Abstract
In mammalian cells, microRNAs regulate the expression of target mRNAs generally by reducing their stability and/or translation, and thereby control diverse cellular processes such as senescence. We recently reported the differential abundance of microRNAs in young (early-passage, proliferating) relative to senescent (late-passage, non-proliferating) WI-38 human diploid fibroblasts. Here we report that the levels of the vast majority of mRNAs were unaltered in senescent compared to young WI-38 cells, while overall mRNA translation was potently reduced in senescent cells. Downregulation of Dicer or Drosha, two major enzymes in microRNA biogenesis, lowered microRNA levels, but, unexpectedly, it also reduced global translation. While a reduction in Dicer levels markedly enhanced cellular senescence, reduction of Drosha levels did not, suggesting that the Drosha/Dicer effects on translation may be independent of senescence, and further suggesting that microRNAs may directly or indirectly enhance mRNA translation in WI-38 cells. We discuss possible scenarios through which Dicer/Drosha/microRNAs could enhance translation.
Introduction
microRNAs are small (∼22 nt long), non-coding RNA molecules that regulate gene expression post-transcriptionally.Citation1–Citation3 The primary microRNA transcript is processed by the RNase III endoribonuclease Drosha to generate a ∼70 nt hairpin-loop precursor (pre-) microRNA. The pre-microRNA is then exported to the cytoplasm, where another RNase III, Dicer, cleaves the loop and the mature single-stranded microRNA is assembled into the RNA-induced silencing complexes (RISC). This complex can target specific mRNAs, resulting either in translational repression or decreased mRNAstability.Citation4–Citation6 However, in some cases microRNAs can enhance mRNA translation; for example, miR-10a was found to bind the 5′UTR of ribosomal protein mRNAs and enhanced their translation,Citation7 and some microRNAs were shown to switch from translation repression to promotion in a cell cycle-dependent manner.Citation8 Several studies have indicated that microRNA levels control cell function in a number of cell types (e.g., immune cellsCitation9 and stem cellsCitation10), cellular processes (e.g., apoptosisCitation11 and senescenceCitation12) and influence numerous diseases such as cancerCitation11,Citation13,Citation14 and neurodegeneration.Citation15
After numerous rounds of division, cells can reach a state known as replicative senescence, in which they cease to divide but remain metabolically active.Citation16,Citation17 Several microRNAs have been reported to be differentially expressed in senescent cells compared to young, proliferating cells. For example, microRNAs miR-146a and miR-146b are upregulated in senescent cells and modulate their inflammatory response by reducing the levels of target interleukin (IL)-1 receptor-associated kinase 1 (IRAK1), which in turn lowers IL-6 and IL-8 secretion.Citation18 In addition, we recently reported subsets of microRNAs upregulated in senescent cells; among them miR-519 regulates translation of the mRNA encoding RNA-binding protein HuR and suppresses tumorigenesis.Citation19–Citation21 Other microRNAs including members of the let-7 family, miR-15b, miR-24, miR-25 and miR-141 decreased during replicative senescence.Citation19,Citation22,Citation23
Global protein translation is known to be reduced in senescent cells.Citation24 While studying this effect, we discovered that the mRNA profiles of senescent cells are vastly similar to those of early-passage cells. Since translation rates can be controlled by post-transcriptional mechanisms, we examined the microRNAs recently reported in ref. Citation19 and enzymes that mediate microRNA biogenesis in young and senescent WI-38 cells. Although we detected differential expression of microRNAs in senescent cells when compared to young cells (both up and downregulation), to our surprise the key enzymes of microRNA biogenesis pathway, Dicer and Drosha, were potently downregulated in senescent cells. Small interfering (si)RNA-mediated downregulation of Dicer or Drosha in young WI-38 cells reduced mature microRNA levels but, unexpectedly, it also reduced global translation, prompting us to envision possible scenarios by which the Drosha→Dicer→microRNA pathway can enhance global translation.
