Abstract
It is widely accepted that DNA damage induces rapid degradation of MDM2 through phosphorylation, resulting in a transient reduction of MDM2 level. Elimination of MDM2 is a logical mechanism that stabilizes p53. This phenomenon has been reproduced by many independent studies and is frequently referenced. Here we present evidence that only phosphorylation-sensitive antibodies SMP14 and 2A10, but not other MDM2 antibodies, can detect robust down-regulation of MDM2 after DNA damage. Therefore, we conclude that DNA damage does not accelerate MDM2 auto-degradation. SMP14 and 2A10 are frequently used to detect human and mouse MDM2, respectively. While it is not clear whether the discrepancy is entirely due to the use of these antibodies, our results suggest that epitope masking by phosphorylation should be an important consideration when interpreting results of MDM2 analysis by SMP14 and 2A10.
Background
The p53 pathway is critical for maintaining genomic stability and preventing development of cancer in higher organisms. A key feature of p53 is its stabilization and activation after exposure to a wide range of stress signals such as oncogene activation, abnormal ribosome biogenesis and DNA damage. These responses may be essential for its tumor suppressor function.Citation1 In normal cells, p53 is present at low levels due to degradation by the ubiquitin-dependent proteasome pathway. p53 turnover is regulated by MDM2, which binds p53 and functions as an E3 ligase to promote p53 ubiquitination.Citation2–Citation4 Additional ubiquitin E3 ligases such as Pirh2 and Cop1 have also been implicated as regulators of p53 turnover.Citation5,Citation6 However, current evidence suggests that MDM2 is a major and indispensable regulator of p53 level.Citation7,Citation8
Elucidating how p53 is stabilized and activated after DNA damage is the subject of many studies. A number of mechanisms have been proposed. p53-MDM2 binding is essential for inhibition of p53 and is a point of regulation by DNA damage signaling. Several studies showed that DNA double strand breaks induce phosphorylation of p53 S15 by DNA-PK and ATM.Citation9–Citation12 ATM also activates Chk2, which in turn phosphorylates p53 on S20 which is part of the MDM2 binding site.Citation9,Citation12–Citation14 These findings suggest that p53 phosphorylation disrupts MDM2 binding and results in p53 stabilization. However, mouse model studies suggested that other mechanisms are needed to mediate robust p53 stabilization after DNA damage.Citation15–Citation17 Other studies have suggested that dephosphorylation of the MDM2 acidic domain,Citation18,Citation19 and phosphorylation of MDM2 C terminal region by c-Abl and ATM regulate MDM2 E3 ligase activity during DNA damage response.Citation20–Citation23
Another potential mechanism of p53 stabilization is through elimination of MDM2. MDM2 undergoes self-ubiquitination and has a short half-life (< 30 min) in cultured cells. It has been reported that MDM2 undergoes accelerated degradation within 1–2 h after DNA damage by Neocarzinostatin (NCS), ultraviolet light and ionizing irradiation (IR),Citation24 suggesting that p53 stabilization is due to elimination of MDM2. Treatment with NCS reduces MDM2 half-live by seven-fold. DNA damage also causes a decrease in the steady state level of MDM2 at early time points. These effects are partially dependent on ATM and other PI-3 family kinases.Citation24 Subsequently, many studies from other groups also showed that DNA damage induces reduction in MDM2 level. Therefore, it is widely accepted that DNA damage promotes MDM2 degradation, which contributes to p53 stabilization. The mechanism of accelerated MDM2 degradation after DNA damage has been addressed in more recent studies.Citation25
However, MDM2 is a well-established transcriptional target for p53.Citation26,Citation27 Numerous studies reported induction of MDM2 level shortly (2–4 h) after DNA damage due to activation of p53. Other studies showed high levels of MDM2 and stabilized p53 coexisting in the cell after gamma irradiation (IR) and found no change in MDM2 half-life.Citation28 Although differences in experimental systems and the means of DNA damage induction may be responsible for the discrepancy, it is note-worthy that an earlier report showed that ATM-dependent phosphorylation of MDM2 causes a loss of reactivity to the antibody SMP14 due to epitope masking.Citation29 SMP14 is widely used for the detection of human MDM2, therefore it may produce results that are interpreted as accelerated MDM2 degradation. Although it is difficult to determine whether all conclusions of MDM2 self-degradation were based on SMP14, it is apparent that most studies do not take into account its sensitivity to MDM2 phosphorylation. Therefore, widespread use of SMP14 can potentially lead to misinterpretation of certain experiments.
