from the proteolytic activity.Citation7
So far, YME1 from Saccharomyces cerevisiae is the best-studied i-AAA protease. Deletion of the gene coding for this protease causes pleiotropic effects, including an increased escape of mitochondrial DNA (mtDNA), heat sensitivity on non-fermentable carbon sources, cold sensitivity on media containing glucose and altered mitochondrial morphology.Citation8,Citation9 Several endogenous substrates of YME1 have been identified, including cytochrome oxidase subunit 2 (COX2), the external mitochondrial NADH dehydrogenase NDE1 and two subunits of the prohibitin complex.Citation10–Citation12 In addition, YME1 has been linked to mitochondrial phospholipid biosynthesis and plays a role in the translocation of the human polynucleotide phosphorylase (PNPase) into the intermembrane space of mitochondria.Citation13,Citation14
The characterization of strains from yeast and the filamentous ascomycete Neurospora crassa, in which the i-AAA protease genes were deleted, identified similar but also different functions of the two homologs, YME1 and IAP-1. In both fungi, deletion of the corresponding genes leads to impaired growth at high temperatures.Citation15 In contrast to yeast, decreased temperatures do not affect growth of the Iap-1 deletion strain. Moreover, IAP-1 can only partially substitute YME1 in S. cerevisiae, pointing to subtle differences in the function of the two i-AAA proteases.Citation15,Citation16 Very little is known about the human homolog YME1L1. Interestingly, YME1L1 is able to rescue the temperature-sensitive phenotype of Yme1-deficient yeast cells underlining the conserved functions of i-AAA proteases.Citation17 Furthermore, YME1L1 seems to be involved in the mitochondrial unfolded protein response, thereby allowing mitochondria to adapt to different types of stress.Citation18
Mitochondria are known to play a key role in aging and lifespan control (reviewed in ref. Citation4 and Citation19–Citation22). One of the best-characterized aging models with a clear mitochondrial etiology of aging is Podospora anserina. In this filamentous fungus, various mitochondrial pathways, including mtDNA reorganization,Citation23 mtDNA repair,Citation24 respiration,Citation25 ROS scavenging,Citation26,Citation27 copper metabolism,Citation28 mitochondrial dynamicsCitation29 and apoptosis,Citation30–Citation32 have been demonstrated to play a role. Recently, an impact of the protein quality control system on aging has been demonstrated.Citation4,Citation33 In this study, the overexpression of PaLon coding for a soluble mitochondrial matrix protease was found to increase lifespan without affecting vital functions.
Here, we report data of investigations analyzing the role of the membrane-bound mitochondrial i-AAA protease PaIAP of P. anserina. We show that deletion of PaIap has a temperature-dependent effect on aging and energy-consuming developmental processes. At 27°C, ΔPaIap showed a healthy wild-type-like growth phenotype and a most remarkable increase in lifespan despite the fact that a component of the protein quality control system was missing. In contrast, at higher temperatures, the ΔPaIap strain was impaired in growth and fertility and displayed a decreased lifespan. These results unravel an important role of the i-AAA protease in heat stress and identify the corresponding pathway as relevant for aging and development.
Results
Identification of the i-AAA protease PaIAP in Podospora anserina.
Within the genome of P. anserina, one open reading frame was reported to encode a putative mitochondrial i-AAA protease (www.podospora.igmors.u-psud.fr). The protein was later termed PaIAP for P. anserina i-AAA protease.Citation34 By in silico analysis, we found that PaIAP shares the typical functional domains of i-AAA proteases from other species, including S. cerevisiae, N. crassa and Homo sapiens (). The AAA domains of the four homologs containing the Walker A and B motif (WA and WB), the so-called second region of homology (SRH), and the metal binding motif (MB) with the proteolytic consensus sequence (HEXXH) are especially well-conserved among the analyzed species ( and S1). The deduced pre-protein of PaIAP is composed of 771 amino acids and contains a putative mitochondrial targeting sequence (MTS). The predicted probability for a mitochondrial localization of PaIAP is approximately 87%. After import into mitochondria, the removal of an N-terminal peptide of 113 amino acids is expected to lead to the mature PaIAP protein of approximately 71 kDa. Consistent with its predicted localization in the inner mitochondrial membrane, a transmembrane domain (TM) was identified in PaIAP by hydrophobicity plot analysis ( and S2).
Ablation and modification of functional PaIAP does not affect vital functions but extends lifespan.
In order to investigate the role of PaIAP as a component of the mitochondrial quality control system for aging of P. anserina, we generated a strain in which its open reading frame was replaced by a phleomycin resistance cassette (ble).Citation35 The successful gene exchange was confirmed in phleomycin-resistant transformants by DNA gel blot analyses using PaIap- and ble- specific probes ( and B). Consistently, protein gel blot analyses with a specific peptide antibody against PaIAP detected the protein only in mitochondria of the wild type but not in the PaIap deletion strain (). Interestingly, compared with the wild type, the PaIap deletion strain was neither impaired in viability, nor did it show any changes in morphology under standard growth conditions (data not shown).
Since the deletion of Yme1 was previously reported to impair mitochondrial function in yeast,Citation36 we investigated the impact of removing PaIap on energy-consuming processes in P. anserina. First, we determined the growth rates of wild-type cultures and descendants of ΔPaIap and their ability to form fruiting bodies. When incubated at 27°C, the standard growth temperature of P. anserina, the growth rates of the PaIap deletion and that of the wild type were identical (). Also, fertilization of female gametangia of the mutant and the wild type by spermatia (male gametes) of the wild type did not reveal any impairment in fruiting body (perithecia) formation. The number of perithecia formed by the two fertilized strains was virtually identical (). The number of ascospores per perithecium (not shown) and the germination rate of ascospores () did not show any obvious deviations from those of the wild type. Next, we measured oxygen consumption of juvenile mycelia of the wild type and ΔPaIap. In contrast to yeast cells lacking YME1 that are characterized by impaired oxygen uptake,Citation36 no change in oxygen consumption was found in mycelia of the ΔPaIap strain in comparison to wild-type strains ().
Finally, we analyzed the effect of the PaIap deletion on aging, a process that in P. anserina has a strong mitochondrial etiology. Surprisingly, although PaIAP as a component of the mitochondrial protein quality control system was missing, the PaIap deletion strain was characterized by a lifespan that was markedly increased compared with the lifespan of the wild type (). This effect could be fully reversed by the ectopic re-introduction of a single wild-type copy of PaIap into the genome of ΔPaIap (Fig. S3A). Expression of the transgenes was confirmed by protein gel blot analyses (Fig. S3B). The lifespan of two independent revertants, termed PaIap_rev1 and PaIap_rev2, was identical to that of the wild type (). In addition, the growth rate (Fig. S3C) and the morphology returned to wild type in the PaIap_rev strains (data not shown).