Results
Young and senescent fibroblasts have similar mRNA expression patterns, but translation is markedly lower in senescent fibroblasts.
It is well established that aging and senescence is associated with lower rates of mRNA translation.Citation24–Citation28 We confirmed this finding in young (Y) and senescent (S) WI-38 human diploid fibroblasts (HDF) that had been incubated in the presence of 35S-labeled methionine and cysteine (Materials and Methods). As anticipated, senescent cells showed lower levels of translation than did Y WI-38 cells (). In parallel, we examined the transcriptome of Y and S HDFs by microarray analysis. Three Y cell groups [population doublings (PDL) 24, 28 and 34 and three S cells (PDLs 47, 50 and 54) were tested. Very few changes in the levels of mRNAs were observed when comparing S and Y cells (). Of all detected transcripts (12,304 total), only 1% were altered (twofold higher or lower) in S relative to Y cells: 0.57% (60 mRNAs) were downregulated in S cells and 0.43% upregulated (47 mRNAs) in S cells, while 98.9% did not change (). lists a subset of upregulated and downregulated mRNAs (shaded areas), as well as some unchanged mRNAs. The complete microarray results are available from the authors. S cells showed higher levels of ankyrin repeat domain (ANKRD1) and endothelin 1 (EDN1) mRNAs, but lower levels of alcohol dehydrogenase 1A (ADH1A) and growth arrest-specific 1 (GAS1) mRNAs. Other transcripts, such as HuR, p53, PTMA, NCL, VHL and SIRT1 mRNAs, did not show significant changes (). Several of these genes were further studied by reverse transcription (RT) followed by real-time quantitative (q)PCR (). These data indicate that although mRNA translation is lower in S cells, the large majority of mRNAs showed similar abundance.
Dicer and Drosha are downregulated in senescent cells.
Our recent analysis of microRNA profiles in Y and S WI-38 fibroblastsCitation19">19">19 prompted us to hypothesize that the difference in translation between Y and S cells could be due to microRNA-mediated post-transcriptional repression. Since Mudhasani and colleagues had previously reported that ablation of Dicer leads to premature senescence in embryonic fibroblasts and in developing and adult mouse tissues,Citation29 we began by examining the expression pattern of Dicer and Drosha in Y and S HDFs. As shown in Dicer and Drosha were downregulated in S HDFs; the levels of HuR (included as positive control) were lower in S cells, as reported in references Citation24 and Citation30 (). MicroRNAs like let-7b, miR-10a and miR-15b () were also lower in S cells, as previously reported.Citation19
Dicer downregulation lowers global translation and induces senescence.
According to the established model of microRNA biosynthesis and function, Dicer or Drosha downregulation would be expected to decrease microRNA levels, which in turn would be predicted to increase translation. However, S cells showed lower levels of Dicer and Drosha and also decreased translation. This prompted us to investigate the effect of Dicer or Drosha downregulation on global translation in Y HDFs. Dicer protein levels were significantly reduced in Dicer siRNA-transfected cells compared to control transfected cells (). Dicer downregulation led to the lowering of several microRNAs (let-7b, miR-10a and miR-15b) as tested by RT-qPCR (). Importantly, Dicer knockdown also led to a global reduction in translation, as determined by using a nascent translation assay (). A reduction in Dicer also triggered a senescent phenotype as determined by assessing increased β-galactosidase activity () and by monitoring changes in gene expression, which were consistent with an increase in senescence (). For example, ADH1A and GAS1 mRNAs were downregulated, while ANKRD1 and EDN1 mRNAs were upregulated in Dicerdeficient cells () following similar patterns as those seen in Y/S cells (compare with ). These results indicate that the increased senescence triggered by Dicer downregulation was accompanied by a lowering of mRNA translation, suggesting that microRNAs enhance translation in this paradigm.