Results and Discussion
In an attempt to address the issue, we tested whether use of different MDM2 antibodies will produce the discrepancy on MDM2 downregulation after DNA damage. We tested several commercially available MDM2 monoclonal antibodies including SMP14,Citation30 2A10, 2A9, 3G9, 4B2 and 4B11.Citation31 Their epitope locations on MDM2 are summarized in . Human osteosarcoma cell line U2OS was treated with 10 Gy IR and analyzed over a wide range of time points for MDM2 level using the 3G9 antibody for western blot. The result showed that IR treatment induced a fraction of MDM2 to migrate faster on SDS-PAGE as previously observed (), which was due to ATM-mediated phosphorylation.Citation23,Citation32 However, no significant decrease of MDM2 level was observed at any time point. MDM2 level increased significantly 4 h after IR with similar kinetics as p53 accumulation, as expected for a p53 target gene. The moderate decrease of MDM2 band intensity at the 0.3, 0.6 and 1.5 h time points were likely due to phosphorylation-induced spreading of the bands rather than a true decrease in total MDM2 level.
When SMP14 was used to detect MDM2, a significant reduction of MDM2 level was observed 0.5–1 h after IR as previously reported ().Citation24 In contrast, antibodies 3G9 and 4B2 showed no significant change or only minor decrease using the same samples. These results suggest that the decrease in MDM2 level was a phenomenon specific to SMP14. To rule out a tumor cell-specific effect, we repeated the experiment using a primary human foreskin fibroblast cell line (HFF). SMP14 also showed a dramatic decrease in MDM2 level shortly after IR, whereas three different antibodies (3G9, 4B2 and 4B11) showed no significant decrease in MDM2 (). Similar results were obtained using NCS to induce DNA damage instead of IR (). Therefore, MDM2 downregulation by DNA damage was only observed using SMP14. We should note that some reduction of band intensity is visible in certain experiments using 4B2 (e.g., and D, 4B2 parts). This may be partially due to phosphorylation-induced band diffusion and, in our experience, is not a robust and tractable phenotype.
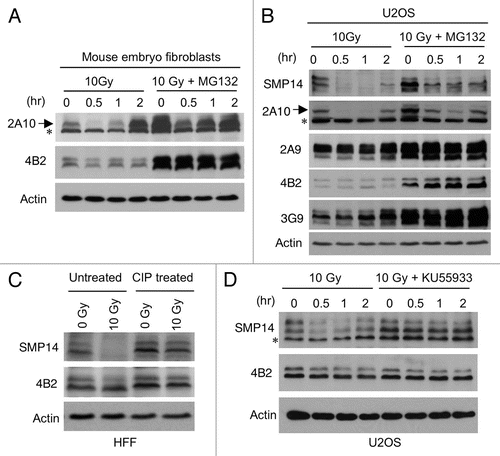
Since the phosphorylation-sensitive MDM2 antibody 2A10 is frequently used for analyzing mouse MDM2,Citation32 we also used 2A10 to detect MDM2 in MEFs after IR. The results showed a reduction of MDM2 in 1–2 h after IR using 2A10, whereas 4B2 showed no significant loss of MDM2 (). The 2A10 antibody also detected a loss of human MDM2 in U2OS after IR (). Therefore, the C terminal 2A10 epitope (a target of ATM mediated phosphorylation) may provide the majority of signal in 2A10 western blot. If SMP14 and 2A10 signal loss after DNA damage was due to degradation of MDM2, inhibition of the proteasome should rescue MDM2 signal. However, we found that the proteasome inhibitor MG132 did not prevent the reduction of MDM2 signal by 2A10 or SMP14 after IR ( and B), suggesting that the signal loss was not due to degradation.