In order to validate these findings and to specifically investigate whether or not the absence of the proteolytic activity and thus the impairment in degradation of specific substrates was critical for the observed phenotype of the deletion strain, we next set out to generate strains in which the active site of PaIAP was inactivated. The proteolytic center of i-AAA proteases is known to contain a consensus metal-binding motif HEXXHCitation6 that is located in an α-helix within the homo-oligomeric complex (Fig. S4). From yeast, it is further known that replacement of glutamate 541 with glutamine ablates the proteolytic activity of YME1.Citation7 Changing this amino acid is not expected to affect the overall structure of the protein (Fig. S4). The phenotype of yeast strains carrying this inactivated form of Yme1 resembles the phenotype of YME1 deletion strains.Citation37
We thus changed the base triplet coding for glutamate at position 540 in the presumed consensus metal-binding motif of PaIAP (HEAGH) to glutamine by in vitro mutagenesis and selected two transformants (E540Q). In addition, one strain was selected in which the introduced glutamine was followed by an additional glycine (E540QG). The corresponding genes were introduced into the PaIap deletion strain, and the transformants were verified by Southern and protein gel blot analyses (Fig. S5A–D). The morphology and growth rates of PaIap_E540QG and PaIap_E540Q were indistinguishable from the wild type (Fig. S5E and F). However, the lifespans of the PaIap_E540QG () and the PaIap_E540Q () transformants were increased and resembled the lifespan of the PaIap deletion strain. This suggested that the absence of the proteolytic activity rather than the loss of a chaperone-like activity in the AAA-domain was responsible for the altered aging phenotype of ΔPaIap.
The ΔPaIap strain is sensitive to elevated temperature.
In both, S. cerevisiae and N. crassa, loss of a functional i-AAA protease leads to a temperature-sensitive phenotype. Hence, we next investigated the effect of an increased cultivation temperature on ΔPaIap. Significantly and in contrast to the increased lifespan at 27°C, the lifespan of the PaIap deletion strain was found to be more than 20% reduced in comparison to the wild type, when the strains were grown at an elevated temperature of 37°C (). In other words, while increasing the growth temperature by 10°C resulted in a ∼2.5-fold shorter lifespan for the wild type, it was reduced by ∼5.5-fold in the ΔPaIap strain. At this temperature, both a decreased growth rate as well as a reduction in ascospore germination was observed in ΔPaIap ( and C). Increased temperature was also found to affect female fertility of both the wild-type and the ΔPaIap strains (). Cultures of the wild type and ΔPaIap incubated at 37°C were spermatized and subsequently incubated at 27°C. After two days of growth at 27°C; both strains developed only very few fruiting bodies. However, their number on plates with the spermatized PaIap deletion strain was slightly but significantly lower than on plates with the fertilized wild type. These results uncover a beneficial role of PaIAP for development and lifespan of P. anserina at higher temperature.
Regulation of PaIap during heat stress and effect of its deletion on components of the mitochondrial protein quality control system.
In order to elucidate the role of PaIAP in more detail, we investigated the protein level of PaIAP in mitochondria from heat-stressed wild-type mycelia. Mitochondrial proteins were isolated from mycelium grown under standard conditions at 27°C for three days and from mycelium incubated for two days at 27°C followed by incubation at 37°C for 24 h (). Protein gel blot analyses revealed a more than 6-fold higher abundance of PaIAP in strains subjected to heat stress, suggesting a role of PaIAP in the heat shock response.
Since PaIap is part of the mitochondrial protein quality control system, we next investigated whether other components of this system were affected in the mutant. Specifically, we analyzed the abundance of mitochondrial PaHSP60 and the proteases PaLON and PaCLPP, for which we previously had generated specific antibodies. PaLON and PaCLPP are soluble proteases, and PaHSP60 is a putative chaperone in the mitochondrial matrix. Protein gel blot analyses of mitochondria isolated from mycelium incubated at 27°C revealed a slight but insignificant decrease in PaCLPP and PaHSP60 in the ΔPaIap strain when compared with the wild type (), and the amount of PaLON was unchanged. Remarkably however, upon incubation at 37°C, mitochondria from the PaIap deletion strain exhibited significantly lower amounts of PaCLPP and PaHSP60 than mitochondria from wild-type cultures (). Whether these changes reflected a compensatory response to the PaIap deletion remains unclear but seems possible. A role of the i-AAA protease in mitochondria-to-nucleus signaling has also been postulated in yeast.Citation38
Deletion of PaIap affects the composition of the mitochondrial respiratory chain complexes.
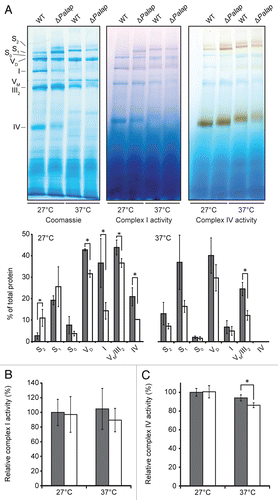
The mitochondrial i-AAA and m-AAA proteases are known to affect the assembly of mitochondrial protein complexes and the degradation of membrane-bound proteins.Citation39 In yeast and the plant Arabidopsis thaliana, mutants lacking the i-AAA protease show a decreased abundance and activity of respiratory chain complexes.Citation10,Citation40 Consequently, we analyzed the oxidative phosphorylation (OXPHOS) complexes of the P. anserina wild-type and the PaIap deletion strains (). Mitochondrial proteins isolated from 14-day-old mycelium of both strains incubated at 27°C or 37°C were solubilized with the mild detergent digitonin and separated by BN-PAGE. Coomassie blue staining of the native complexes revealed the typical composition of the respiratory chain complexes of P. anserina.Citation41 However at 27°C in mitochondria of the PaIap deletion strain, significant changes in the abundances of the individual complexes were observed (). In the mutant strain, the amounts of the monomers of complex I and complex IV and the dimer of complex V (VD) were found to be clearly lower than in the wild type. The monomer of complex V (VM) and the dimer of complex III (III2) could not be separated by BN-PAGE, but the intensity of the band containing these two complexes was also slightly decreased. Compensating these changes, the abundance of supercomplex I1III2IV2 (S2) was markedly increased in the mutants, and the amount of supercomplex I1III2IV1 (S1) also tended to be elevated. This suggested that the slight decrease in the VM + III2 band, in fact, most likely reflected a much stronger decrease of III2 along with an increase in VM compensating the decrease in VD. Taken together, the total abundance of the OXPHOS complexes seemed essentially unchanged, but a shift toward the formation of stable respiratory supercomplexes and a destabilization of complex V dimers was evident. Strikingly, a similar shift was observed upon incubation at 37°C in the wild type as well, along with an overall reduction in the total amounts of the respiratory complexes that was somewhat more pronounced in the ΔPaIap strain. Interestingly, and in contrast to normal temperature, at 37°C the pattern of OXPHOS complexes was rather similar for the wild type and ΔPaIap strain, suggesting a function of supercomplex formation as part of the cellular stress response.