Drosha downregulation lowers global translation but does not induce senescence.
To further investigate if the effects of Dicer on translation are carried out by microRNAs, we examined the role of Drosha, another major enzyme in microRNA biogenesis. As mentioned above, Drosha is downregulated in S cells (). Drosha downregulation in Y cells using Drosha-directed siRNA () resulted in lower levels of several microRNAs (let-7b, miR-10a and miR-15b) as shown in . Cells with reduced Drosha also showed a suppression of global translation, as measured by 35S labeling (). However, unlike the effect of Dicer downregulation, silencing Drosha did not induce senescence as determined by β-galactosidase staining (.). Likewise, RT-qPCR analysis revealed that Drosha deficiency did not recapitulate the changes in senescenc-especific mRNAs seen in S cells or Dicer-silenced cells (). Taken together, these findings indicate that Dicer may influence senescence via mechanisms other than by inhibiting microRNA biosynthesis and further suggest that microRNAs promote global protein translation.
Discussion
Over the past few years, microRNAs have emerged as pivotal regulators of gene expression in mammalian cells. Many reports have indicated that microRNAs are capable of suppressing the translation of target mRNAs. Other reports have shown that microRNAs can induce the decay of target mRNAs. Additionally, there are examples of microRNAs reducing both the stability and the translation of target mRNAs.Citation5 Furthermore, other studies have shown that microRNAs can work in the opposite direction, enhancing translation.Citation7,Citation8 The net effect of an individual microRNA upon a specific target mRNA may in fact depend on the cell type, the specific metabolic state of the cell and the intrinsic stability of the mRNA. However, few studies to-date have examined the overall effect of microRNAs on cellular processes such as mRNA stability or translation. Our results showed that Dicer or Drosha downregulation in WI-38 cells significantly reduced whole-cell translation, as monitored by 35S labeling of nascent protein. These data suggest that Dicer/Drosha-dependent microRNAs promote global translation.
MicroRNAs and senescence.
Our lab reported subsets of microRNAs that were differentially expressed in young and senescent fibroblastsCitation19,Citation31 and several other studies have described changes in individual microRNAs during cellular senescence.Citation32,Citation33 For example, four microRNAs (miR-15b, miR-24, miR-25 and miR-141) shown to jointly regulate MKK4 expression are downregulated in senescent cells.Citation22 By contrast, many other microRNAs are upregulated with senescence. On the other hand, while Dicer and Drosha are downregulated in senescent cells (), many microRNAs are instead upregulated.Citation19 miR-146a and miR-146b, which downregulate the senescence-associated inflammatory mediators IL-6 and IL-8, are elevated in senescent HCA2 fibroblasts;Citation18 miR-519, which represses translation of RNA-binding protein HuR, is upregulated in senescent cells;Citation19,Citation21 and miR-34a, a microRNA upregulated in senescent umbilical cord vein endothelial cells, reduces SIRT1 expression.Citation34,Citation35 The increased or maintained expression of these microRNAs might be due to the presence of other enzymes that process these microRNAs even in the absence of or reduced abundance of Dicer or Drosha. Or perhaps the residual Drosha and/ or Dicer can selectively process certain microRNAs, maintaining or even enhancing their expression. Although relatively few microRNAs appear to increase in levels during senescence, it is widely believed that a single microRNA is capable of targeting multiple mRNAs that could be involved in different pathways driving the cell fate into senescence.
MicroRNAs and translation.