To test whether phosphorylation of MDM2 after DNA damage prevents detection by SMP14, the irradiated cell extract was treated with phosphatase (CIP). SMP14 signal was effectively restored by the phosphatase treatment (). Reprobing of the same membrane using 4B2 clearly showed the inability of SMP14 to detect MDM2 in the irradiated sample prior to phosphatase treatment (). Furthermore, inhibition of ATM kinase using a specific inhibitor KU55933 prevented the loss of SMP14 signal after IR, without changing MDM2 level as detected by 4B2. These results demonstrated that the reduction in SMP14 signal after DNA damage was caused by phosphorylation-dependent epitope masking.
Previous study showed that one of the two 2A10 epitopes on MDM2 (255-DSEDYSLS-262 and 390-ESDDYSQ-396) was blocked by ATM phosphorylation of S395.Citation20,Citation32 The SMP14 epitope (154-SRPSTSSRRRAISE-167) contains multiple S and T residues, with S166 being a known AKT target site.Citation33,Citation34 Analysis of MDM2 using a commercial PS166 antibody showed no change in reactivity after IR (data not shown), ruling out S166 phosphorylation as the cause of SMP14 epitope masking. Our mass spectrometric analysis of MDM2 purified from irradiated cells was uninformative due to poor coverage of the SMP14 epitope. Therefore, it is unclear which residue within or near the SMP14 epitope is phosphorylated after DNA damage that interferes detection by SMP14. Although SMP14 was generated using a human MDM2 peptide,Citation30 it has been shown to coincidentally cross-recognize mouse MDM2 through a different epitope that includes T216. Interestingly, Cyclin A-CDK2 mediated phosphorylation of T216 also blocked mouse MDM2 detection by SMP14.Citation35 Therefore, SMP14 binding appears to be inherently sensitive to phosphorylation of its epitopes.
Together, our findings reinforce prior reports that MDM2 detection by SMP14 and 2A10 are blocked by DNA-damage induced epitope masking.Citation29,Citation36 Our results showed that DNA damage does not cause accelerated MDM2 degradation to a significant degree. Since there is no decrease in MDM2 level immediately after DNA damage, and there is robust induction of MDM2 at later time points that coexist with stabilized p53, other mechanisms such as regulation of MDM2 E3 ligase activity or p53 de-ubiquitination may be responsible for p53 stabilization.
How can we reconcile our finding with many reports that DNA damage induces MDM2 degradation or downregulation? We speculate that this discrepancy arises because SMP14 and 2A10 are widely used to detect human and mouse MDM2, often individually or in antibody cocktails. Unfortunately, commercial venders do not provide adequate information on the phosphorylation-sensitive nature of these antibodies, and the original publications on these antibodies are dated and may be overlooked. As such, the results of MDM2 analysis using these antibodies are rarely discussed in terms of their sensitivity to epitope masking. Although neglecting this information is inconsequential in most cases when analyzing MDM2 level at late time points, it may compromise the conclusion of certain experiments. Our results suggest that MDM2 detection using SMP14 and 2A10 should be interpreted with caution.
Materials and Methods
Cell lines and antibodies.
Human foreskin fibroblast (HFF) was provided by Dr. Jack Pledger. Primary mouse embryo fibroblasts were produced in a previous study in reference 37. DNA damaging agent Neocarzinostatin was provided by the NCI. ATM inhibitor KU55933 was purchased from Calbiochem and used at 10 µM. SMP14 was purchased from Santa Cruz Biotechnology. DO-1 was purchased from BD Bioscience. PS166 antibody was purchased from Cell Signaling. MDM2 antibodies 2A9, 2A10, 4B2, 4B11 and 3G9 were produced from hybridomas generated in a previous study in reference Citation31.
Protein analysis.