Supporting the notion that the organization but not the total amount of the OXPHOS complexes was altered in the PaIap deletion strain at 27°C, the total in-gel enzyme activities of neither complex I nor of complex IV were found to be changed as compared with the wild type ( and C); i.e., the optical densities of all bands containing mitochondrial complex I (I, S0, S1 and S2) and complex IV (IV, S1 and S2) were similar in mitochondrial extracts from the wild type and ΔPaIap. Moreover, the NADH:HAR and dNADH:DBQ oxidoreductase activities of complex I in mitochondrial membranes of the PaIap deletion strain were unchanged (data not shown). At 37°C, the total in-gel activity of complex IV was slightly reduced in mitochondria of both the wild type and the PaIap deletion strain, with a stronger effect on the deletion strain, while the total in-gel activity of complex I was not significantly affected at all.
Discussion
Mitochondria, the organelles of eukaryotic cells involved in a variety of fundamental processes, are the major source of cellular ROS and, at the same time, the target of these aggressive compounds. Accumulation of mitochondrial damage contributes to cellular degeneration leading to disease and aging.Citation21,Citation22,Citation42 In the past, it has been repeatedly demonstrated that strengthening of ROS scavenging systems or different surveillance pathways can keep biological systems functional and healthy for increased periods of time. For instance, intervening into mitochondrial ROS generation via the overexpression of a gene coding for a mitochondrial methyltransferase (PaMTH1) lowers the time-dependent accumulation of damaged mitochondrial proteins and results in increased stress resistance and healthspan of transgenic strains of P. anserina.Citation43 Consistently, the deletion of PaMth1 was found to lead to accelerated aging.Citation44 Recently, similar effects were obtained when the abundance of components of the mitochondrial protein quality control system were changed by different specific genetic manipulations of P. anserina. In one study, accelerated aging was observed in strains overexpressing a mitochondrial superoxide dismutase gene. This unexpected result could be linked to a reduced abundance of mitochondrial peroxiredoxin (PaPRX), mitochondrial PaHSP60 and PaCLPP.Citation26 In another study, the constitutive overexpression of the gene coding for the mitochondrial matrix protease PaLON was found to lower mitochondrial protein damage, to improve mitochondrial function and to increase healthspan.Citation33
Unexpectedly and counterintuitively to the results of these and other earlier investigations, deletion of PaIap or ablating the proteolytic activity of the corresponding protein led to an increased lifespan under standard growth conditions. At first glance, these results are contradictory to those reported for the S. cerevisiae Yme1 deletion strain that, as compared with the wild type, shows a decreased chronological lifespan.Citation45 However, at this point it needs to be emphasized that the aging process studied in P. anserina is not comparable to chronological aging in yeast, which is a model of aging of post-mitotic cells. Aging processes in P. anserina are dependent on hyphal tip growth and on nuclear divisions and, as such, resemble replicative aging of yeast. It thus will be interesting to see, whether or not replicative aging is affected in the Yme1 deletion strain.
Remarkably, the surprising beneficial effect on lifespan of the PaIap deletion is only observed at lower temperature. Increasing the incubation temperature from 27°C to 37°C disclosed protective functions of PaIAP as they are expected for a component of the protein quality control system. Both, developmental processes, like spore germination and fruiting body formation, and aging are impaired in the PaIap deletion strain. While such a temperature-dependent effect of an i-AAA protease has not been reported in any other system so far, alterations in leaf rosette development and morphology under long- and short-day conditions, respectively, have been demonstrated in strains lacking the PaIAP homolog AtFtsH4 in A. thaliana and have been linked to structural changes in mitochondria and chloroplasts.Citation46 In yeast and in N. crassa, a temperature-dependent function of the i-AAA protease is associated with growth impairments.Citation15,Citation47 At 37°C, Yme1 deletion strains of yeast are unable to grow on non-fermentable carbon sources but do so at 14°C. On rich medium containing glucose or galactose, the mutant shows decreased growth rates at 14°C. Similarly, disruption of Iap-1 in N. crassa results in a decreased growth rate on non-fermentable carbon sources.
Currently, the mechanistic basis of the observed temperature-dependent functions of i-AAA proteases in biological systems is only poorly understood. In yeast, it is known that YME1 is involved in the degradation of membrane-bound proteins like COX2, PHB1 and PHB2.Citation10,Citation12 Interestingly, the N. crassa homolog, IAP-1, is unable to degrade COX2 and PHB1 when expressed in the yeast Yme1 deletion strain, pointing to subtle differences in the substrate specificities of i-AAA proteases from different organisms.Citation16 In P. anserina, initial experiments using antibodies against the yeast proteins COX2, PHB1 and PHB2 failed to identify these proteins as substrates of PaIAP (data not shown).
In addition to the degradation of membrane-bound proteins, YME1 was also found to be active in the degradation of proteins in the intermembrane space. Defects in YME1 activity were reported to lead to an accumulation of phosphatidylethanolamine and were linked to impaired degradation of PDS1, UPS1 and UPS2, proteins active in mitochondrial phospholipid metabolism.Citation14,Citation48 Although not explicitly demonstrated yet, an impact of this metabolism and the composition of the membranes on the assembly and/or stability of membrane-embedded protein complexes may be of general relevance and, at least in part, may be controlled by i-AAA proteases. The observed decreased activity of respiratory complexes IV and V in an A. thaliana AtFtsH4 deletion mutants may be an example for such a scenario.Citation40 Also, the various changes in respiratory complex and supercomplex organization we observed in the PaIap deletion strain of P. anserina support such an indirect effect of PaIAP function. However, at this time, we cannot exclude more direct effects of i-AAA proteases on the observed changes in protein complex composition. The shift toward the formation of supercomplexes observed in the PaIap deletion strain seems to be part of a stress response, as the wild type showed a similar shift when incubated at 37°C. This suggests that the loss of PaIAP causes a mild stress in P. anserina and results in a longevity-promoting hormetic effect at 27°C. For the PaIap deletion strain, the drawback of this reorganization of the OXPHOS complexes is that, while the wild type is still able to react to a new stress situation (37°C), the strains lacking PaIAP cannot induce their stress response any further. Thus, physiological functions of the PaIap deletion strain are more affected at 37°C.
In this study, we demonstrated an essential role of the proteolytic activity for wild type-specific development and aging. Although we found a decrease in abundance of PaHSP60 and PaCLPP in mitochondria of the PaIap deletion strain grown at elevated temperature, it is rather unlikely that these two proteins are direct interaction partners of PaIAP. Instead, PaIAP may affect the abundance of these proteins by other processes, like the translocation of proteins across the outer mitochondrial membrane or via the generation of peptides acting in mitochondria-to-nucleus signaling leading to a readjustment of mitochondrial gene expression. Such functions have been attributed to YME1 in yeast.Citation13,Citation38 Whatever the pathways are, the changes in PaHSP60 and PaCLPP abundance most certainly contribute to the observed impairments of the PaIap deletion strain grown at 37°C.