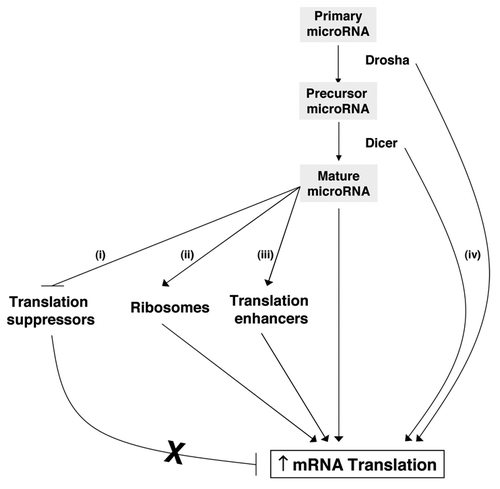
As mentioned above, microRNAs can suppress the translation of individual mRNAs. The absence of Dicer or Drosha usually results in microRNA downregulation, thus one would expect more translation of target mRNAs when one of these components of microRNA biogenesis is downregulated. In contrast, silencing either Dicer or Dosha instead caused reduced global translation in WI-38 cells. How might microRNAs enhance translation? A number of scenarios can be envisioned to explain this paradoxical phenomenon, although experimental support is not available in all instances. One possibility is that Drosha or Dicer silencing lowers the production of microRNAs that negatively regulate the expression of general translation suppressor proteins (e.g., microRNAs directed to 4E-BP and TIAR ), as this would have a net overall effect of promoting translation. In some cases, microRNAs such as miR-10a can stimulate the translation of mRNAs encoding ribosomal proteins,Citation7 which favors ribosome biogenesis and overall protein translation (). Additional mechanisms might include the ability of some microRNAs of switching from repressing to activating the translation of target mRNAs in a cell cycle-dependent manner;Citation8,Citation36 if a subset of the positively regulated mRNAs encodes translation enhancers, this would result in a net increase in overall protein biosynthesis (). If one or several of these pathways are in operation, one can explain how microRNAs are required to maintain a high level of translation in the cell. Conversely, we can understand how downregulating Drosha or Dicer causes the striking decreases in global translation shown in and .
Possible microRNA-independent role of Dicer/Drosha.
Although the primary known function of Drosha and Dicer is in the biogenesis of microRNAs and other small ncRNAs (e.g., siRNAs and xiRNAs),Citation37 they may have other functions that are yet to be described, perhaps acting directly upon mRNAs. Future studies will be needed to reveal how these and other processes may affect global translation in fibroblasts. Additional work will be required to elucidate if reducing Drosha/Dicer levels also lowers translation in transformed cells and whether such influence can be exploited in diseases such as cancer, where translation is aberrant.
Materials and Methods
Cell culture, transfections and β-galactosidase staining.
Early-passage, proliferating (young, Y) and late-passage (senescent, S) WI-38 human diploid fibroblasts (HDFs, from Coriell Cell Repositories) were cultured in Dulbecco's modified Eagle's medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum and 0.1 mM non-essential amino acids (Invitrogen). Cells were transfected either with control siRNA (Qiagen) or Dicer siRNA (Santa Cruz) using Lipofectamine-2000 (Invitrogen) according to the manufacturer's protocol. WI-38 cells were stained with senescence associated β-galactosidase (Cell Signaling) following the manufacturer's protocol.
RNA isolation, microarrays, RT-qPCR and microRNA measurements.