To detect proteins by western blot, cells were lysed in lysis buffer [50 mM Tris-HCl (pH 8.0), 5 mM EDTA, 150 mM NaCl, 0.5% NP40, 1 mM PMSF, 50 mM NaF] and centrifuged for 5 min at 10,000 g. Cell lysate (10–50 µg protein) was fractionated by SDS-PAGE using gradient gel and transferred to Immobilon P filters (Millipore). The filter was blocked for 1 h with phosphate-buffered saline (PBS) containing 5% non-fat dry milk, 0.1% Tween-20. The filter was developed using ECL-plus reagent (Amersham). Dephosphorylation reaction was performed by incubating 30 µg whole cell extract with 1 U calf intestinal phosphatase for 1 h at 37°C. Control was performed by incubation without phosphatase.
Figures and Tables
Figure 1 DNA damage reduces MDM2 reactivity to SMP14 antibody. (A) Diagram of MDM2 and epitope locations of the monoclonal antibodies used in this study. (B) U2OS osteosarcoma cells were treated with 10 Gy IR and analyzed for MDM2 level at indicated time points by western blot using 3G9. Identical amount of whole cell extract was used in each lane. (C and D) U2OS and normal human fibroblasts were treated with 10 Gy IR and analyzed for MDM2 level at indicated time points by western blot using different antibodies. (E) U2OS cells were incubated with Neocarzinostatin and analyzed for MDM2 level at indicated time points by western blot.
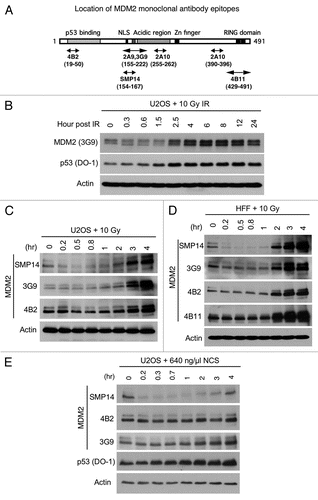
Figure 2 Loss of SMP14 signal after DNA damage due to epitope masking. (A and B) Mouse embryo fibroblasts and U2oS were treated with 10 Gy IR in the absence or presence of 30 µM MG132. MDM2 level was analyzed by western blot at indicated time points using 2A10. Arrow indicates MDM2 band detected by 2A10. A background band is marked by *. (C) Normal human fibroblasts were irradiated with 10 Gy and after 1 h whole cell extract was treated with phosphatase (CIP) or incubated without CIP. Samples were analyzed by SMP14 western blot. The membrane was stripped and reprobed using 4B2. (D) U2OS cells were irradiated with 10 Gy in the absence or presence of 10 µM ATM inhibitor KU55933. Samples were analyzed by SMP14 western blot. A background band is marked by *.
Acknowledgements
We would like to thank Dr. Wei Gu and Dr. Yanping Zhang for helpful discussions. This work was supported in part by grants from the National Institutes of Health to J.C. (CA141244).
References
- Harris SL, Levine AJ. The p53 pathway: Positive and negative feedback loops. Oncogene 2005; 24:2899 - 2908
- Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett 1997; 420:25 - 27
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature 1997; 387:296 - 269
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature 1997; 387:299 - 303
- Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 2004; 429:86 - 92
- Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, et al. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell 2003; 112:779 - 791
- Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 1995; 378:203 - 206
- Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 1995; 378:206 - 208
- Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 1997; 91:325 - 334
- Prives C, Hall PA. The p53 pathway. J Pathol 1999; 187:112 - 126
- Mayo LD, Turchi JJ, Berberich SJ. Mdm-2 phosphorylation by DNA-dependent protein kinase prevents interaction with p53. Cancer Res 1997; 57:5013 - 5016
- Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 1998; 281:1674 - 1677
- Chehab NH, Malikzay A, Appel M, Halazonetis TD. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev 2000; 14:278 - 288