Overall, our study identified a novel and remarkable role of a component of the mitochondrial protein quality control system on aging and development. The surprising effect on the phenotype of the PaIap deletion mutant, with an increased lifespan at 27°C and a shortened lifespan at an incubation temperature of 37°C, identified a differential function of a single protein in aging. This protease, as well as other yet-to-be-analyzed proteins, appears to be adapted to the changing conditions in nature with regularly changing temperatures. Our analysis also demonstrates that the investigations under well-selected laboratory conditions are prone to miss important parameters, pathways and mechanisms that are relevant for complex processes like aging and development, as they have evolved over the long time of evolution of any given species. It will be interesting to see whether temperature-associated effects also play a role in higher biological systems, including humans. Although humans are homeothermic, body temperature does increase at least under certain conditions, like in the skin during sun exposure or during febrile diseases.Citation49 At least for human cell cultures, it has been demonstrated that a mild heat stress (≥ 38.5°C) is already sufficient to activate heat shock factor HSF-1, which leads to the upregulation of target genes like Hsp72 coding for a chaperone that is translocated into the nucleus upon heat stress.Citation50,Citation51
Materials and Methods
Strains and culture conditions.
In this study, the wild-type “s”Citation52 strain of P. anserina was used. The deletion strain ΔPaIap was generated in the genetic background of this wild-type strain. The PaIap wild-type gene and its modified versions were reintroduced into ΔPaIap. All strains were grown on M2 medium as described in reference Citation53. Cultures of defined age were obtained as described in reference Citation30. Mitochondria were isolated from mycelium of 11-day-old wild type and ΔPaIap grown on M2 agar plates for three days at 27°C. Heat-stressed mycelium was incubated at 27°C for only two days following a 24 h exposure to 37°C.
Growth rate and lifespan determination.
Lifespan and growth rate on M2 medium were determined as described in reference Citation43. To determine the lifespan at 37°C, the monokaryotic isolates were placed on race tubes with M2 and incubated at 27°C for two days. Subsequently, the strains were transferred to 37°C and incubated until senescence. The time period of linear growth was used to measure the growth rate in centimeters per day.
Spore germination and growth of the cultures was determined for dikaryotic isolates. Dikaryotic spores of the wild type and ΔPaIap were incubated on agar plates containing corn meal extract (BMM) complete medium at 27°C or 37°C in the dark.Citation54 After three days, germination was determined macroscopically.
Fertility analysis.
To assess female fertility, freshly isolated monokaryotic isolates of the wild type or ΔPaIap of both mating types were allowed to overgrow the surface of M2 agar plates at 27°C under constant light. To determine the ability to form fruiting bodies after heat stress, a set of strains was transferred to 37°C after eight days at 27°C. After five days at 37°C, the strains were spermatized. Spermatia of a wild type with the opposite mating type were harvested by flooding the plates with 5 mL of sterile water. The suspension was diluted 1:1 with sterile H2O, and six drops à 300 µL were pipetted onto mycelium of each wild type and ΔPaIap of the opposite mating type. After 5 min, the drops were removed carefully, and the strains were incubated for another two days at 27°C. Subsequently, the number of perithecia was counted. The resulting values were divided by the area of the drop. The number of perithecia developing on plates overgrown with the wild-type “s” were set to 100% fertility.
Construction of the deletion strain ΔPaIap.
Deletion of PaIap in the wild-type strain ‘s’ (mating type -) was performed as described in reference Citation35. Briefly, small flanking regions of PaIap were amplified using the 5′-flank oligonucleotides pKO4-Yme1-1 (5′-TAG GTA CCC CGC ATT CCT TTT TCT AG-3′) and pKO4-Yme1-2 (5′-GGA AGC TTG GTC GTC GAG A-3′), introducing KpnI and HindIII restriction sites. To amplify the 3′-flank region, the oligonucleotides pKO4-Yme1-3 (5′-GTA CTA GTG GGT TGG TTG GGT GTG TT-3′) and pKO4-Yme1-4 (5′-TAG CGG CCG CAC TCA TCT CAG CAG CCA T-3′) were used, introducing BcuI and NotI restriction sites. Homologous recombination of PaIap was achieved as described in reference Citation30, using a cosmid from the cosmid library of P. anserina wild-type strain ‘s’Citation55 containing the PaIap locus.
Construction of PaIap_rev and PaIap_E540Q(G).
To reintroduce the PaIap wild-type gene into the genome of ΔPaIap, the open reading frame (ORF) encoding for PaIap including a ∼1,000 bp promoter region and a ∼500 bp terminator region was amplified from the cosmid containing the PaIap locus by PCR with Phusion polymerase (Finnzymes). The oligonucleotides PaIap-KpnI-for (5′-ATG GTA CCG AGA TGG CTG TAG AGG CG-3′) and PaIap-EcoRV-rev (5′-CTG ATA TCC AAG TGT TGC CCT CTC TGC-3′) were used to introduce KpnI and EcoRV restriction sites. The generated PCR product was digested with KpnI and EcoRV and cloned into the KpnIEcoRV-digested plasmid pKO7.Citation43 The nucleotide sequence was verified by sequencing (SRD GmbH), and the vector pPaIap_rev was transformed into P. anserina protoplasts of the strain ΔPaIap. The strains containing the ORF of PaIap were selected by hygromycin B (Hyg) resistance.
To express a modified form of PaIap in P. anserina, a point mutation was introduced in the ORF of wild-type PaIap. Using pPaIap_rev as template, the oligonucleotides PaIap_MunI_for (5′-TTC AAT TGC CGC TCA GCG CAA CGG-3′) and PaIap_PvuII_rev (5′-TTC AGC TGG TGG TAG GCA GTC ATC TC-3′) were used to introduce MunI and PvuII restriction sites. The oligonucleotides PaIap_EheI_for (5′-TTT GGC GCC GGC CAC GCC CTT GTT G-3′) and the above-mentioned PaIap-EcoRV-rev were used to introduce EheI and EcoRV restriction sites. The two fragments were cloned into the digested pPaIap_rev to generate the vector pPaIap_E540Q. The modified nucleotide sequence was verified by sequencing (SRD GmbH) and the vector was transformed into ΔPaIap protoplasts. Transformants were selected on Hyg-containing medium.
DNA gel blot analysis.
Isolation of total DNA, DNA digestion, gel electrophoresis and DNA gel blotting were performed as described in reference Citation30.
Isolation of mitochondria from P. anserina.
Mitochondria of P. anserina cultures were isolated by differential centrifugation and purified using a 20–50% discontinuous sucrose gradient as previously described except for a final centrifugation at 100,000 g for 15 min.Citation43
SDS-PAGE and protein gel blot analyses.
SDS-PAGE and protein gel blot analyses were performed as described in reference Citation26. 30 µg or 100 µg of mitochondrial protein were analyzed. The antibody Anti-PaIAP (1:5,000 dilution) against a PaIAP-specific synthetic peptide was newly generated ([H]-KAENQKARFSDVHGC-[OH]; Sigma-Genosys).
BN-PAGE.