Total RNA was isolated directly from cells using Trizol (Invitrogen). RNA was used to probe Illumina microarrays (the complete array results are available from the authors). RNAs isolated from Y and S cells were used to validate the microarray data. After reverse transcription (RT) with random hexamers and SSII reverse transcriptase (Invitrogen), real-time, quantitative (q)PCR analysis was performed with gene-specific primer pairs (below) and SYBR Green PCR master mix (Kapa Biosystems). The oligomer pairs (each forward and reverse) used for the amplification of PCR products were: TGC ACC ACC AAC TGC TTA GC and GGC ATG GAC TGT GGT CAT GAG for GAPDH, TTC CTC ACC AAT GGG TCC TTT and GCT TCA AGC AGT TCA ACC TGA T for Dicer, GTG ACA TCG GGA GAA CGA AT and GCG GTC ACG TAG TTC ACA AA for HuR, CCC CAG CCA AAG AAG AAA C and AAC ATC TCG AAG CGC TCA C for p53, CAG GTT GCG GGA ATC CAA AG and GCT GGG CAC CTA GGA CAT CG for SIRT1, CCA ACC CAA ACC ATG AGA A and GGT CAC ACC ACA AGT AAA GTC AG for PTMA, GAA GGA AAT GGC CAA ACA GA and A CGC TTT CTC CAG GTC TTC A for NCL, AGA AGG TGG TGG CAT TTT TG and (inC AGT CTT CCC AAA GCA GGA G for VHL, GAG TGC AGC TAC GCC TAC AAC and GGA CTT GCA GTT CTC GTC CTG for GAS1, AGT CAT CCC ACT CGC TAT TCC and GTC CCC TGA GGA TTG CTT ACA for ADH1A, CAG CAG TCT TAG GCG CTG AG and ACT CTT TAT CCA TCA GGG ACG AG for EDN1, AGT AGA GGA ACT GGT CAC TGG and TGG GCT AGA AGT GTC TTC AGA T for ANKRD1, GAC ACC ACT GGA GGG TGA CT and CAG GTC CAC ATG GTC TTC CT for p21 and CCC TAT CAA CTT TCG ATG GTA GTC G and CCA ATG GAT CCT CGT TAA AGG ATT T for 18S rRNA.
MicroRNAs were measured using microRNA-specific forward primers and a universal reverse primer (System biosciences, SBI) following the manufacturer's protocol. The microRNA primers used for PCR amplification were: TGA GGT AGT AGG TTG TGT GGT T for let-7b, TAC CCT GTA GAA CCG AAT TTG TG for hsa-miR-10a, TAG CAG CAC ATC ATG GTT TAC A for miR-15b and CGA CTG CAT AAT TTG TGG TAG TGG for the snRNA U1, included for normalization.
Western blotting.
Whole-cell lysates were prepared with RIPA buffer [10 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% NP-40, 1 mM EDTA, 0.1% SDS and 1 mM dithiothreitol]. Proteins were resolved by SDS-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride (PVDF) membranes (Invitrogen). After incubation with primary antibodies recognizing Dicer, Drosha or HuR (Santa Cruz) and β-actin (Abcam), membranes were incubated with the appropriate secondary antibodies and signals were detected by ECL Plus (Amersham).
Analysis of newly translated proteins.
WI-38 cells were incubated with methionine- and cysteine-free medium for 1 hr and then with L-[35S]methionine and L-[35S]cysteine (EasyTag™ EXPRESS, NEN/Perkin Elmer) for 15 min. Cells were lysed in RIPA buffer and equal protein amounts were resolved by SDS-PAGE, transferred onto PVDF filters and visualized with a PhosphorImager (Molecular Dynamics).
Abbreviations
| RISC | = | RNA-induced silencing complex |
| UTR | = | untranslated region |
Figures and Tables
Figure 1 Low translation levels in senescent cells are not due to global changes in mRNA abundance. (A) Young (Y) and senescent (S) WI-38 cells were labeled with L-[35S]methionine and L-[35S]cysteine for 15 min and whole-cell lysates were resolved by SDS-PAGE (12% polyacrylamide), transferred onto filters and visualized using a PhosphorImager. (B) Percentages of unchanged, upregulated and downregulated mRNAs in S relative to Y cells, as revealed by microarray analysis. (C) Partial list of genes identified by microarray analysis of Y and S cells; after isolation of total RNA, mRNAs were identified by Illumina microarray analysis. The threshold considered was ±2 fold. (D) Validation of expressed mRNAs by RT-qPCR amplification using gene-specific primer pairs. Transcript abundance was normalized to GAPDH mRNA.