- Shieh SY, Ahn J, Tamai K, Taya Y, Prives C. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev 2000; 14:289 - 300
- Chao C, Herr D, Chun J, Xu Y. Ser18 and 23 phosphorylation is required for p53-dependent apoptosis and tumor suppression. EMBO J 2006; 25:2615 - 2622
- MacPherson D, Kim J, Kim T, Rhee BK, Van Oostrom CT, DiTullio RA, et al. Defective apoptosis and B-cell lymphomas in mice with p53 point mutation at Ser 23. EMBO J 2004; 23:3689 - 3699
- Chao C, Hergenhahn M, Kaeser MD, Wu Z, Saito S, Iggo R, et al. Cell type- and promoter-specific roles of Ser18 phosphorylation in regulating p53 responses. J Biol Chem 2003; 278:41028 - 41033
- Blattner C, Hay T, Meek DW, Lane DP. Hypophosphorylation of Mdm2 augments p53 stability. Mol Cell Biol 2002; 22:6170 - 6182
- Kulikov R, Winter M, Blattner C. Binding of p53 to the central domain of Mdm2 is regulated by phosphorylation. J Biol Chem 2006; 281:28575 - 28583
- Maya R, Balass M, Kim ST, Shkedy D, Leal JF, Shifman O, et al. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev 2001; 15:1067 - 1077
- Shinozaki T, Nota A, Taya Y, Okamoto K. Functional role of Mdm2 phosphorylation by ATR in attenuation of p53 nuclear export. Oncogene 2003; 22:8870 - 8880
- Sionov RV, Coen S, Goldberg Z, Berger M, Bercovich B, Ben-Neriah Y, et al. c-Abl regulates p53 levels under normal and stress conditions by preventing its nuclear export and ubiquitination. Mol Cell Biol 2001; 21:5869 - 5878
- Cheng Q, Chen L, Li Z, Lane WS, Chen J. ATM activates p53 by regulating MDM2 oligomerization and E3 processivity. EMBO J 2009; 28:3857 - 3867
- Stommel JM, Wahl GM. Accelerated MDM2 auto-degradation induced by DNA-damage kinases is required for p53 activation. EMBO J 2004; 23:1547 - 1556
- Inuzuka H, Tseng A, Gao D, Zhai B, Zhang Q, Shaik S, et al. Phosphorylation by casein kinase I promotes the turnover of the Mdm2 oncoprotein via the SCF(beta-TRCP) ubiquitin ligase. Cancer Cell 2010; 18:147 - 159
- Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev 1993; 7:1126 - 1132
- Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J 1993; 12:461 - 468
- Chen L, Gilkes DM, Pan Y, Lane WS, Chen J. ATM and Chk2-dependent phosphorylation of MDMX contribute to p53 activation after DNA damage. EMBO J 2005; 24:3411 - 3422
- de Toledo SM, Azzam EI, Dahlberg WK, Gooding TB, Little JB. ATM complexes with HDM2 and promotes its rapid phosphorylation in a p53-independent manner in normal and tumor human cells exposed to ionizing radiation. Oncogene 2000; 19:6185 - 6193
- Picksley SM, Vojtesek B, Sparks A, Lane DP. Immunochemical analysis of the interaction of p53 with MDM2;—fine mapping of the MDM2 binding site on p53 using synthetic peptides. Oncogene 1994; 9:2523 - 2529
- Chen J, Marechal V, Levine AJ. Mapping of the p53 and mdm-2 interaction domains. Mol Cell Biol 1993; 13:4107 - 4114
- Khosravi R, Maya R, Gottlieb T, Oren M, Shiloh Y, Shkedy D. Rapid ATM-dependent phosphorylation of MDM2 precedes p53 accumulation in response to DNA damage. Proc Natl Acad Sci USA 1999; 96:14973 - 14977
- Mayo LD, Donner DB. A phosphatidylinositol-3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc Natl Acad Sci USA 2001; 98:11598 - 11603
- Ashcroft M, Ludwig RL, Woods DB, Copeland TD, Weber HO, MacRae EJ, et al. Phosphorylation of HDM2 by Akt. Oncogene 2002; 21:1955 - 1962
- Zhang T, Prives C. Cyclin a-CDK phosphorylation regulates MDM2 protein interactions. J Biol Chem 2001; 276:29702 - 29710
- Maya R, Oren M. Unmasking of phosphorylation-sensitive epitopes on p53 and Mdm2 by a simple western-phosphatase procedure. Oncogene 2000; 19:3213 - 3215