BN-PAGE was performed as previously described in reference Citation56. 100 µg of mitochondrial protein was solubilized using a digitonin/protein ratio of 3 g/g. BN-PAGE and in-gel enzyme activity were performed as described in reference Citation33. In-gel quantification of complex I and IV was performed densitometrically. The optical densities of all bands presenting complex I activity (I, S0, S1, S2) and complex IV activity (IV, S1, S2) were evaluated. The values of the relative activity were calculated as a percentage of the mean volume determined for the wild type at 27°C.
Oxygen consumption.
Respiration rates of P. anserina wild-type and ΔPaIap strains were determined using a Clark-type electrode (Rank Bros.) as previously described in reference Citation57.
Complex I oxidoreductase activity.
NADH:HAR and dNADH:DBQ oxidoreductase activities of mitochondrial membranes isolated from P. anserina wild-type and ΔPaIap strains were analyzed spectrophotometrically using a SpectraMax Plus384 microplate reader (Molecular Devices) at 27°C as detailed previously in reference Citation58.
Alignments and protein modeling.
ClustalW was used for the alignment of the amino acid sequences (www.ch.embnet.org/software/ClustalW.html). To generate structural models of proteins, the alignments were processed by Swiss-Model (www.swissmodel.expasy.org). UCSF Chimera was used to visualize the molecular graphics (version 1.5.3, www.cgl.ucsf.edu/chimera).
Statistical analyses.
For statistical analyses of protein gel blot and BN-PAGE data, the Students t-test (two-tailed) was used. For statistical analyses of growth rates, female fertility, germination of ascospores, oxygen consumption and lifespan, the Wilcoxon test (two-tailed) was used. p < 0.05 was considered statistically significant. The standard deviation is shown in all graphs.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Figures and Tables
Figure 1 Domain structure and homology of i-AAA proteases. The protein sequence of i-AAA proteases from P. anserina (PaIAP), S. cerevisiae (YME1), N. crassa (IAP-1) and H. sapiens (YME1L1) are composed of a mitochondrial targeting sequence (MTS), one transmembrane domain (TM), the conserved AAA domain, including the Walker A and B motif and the second region of homology, and a metal binding motif (MB) representing the catalytic center. The amino acid sequence of the proteolytic domain of the P. anserina protein is indicated (HEAGH). The length of the amino acid sequence of the protein varies from 738 amino acids in N. crassa to 773 amino acids in H. sapiens. Similarities of the P. anserina protein to the other proteases are 59%, 78% and 45%, respectively.
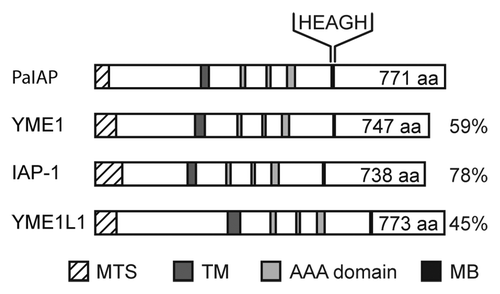
Figure 2 Deletion of PaIap does not change the phenotype of ΔPaIap at 27°C but increases lifespan. (A) DNA gel blot analyses of BglII digested DNA from the wild-type ‘s’ (WT) and the PaIap deletion strain (ΔPaIap). The PaIap-specific probe (upper part) detects the ∼4.3 kbp fragment containing PaIap only in the genome of the wild-type. The gene encoding the phleomycin resistance gene (ble) is present as a ∼2.4 kbp fragment only in ΔPaIap (lower part). (B) Location of the sequence used as a PaIap-specific probe (line). The genomic sequences flanking PaIap are indicated by punctuation. (C) Protein gel blot analyses of mitochondrial proteins from the WT and the PaIap deletion. The PaIAP-specific antibody detects PaIAP only in the sample of the WT (upper part). PaPORIN (PaPOR) was used as loading control (lower part). (D) Growth rates of the WT (gray; n = 71) and the PaIap deletion strain (white; n = 73; p = 5E−1). (E) Fruiting body formation of the PaIap deletion (n = 12) strain and the WT (n = 12; p = 9E−1) after spermatization. (F) Germination rate of ascospores from perithecia of fertilized ΔPaIap (n = 19) and WT cultures (n = 19). (G) Oxygen consumption of mycelium of juvenile WT (n = 4) and ΔPaIap strains (n = 4; p = 5.6E−1). (H) Lifespan of ΔPaIap (n = 73; p = 1.8E−16), the WT (n = 71) and of the two revertants PaIap_rev1 (n = 59) and PaIap_rev2 (n = 40).
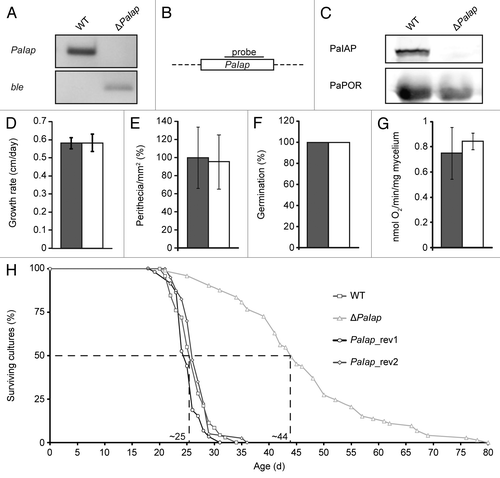
Figure 3 Manipulation of the proteolytic activity of PaIAP [PaIAP_E540Q(G)]. (A) Lifespans of PaIap_E540QG (E540QG; n = 30; p = 1.7E−7), the WT (n = 21) and of ΔPaIap (n = 34; p = 6.9E−10). (B) Lifespans of PaIap_E540Q1 (E540Q1; n = 35; p = 7.1E−11), of PaIap_E540Q2 (E540Q2; n = 27; p = 7.4E−10), the WT (n = 24) and of ΔPaIap (n = 24; p = 2.1E−9).
![Figure 3 Manipulation of the proteolytic activity of PaIAP [PaIAP_E540Q(G)]. (A) Lifespans of PaIap_E540QG (E540QG; n = 30; p = 1.7E−7), the WT (n = 21) and of ΔPaIap (n = 34; p = 6.9E−10). (B) Lifespans of PaIap_E540Q1 (E540Q1; n = 35; p = 7.1E−11), of PaIap_E540Q2 (E540Q2; n = 27; p = 7.4E−10), the WT (n = 24) and of ΔPaIap (n = 24; p = 2.1E−9).](/cms/asset/20a24c6d-c3c1-4d97-bf3f-a0c13723b56e/kccy_a_10918560_f0003.gif)
Figure 4 Vital functions of the PaIap deletion strain are impaired at 37°C. (A) Lifespans of ΔPaIap (n = 45) and the WT (n = 45; p = 4.8E−9). (B) Growth rates of ΔPaIap (n = 45) and the WT (n = 45; p = 1.4E−2). (C) Germination of ascospores after incubation for three days at 37°C (ΔPaIap n = 20, WT n = 20). (D) Fruiting body formation of ΔPaIap (n = 8) and the WT (n = 8; p = 2E−2).