![Figure 1 Low translation levels in senescent cells are not due to global changes in mRNA abundance. (A) Young (Y) and senescent (S) WI-38 cells were labeled with L-[35S]methionine and L-[35S]cysteine for 15 min and whole-cell lysates were resolved by SDS-PAGE (12% polyacrylamide), transferred onto filters and visualized using a PhosphorImager. (B) Percentages of unchanged, upregulated and downregulated mRNAs in S relative to Y cells, as revealed by microarray analysis. (C) Partial list of genes identified by microarray analysis of Y and S cells; after isolation of total RNA, mRNAs were identified by Illumina microarray analysis. The threshold considered was ±2 fold. (D) Validation of expressed mRNAs by RT-qPCR amplification using gene-specific primer pairs. Transcript abundance was normalized to GAPDH mRNA.](/cms/asset/7e40bbdd-3034-4fd5-bc33-8db7c140f2b6/kccy_a_10914825_f0001.gif)
Figure 2 Dicer downregulation suppresses translation and induces senescence. (A) Levels of Dicer, Drosha, HuR and loading control β-actin in Y and S WI-38 cells, as assessed by western blot analysis. (B) The levels of let-7b, miR-10a and miR-15b were quantified in Y and S cells by RT-qPCR analysis and normalized to the levels of U1. (C) Two days after siRNA transfection, western blot analysis was performed to monitor the levels of Dicer and β-actin. (D) Forty-eight hours after siRNA transfections, the levels of let-7b, miR-10a and miR-15b were quantified by RT-qPCR analysis, normalized to the levels of U1, and plotted relative to the microRNA levels measured in the Ctrl siRNA transfection group. (E) Forty-eight hours after siRNA transfections, cells were labeled with L-[35S]methionine and L-[35S]cysteine for 15 min and whole-cell lysates were resolved by SDS-PAGE (12% polyacrylamide), transferred onto filters and visualized using a PhosphorImager. (F) Three days after transfection with the siRNAs indicated, the activity of senescence-associated-β-galactosidase was assessed in cultured cells. (G) Forty-eight hours after siRNA transfection, the levels of the indicated mRNAs were measured by RT-qPCR and normalized to the levels of GAPDH mRNA.
![Figure 2 Dicer downregulation suppresses translation and induces senescence. (A) Levels of Dicer, Drosha, HuR and loading control β-actin in Y and S WI-38 cells, as assessed by western blot analysis. (B) The levels of let-7b, miR-10a and miR-15b were quantified in Y and S cells by RT-qPCR analysis and normalized to the levels of U1. (C) Two days after siRNA transfection, western blot analysis was performed to monitor the levels of Dicer and β-actin. (D) Forty-eight hours after siRNA transfections, the levels of let-7b, miR-10a and miR-15b were quantified by RT-qPCR analysis, normalized to the levels of U1, and plotted relative to the microRNA levels measured in the Ctrl siRNA transfection group. (E) Forty-eight hours after siRNA transfections, cells were labeled with L-[35S]methionine and L-[35S]cysteine for 15 min and whole-cell lysates were resolved by SDS-PAGE (12% polyacrylamide), transferred onto filters and visualized using a PhosphorImager. (F) Three days after transfection with the siRNAs indicated, the activity of senescence-associated-β-galactosidase was assessed in cultured cells. (G) Forty-eight hours after siRNA transfection, the levels of the indicated mRNAs were measured by RT-qPCR and normalized to the levels of GAPDH mRNA.](/cms/asset/172c94d1-9904-4699-9020-f22e380737f7/kccy_a_10914825_f0002.gif)
Figure 3 Drosha downregulation suppresses translation but does not increase senescence. (A) The levels of Drosha and β-actin were assessed by western blot analysis two days after transfection with the indicated siRNAs. (B) Forty-eight hours after transfecting the indicated siRNAs, the levels of let-7b, miR-10a and miR-15b were quantified by RT-qPCR analysis, normalized to the levels of 18S rRNA and plotted relative to the microRNA levels measured in the Ctrl siRNA transfection group. (C) Forty-eight hours after siRNA transfection, cells were incubated with L-[35S]methionine and L-[35S]cysteine for 15 min and whole-cell lysates were resolved by SDS-PAGE (12% polyacrylamide), transferred onto filters and visualized using a PhosphorImager. (D) Three days after transfection with the siRNAs indicated, the senescence-associated-β-galactosidase activity was assessed in cultured cells. (E) Forty-eight hours after transfection of siRNAs, the levels of the indicated mRNAs were measured by RT-qPCR and normalized to the levels of GAPDH mRNA.