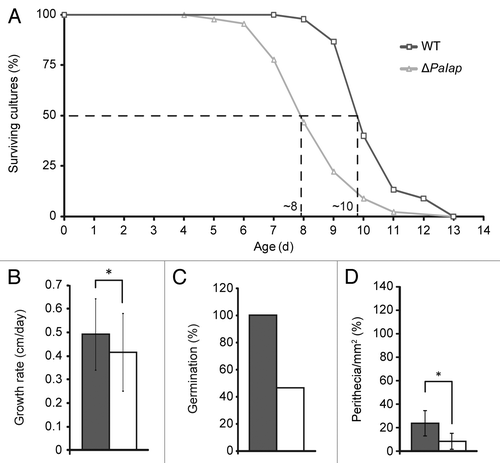
Figure 5 PaIAP is heat-inducible, and its loss affects other components of the mitochondrial quality control system. (A) Protein gel blot analyses of mitochondrial proteins of 14-day-old WT cultures (n = 4) after incubation at 27°C or for 24 h at 37°C. The protein levels of PaIAP were normalized to the level of PaPOR and the PaIAP amount at 27°C set to 1. Twenty-four h at 37°C led to a 6.7-fold increased protein amount (p = 1.8E−5). PaPOR serves as loading control. Protein gel blot analyses of mitochondrial proteins of 14-day-old WT (27°C: n = 3; 24 h 37°C: n = 4) and ΔPaIap cultures (n = 4) after (B) incubation at 27°C and (C) 24 h of incubation at 37°C. The protein levels of PaLON, PaCLPP and PaHSP60 were normalized to the level of PaPOR, and the protein amount present in the WT strains was set to 1. At 27°C, there are no significant differences in the abundances of PaLON (p = 8E−1), PaCLPP (p = 6E−2) and PaHSP60 (p = 4E−1). Incubation of the cultures for 24 h at 37°C leads to significant differences in the protein amounts of PaCLPP (p = 4E−2) and PaHSP60 (p = 1E−2) between the WT and ΔPaIap strains. In contrast, the level of PaLON (p = 5E−1) is not changed.
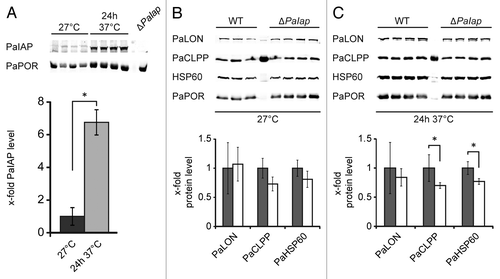
Figure 6 Composition and activity of mitochondrial respiratory chain complexes. (A) BN-PAGE of mitochondrial proteins from mycelium of 14-day-old WT (n = 3) and ΔPaIap (n = 3) cultures incubated at 27°C and 24 h at 37°C prior to protein isolation. The Coomassie blue staining of the complexes was quantified densitometrically and normalized using the total protein staining of each lane. Amounts of the complexes IV (p = 1.2E−2), VM/III2 (p = 3.6E−2), I (p = 3.3E−2), VD (p = 3.5E−4) and of the supercomplexes S0 (I1III2IV0; p = 1.8E−1), S1 (I1III2IV1; p = 3.2E−1) and S2 (I1III2IV2; p = 2.6E−2) at 27°C. Heat stress leads to temperature-dependent changes in the abundances of the OXPHOS complexes. The amount of complex VM/III2 is significantly different (p = 4.8E−3). In-gel activities of complex I and IV. (B) In-gel quantification of the total complex I activity of the WT (n = 3) and ΔPaIap (n = 3; p = 8E−1) at 27°C and of the WT (n = 3) and ΔPaIap (n = 3; p = 4E−1) at 37°C. (C) In-gel quantification of total complex IV activity of the WT (n = 3) and ΔPaIap (n = 3; p = 8E−1) at 27°C. At 37°C, total complex IV activity of ΔPaIap (n = 3) is slightly decreased compared with the WT (n = 3; p = 3E−2).
Additional material
Download Zip (5.3 MB)Acknowledgments
We thank T. Langer (University of Cologne) for providing COX2, PHB1 and PHB2 antibodies. The project was supported by the European Commission (FP6-518230: Proteomage), the Deutsche Forschungsgemeinschaft (EXC115 and SFB815/Z1), the BMBF (0315584A: GerontoMitoSys and 01GM0863: mitoNET) and the Förderfonds der Goethe-Universität. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization and Informatics at the University of California, San Francisco (supported by NIH P41 RR001081). The authors thank H. Schägger for valuable advice and I. Siebels for excellent technical assistance.
References
- Brand MD. The sites and topology of mitochondrial superoxide production. Exp Gerontol 2010; 45:466 - 472; PMID: 20064600; http://dx.doi.org/10.1016/j.exger.2010.01.003
- Luce K, Weil AC, Osiewacz HD. Tavernarakis N. Mitochondrial protein quality control systems in aging and disease. Protein metabolism and homeostasis in aging 2010; New York Springer US 108 - 125
- Tatsuta T, Langer T. Quality control of mitochondria: protection against neurodegeneration and ageing. EMBO J 2008; 27:306 - 314; PMID: 18216873; http://dx.doi.org/10.1038/sj.emboj.7601972
- Osiewacz HD. Mitochondrial quality control in aging and lifespan control of the fungal aging model Podospora anserina. Biochem Soc Trans 2011; 39:1488 - 1492; PMID: 21936839
- Juhola MK, Shah ZH, Grivell LA, Jacobs HT. The mitochondrial inner membrane AAA metalloprotease family in metazoans. FEBS Lett 2000; 481:91 - 95; PMID: 10996302; http://dx.doi.org/10.1016/S0014-5793(00)01989-X
- Leonhard K, Herrmann JM, Stuart RA, Mannhaupt G, Neupert W, Langer T. AAA proteases with catalytic sites on opposite membrane surfaces comprise a proteolytic system for the ATP-dependent degradation of inner membrane proteins in mitochondria. EMBO J 1996; 15:4218 - 4229; PMID: 8861950
- Leonhard K, Stiegler A, Neupert W, Langer T. Chaperone-like activity of the AAA domain of the yeast Yme1 AAA protease. Nature 1999; 398:348 - 351; PMID: 10192337; http://dx.doi.org/10.1038/18704
- Thorsness PE, Fox TD. Nuclear mutations in Saccharomyces cerevisiae that affect the escape of DNA from mitochondria to the nucleus. Genetics 1993; 134:21 - 28; PMID: 8514129
- Campbell CL, Tanaka N, White KH, Thorsness PE. Mitochondrial morphological and functional defects in yeast caused by yme1 are suppressed by mutation of a 26S protease subunit homologue. Mol Biol Cell 1994; 5:899 - 905; PMID: 7803857
- Pearce DA, Sherman F. Degradation of cytochrome oxidase subunits in mutants of yeast lacking cytochrome c and suppression of the degradation by mutation of yme1. J Biol Chem 1995; 270:20879 - 20882; PMID: 7673107; http://dx.doi.org/10.1074/jbc.270.36.20879
- Augustin S, Nolden M, Müller S, Hardt O, Arnold I, Langer T. Characterization of peptides released from mitochondria: evidence for constant proteolysis and peptide efflux. J Biol Chem 2005; 280:2691 - 2699; PMID: 15556950; http://dx.doi.org/10.1074/jbc.M410609200
- Kambacheld M, Augustin S, Tatsuta T, Müller S, Langer T. Role of the novel metallopeptidase Mop112 and saccharolysin for the complete degradation of proteins residing in different subcompartments of mitochondria. J Biol Chem 2005; 280:20132 - 20139; PMID: 15772085; http://dx.doi.org/10.1074/jbc.M500398200
- Rainey RN, Glavin JD, Chen HW, French SW, Teitell MA, Koehler CM. A new function in translocation for the mitochondrial i-AAA protease Yme1: import of polynucleotide phosphorylase into the intermembrane space. Mol Cell Biol 2006; 26:8488 - 8497; PMID: 16966379; http://dx.doi.org/10.1128/MCB.01006-06
- Nebauer R, Schuiki I, Kulterer B, Trajanoski Z, Daum G. The phosphatidylethanolamine level of yeast mitochondria is affected by the mitochondrial components Oxa1p and Yme1p. FEBS J 2007; 274:6180 - 6190; PMID: 17976194; http://dx.doi.org/10.1111/j.1742-4658.2007.06138.x
- Klanner C, Prokisch H, Langer T. MAP-1 and IAP-1, two novel AAA proteases with catalytic sites on opposite membrane surfaces in mitochondrial inner membrane of Neurospora crassa. Mol Biol Cell 2001; 12:2858 - 2869; PMID: 11553723
- Graef M, Seewald G, Langer T. Substrate recognition by AAA+ ATPases: Distinct substrate binding modes in the ATP-dependent protease Yme1 of the mitochondrial intermembrane space. Mol Cell Biol 2007; 27:2476 - 2485; PMID: 17261594; http://dx.doi.org/10.1128/MCB.01721-06
- Shah ZH, Hakkaart GA, Arku B, de Jong L, van der Spek H, Grivell LA, et al. The human homologue of the yeast mitochondrial AAA metalloprotease Yme1p complements a yeast yme1 disruptant. FEBS Lett 2000; 478:267 - 270; PMID: 10930580; http://dx.doi.org/10.1016/S0014-5793(00)01859-7
- Aldridge JE, Horibe T, Hoogenraad NJ. Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements. PLoS ONE 2007; 2:874; PMID: 17849004; http://dx.doi.org/10.1371/journal.pone.0000874
- Edgar D, Trifunovic A. The mtDNA mutator mouse: Dissecting mitochondrial involvement in aging. Aging (Albany NY) 2009; 1:1028 - 1032; PMID: 20157586
- Hur JH, Cho J, Walker DW. Aging: Dial M for Mitochondria. Aging (Albany NY) 2010; 2:69 - 73; PMID: 20228940
- Osiewacz HD. Mitochondrial functions and aging. Gene 2002; 286:65 - 71; PMID: 11943461; http://dx.doi.org/10.1016/S0378-1119(01)00804-6
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging and cancer: a dawn for evolutionary medicine. Annu Rev Genet 2005; 39:359 - 407; PMID: 16285865; http://dx.doi.org/10.1146/annurev.genet.39.110304.095751
- Osiewacz HD, Hermanns J, Marcou D, Triffi M, Esser K. Mitochondrial DNA rearrangements are correlated with a delayed amplification of the mobile intron (plDNA) in a long-lived mutant of Podospora anserina. Mutat Res 1989; 219:9 - 15; PMID: 2911274
- Soerensen M, Gredilla R, Müller-Ohldach M, Werner A, Bohr VA, Osiewacz HD, et al. A potential impact of DNA repair on ageing and lifespan in the ageing model organism Podospora anserina: decrease in mitochondrial DNA repair activity during ageing. Mech Ageing Dev 2009; 130:487 - 496; PMID: 19486911; http://dx.doi.org/10.1016/j.mad.2009.05.003
- Gredilla R, Grief J, Osiewacz HD. Mitochondrial free radical generation and lifespan control in the fungal aging model Podospora anserina. Exp Gerontol 2006; 41:439 - 447; PMID: 16530367; http://dx.doi.org/10.1016/j.exger.2006.01.010
- Zintel S, Schwitalla D, Luce K, Hamann A, Osiewacz HD. Increasing mitochondrial superoxide dismutase abundance leads to impairments in protein quality control and ROS scavenging systems and to lifespan shortening. Exp Gerontol 2010; 45:525 - 532; PMID: 20080171; http://dx.doi.org/10.1016/j.exger.2010.01.006
- Zintel S, Bernhardt D, Rogowska-Wrzesinska A, Osiewacz HD. PaCATB, a secreted catalase protecting Podospora anserina against exogenous oxidative stress. Aging (Albany NY) 2011; 3:768 - 781; PMID: 21865610
- Stumpferl SW, Stephan O, Osiewacz HD. Impact of a disruption of a pathway delivering copper to mitochondria on Podospora anserina metabolism and life span. Eukaryot Cell 2004; 3:200 - 211; PMID: 14871950; http://dx.doi.org/10.1128/EC.3.1.200-211.2004
- Scheckhuber CQ, Erjavec N, Tinazli A, Hamann A, Nyström T, Osiewacz HD. Reducing mitochondrial fission results in increased life span and fitness of two fungal ageing models. Nat Cell Biol 2007; 9:99 - 105; PMID: 17173038; http://dx.doi.org/10.1038/ncb1524
- Brust D, Daum B, Breunig C, Hamann A, Kühlbrandt W, Osiewacz HD. Cyclophilin D links programmed cell death and organismal aging in Podospora anserina. Aging Cell 2010; 9:761 - 775; PMID: 20626725; http://dx.doi.org/10.1111/j.1474-9726.2010.00609.x
- Hamann A, Brust D, Osiewacz HD. Deletion of putative apoptosis factors leads to lifespan extension in the fungal ageing model Podospora anserina. Mol Microbiol 2007; 65:948 - 958; PMID: 17627766; http://dx.doi.org/10.1111/j.1365-2958.2007.05839.x
- Osiewacz HD. Regulation of the mitochondrial transition pore: impact on mammalian aging. Aging (Albany NY) 2011; 3:10 - 11; PMID: 21248375
- Luce K, Osiewacz HD. Increasing organismal healthspan by enhancing mitochondrial protein quality control. Nat Cell Biol 2009; 11:852 - 858; PMID: 19543272; http://dx.doi.org/10.1038/ncb1893
- Osiewacz HD, Brust D, Hamann A, Kunstmann B, Luce K, Müller-Ohldach M, et al. Mitochondrial pathways governing stress resistance, life and death in the fungal aging model Podospora anserina. Ann NY Acad Sci 2010; 1197:54 - 66; PMID: 20536834; http://dx.doi.org/10.1111/j.1749-6632.2010.05190.x
- Hamann A, Krause K, Werner A, Osiewacz HD. A two-step protocol for efficient deletion of genes in the filamentous ascomycete Podospora anserina. Curr Genet 2005; 48:270 - 275; PMID: 16160832; http://dx.doi.org/10.1007/s00294-005-0018-1