![Figure 3 Drosha downregulation suppresses translation but does not increase senescence. (A) The levels of Drosha and β-actin were assessed by western blot analysis two days after transfection with the indicated siRNAs. (B) Forty-eight hours after transfecting the indicated siRNAs, the levels of let-7b, miR-10a and miR-15b were quantified by RT-qPCR analysis, normalized to the levels of 18S rRNA and plotted relative to the microRNA levels measured in the Ctrl siRNA transfection group. (C) Forty-eight hours after siRNA transfection, cells were incubated with L-[35S]methionine and L-[35S]cysteine for 15 min and whole-cell lysates were resolved by SDS-PAGE (12% polyacrylamide), transferred onto filters and visualized using a PhosphorImager. (D) Three days after transfection with the siRNAs indicated, the senescence-associated-β-galactosidase activity was assessed in cultured cells. (E) Forty-eight hours after transfection of siRNAs, the levels of the indicated mRNAs were measured by RT-qPCR and normalized to the levels of GAPDH mRNA.](/cms/asset/34fdf26c-baea-46f6-b0a8-7eeaf59b64d3/kccy_a_10914825_f0003.gif)
Figure 4 Schematic depicting possible scenarios whereby the Drosha→Dicer→microRNA might promote global translation. Plausible mechanisms include the microRNA-mediated (i) repression of translation inhibitors, (ii) increase in ribosome biosynthesis and (iii) increase and/or activation of translation enhancers. Additional microRNA-independent effects (iv) could involve the direct activation of translation via Dicer- and/or Drosha-controlled mechanisms.
Acknowledgements
This research was funded by the Intramural Research Program of the National Institute on Aging, National Institutes of Health.
References
- Sevignani C, Calin GA, Siracusa LD, Croce CM. Mammalian microRNAs: A small world for fine-tuning gene expression. Mamm Genome 2006; 17:189 - 202
- Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol 2009; 21:452 - 460
- Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell 2009; 8:215 - 33
- Du T. Zamore PD, microPrimer: The biogenesis and function of microRNA. Development 2005; 132:4645 - 4652
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 2006; 20:515 - 524
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010; 466:835 - 840
- Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell 2008; 30:460 - 471
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can upregulate translation. Science 2007; 318:1931 - 1934
- Lindsay MA. microRNAs and the immune response. Trends Immunol 2008; 29:343 - 351
- Wang Y, Russell I, Chen C. MicroRNA and stem cell regulation. Curr Opin Mol Ther 2009; 11:292 - 298
- Lynam-Lennon N, Maher SG, Reynolds JV. The roles of microRNA in cancer and apoptosis. Biol Rev Camb Philos Soc 2009; 84:55 - 71
- Chen LH, Chiou GY, Chen YW, Li HY, Chiou SH. microRNA and aging: A novel modulator in regulating the aging network. Ageing Res Rev 2010; 9:59 - 66
- Deng S, Calin GA, Croce CM, Coukos G, Zhang L. Mechanisms of microRNA deregulation in human cancer. Cell Cycle 2008; 7:2643 - 2646
- Tong AW, Nemunaitis J. Modulation of miRNA activity in human cancer: A new paradigm for cancer gene therapy?. Cancer Gene Ther 2008; 15:341 - 355
- Nelson PT, Wang WX, Rajeev BW. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol 2008; 18:130 - 138
- Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res 1965; 37:614 - 636
- Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes Dev 2010; 24:2463 - 2479
- Bhaumik D, Scott GK, Schokrpur S, Patil CK, Orjalo AV, Rodier F, et al. MicroRNAs miR-146a/b negatively modulate the senescence-associated inflammatory mediators IL-6 and IL-8. Aging 2009; 1:402 - 411
- Marasa BS, Srikantan S, Martindale JL, Kim MM, Lee EK, Gorospe M, et al. MicroRNA profiling in human diploid fibroblasts uncovers miR-519 role in replicative senescence. Aging 2010; 2:333 - 343
- Abdelmohsen K, Kim MM, Srikantan S, Mercken EM, Brennan SE, Wilson GM, et al. miR-519 suppresses tumor growth by reducing HuR levels. Cell Cycle 2010; 9:1354 - 1359
- Abdelmohsen K, Srikantan S, Kuwano Y, Gorospe M. miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels. Proc Natl Acad Sci USA 2008; 105:20297 - 20302
- Marasa BS, Srikantan S, Masuda K, Abdelmohsen K, Kuwano Y, Yang X, et al. Increased MKK4 abundance with replicative senescence is linked to the joint reduction of multiple microRNAs. Sci Signal 2009; 2:69
- Lal A, Kim HH, Abdelmohsen K, Kuwano Y, Pullmann R Jr, Srikantan S, et al. p16(INK4a) translation suppressed by miR-24. PLoS One 2008; 3:1864
- Abdelmohsen K, Pullmann R Jr, Lal A, Kim HH, Galban S, Yang X, et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell 2007; 25:543 - 557
- Macieira-Coelho A, Loria E, Berumen L. Relationship between cell kinetic changes and metabolic events during cell senescence in vitro. Adv Exp Med Biol 1975; 53:51 - 65
- Blazejowskia CA, Webster GC. Decreased rates of protein synthesis by cell-free preparations from different organs of aging mice. Mech Ageing Dev 1983; 21:345 - 356
- Makrides SC. Protein synthesis and degradation during aging and senescence. Biological Rev 1983; 58:343 - 422
- Makrides SC, Goldthwaite J. The content and size distribution of membrane-bound and free polyribosomes in mouse liver during aging. Mech Ageing Dev 1984; 27:111 - 134
- Mudhasani R, Zhu Z, Hutvagner G, Eischen CM, Lyle S, Hall LL, et al. Loss of miRNA biogenesis induces p19Arf-p53 signaling and senescence in primary cells. J Cell Biol 2008; 181:1055 - 1063
- Wang W, Yang X, Cristofalo VJ, Holbrook NJ, Gorospe M. Loss of HuR is linked to reduced expression of proliferative genes during replicative senescence. Mol Cell Biol 2001; 21:5889 - 5898
- Noren Hooten N, Abdelmohsen K, Gorospe M, Ejiogu N, Zonderman AB, Evans MK. microRNA expression patterns reveal differential expression of target genes with age. PLoS One 2010; 5:10724
- Lanceta J, Prough RA, Liang R, Wang E. MicroRNA group disorganization in aging. Exp Gerontol 2010; 45:269 - 278
- Bonifacio LN, Jarstfer MB. MiRNA profile associated with replicative senescence, extended cell culture and ectopic telomerase expression in human foreskin fibroblasts. PLoS One 2010; 5:12519
- Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci USA 2008; 105:13421 - 13426
- Ito T, Yagi S, Yamakuchi M. MicroRNA-34a regulation of endothelial senescence. Biochem Biophys Res Commun 2010; 398:735 - 740
- Vasudevan S, Tong Y, Steitz JA. Cell cycle control of microRNA-mediated translation regulation. Cell Cycle 2008; 7:1545 - 1549
- Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: Regulators of disease. J Pathol 2010; 220:126 - 139