- Campbell CL, Thorsness PE. Escape of mitochondrial DNA to the nucleus in yme1 yeast is mediated by vacuolar-dependent turnover of abnormal mitochondrial compartments. J Cell Sci 1998; 111:2455 - 2464; PMID: 9683639
- Weber ER, Hanekamp T, Thorsness PE. Biochemical and functional analysis of the YME1 gene product, an ATP and zinc-dependent mitochondrial protease from S. cerevisiae. Mol Biol Cell 1996; 7:307 - 317; PMID: 8688560
- Arnold I, Wagner-Ecker M, Ansorge W, Langer T. Evidence for a novel mitochondria-to-nucleus signalling pathway in respiring cells lacking i-AAA protease and the ABC-transporter Mdl1. Gene 2006; 367:74 - 88; PMID: 16403607; http://dx.doi.org/10.1016/j.gene.2005.09.044
- Leonhard K, Guiard B, Pellecchia G, Tzagoloff A, Neupert W, Langer T. Membrane protein degradation by AAA proteases in mitochondria: extraction of substrates from either membrane surface. Mol Cell 2000; 5:629 - 638; PMID: 10882099; http://dx.doi.org/10.1016/S1097-2765(00)80242-7
- Kolodziejczak M, Gibala M, Urantowka A, Janska H. The significance of Arabidopsis AAA proteases for activity and assembly/stability of mitochondrial OXPHOS complexes. Physiol Plant 2007; 129:135 - 142; http://dx.doi.org/10.1111/j.1399-3054.2006.00835.x
- Krause F, Scheckhuber CQ, Werner A, Rexroth S, Reifschneider NH, Dencher NA, et al. Supramolecular organization of cytochrome c oxidase- and alternative oxidase-dependent respiratory chains in the filamentous fungus Podospora anserina. J Biol Chem 2004; 279:26453 - 26461; PMID: 15044453; http://dx.doi.org/10.1074/jbc.M402756200
- Harman D. Free radical theory of aging: dietary implications. Am J Clin Nutr 1972; 25:839 - 843; PMID: 5046729
- Kunstmann B, Osiewacz HD. Overexpression of an S-adenosylmethionine-dependent methyltransferase leads to an extended lifespan of Podospora anserina without impairments in vital functions. Aging Cell 2008; 7:651 - 662; PMID: 18616635; http://dx.doi.org/10.1111/j.1474-9726.2008.00412.x
- Kunstmann B, Osiewacz HD. The S-adenosylmethionine dependent O-methyltransferase PaMTH1: a longevity assurance factor protecting Podospora anserina against oxidative stress. Aging (Albany NY) 2009; 1:328 - 334; PMID: 20157520
- Palermo V, Falcone C, Mazzoni C. Apoptosis and aging in mitochondrial morphology mutants of S. cerevisiae. Folia Microbiol (Praha) 2007; 52:479 - 483; PMID: 18298044; http://dx.doi.org/10.1007/BF02932107
- Gibala M, Kicia M, Sakamoto W, Gola EM, Kubrakiewicz J, Smakowska E, et al. The lack of mitochondrial AtFtsH4 protease alters Arabidopsis leaf morphology at the late stage of rosette development under short-day photoperiod. Plant J 2009; 59:685 - 699; PMID: 19453455; http://dx.doi.org/10.1111/j.1365-313X.2009.03907.x
- Thorsness PE, White KH, Fox TD. Inactivation of YME1, a member of the ftsH-SEC18-PAS1-CDC48 family of putative ATPase-encoding genes, causes increased escape of DNA from mitochondria in Saccharomyces cerevisiae. Mol Cell Biol 1993; 13:5418 - 5426; PMID: 8355690
- Potting C, Wilmes C, Engmann T, Osman C, Langer T. Regulation of mitochondrial phospholipids by Ups1/PRELI-like proteins depends on proteolysis and Mdm35. EMBO J 2010; 29:2888 - 2898; PMID: 20657548; http://dx.doi.org/10.1038/emboj.2010.169
- Chen Z, Seo JY, Kim YK, Lee SR, Kim KH, Cho KH, et al. Heat modulation of tropoelastin, fibrillin-1 and matrix metalloproteinase-12 in human skin in vivo. J Invest Dermatol 2005; 124:70 - 78; PMID: 15654955; http://dx.doi.org/10.1111/j.0022-202X.2004.23550.x
- Tulapurkar ME, Asiegbu BE, Singh IS, Hasday JD. Hyperthermia in the febrile range induces HSP72 expression proportional to exposure temperature but not to HSF-1 DNA-binding activity in human lung epithelial A549 cells. Cell Stress Chaperones 2009; 14:499 - 508; PMID: 19221897; http://dx.doi.org/10.1007/s12192-009-0103-3
- Ellis S, Killender M, Anderson RL. Heat-induced alterations in the localization of HSP72 and HSP73 as measured by indirect immunohistochemistry and immunogold electron microscopy. J Histochem Cytochem 2000; 48:321 - 332; PMID: 10681386; http://dx.doi.org/10.1177/002215540004800302
- Rizet G. Sur l'impossibilité d'obtenir la multiplication végétative ininterrompue illimitée de l'ascomycète Podospora anserina. C R Acad Sci Paris 1953; 237:838 - 840; PMID: 13107134
- Müller-Ohldach M, Brust D, Hamann A, Osiewacz HD. Overexpression of PaParp encoding the poly(ADP-ribose) polymerase of Podospora anserina affects organismal aging. Mech Ageing Dev 2011; 132:33 - 42; PMID: 21145908; http://dx.doi.org/10.1016/j.mad.2010.11.003
- Esser K. King RC. Podospora anserina. Handbook of Genetics 1974; New York Plenum Press 531 - 551
- Osiewacz HD. A versatile shuttle cosmid vector for the efficient construction of genomic libraries and for the cloning of fungal genes. Curr Genet 1994; 26:87 - 90; PMID: 7954902; http://dx.doi.org/10.1007/BF00326309
- Wittig I, Braun HP, Schägger H. Blue native PAGE. Nat Protoc 2006; 1:418 - 428; PMID: 17406264; http://dx.doi.org/10.1038/nprot.2006.62
- Borghouts C, Werner A, Elthon T, Osiewacz HD. Copper-modulated gene expression and senescence in the filamentous fungus Podospora anserina. Mol Cell Biol 2001; 21:390 - 399; PMID: 11134328; http://dx.doi.org/10.1128/MCB.21.2.390-399.2001
- Tocilescu MA, Fendel U, Zwicker K, Kerscher S, Brandt U. Exploring the ubiquinone binding cavity of respiratory complex I. J Biol Chem 2007; 282:29514 - 29520; PMID: 17681940; http://dx.doi.org/10.1074/jbc.M704519200