Abstract
The continued turn over of human embryonic stem cells (hESC) while maintaining an undifferentiated state is dependent on the regulation of the cell cycle. Here we asked the question if a single cell cycle gene could regulate the self-renewal or pluripotency properties of hESC. We identified that the protein expression of the p27Kip1 cell cycle inhibitor is low in hESC cells and increased with differentiation. By adopting a gain and loss of function strategy we forced or reduced its expression in undifferentiating conditions to define its functional role in self-renewal and pluripotency. Using undifferentiation conditions, overexpression of p27Kip1 in hESC lead to a G1 phase arrest with an enlarged and flattened hESC morphology and consequent loss of self-renewal ability. Loss of p27Kip1 caused an elongated/scatter cell-like phenotype involving upregulation of Brachyury and Twist gene expression. We demonstrate the novel finding that p27Kip1 protein occupies the Twist1 gene promoter and manipulation of p27Kip1 by gain and loss of function is associated with Twist gene expression changes. These results define p27Kip1 expression levels as critical for self-renewal and pluripotency in hESC and suggest a role for p27Kip1 in controlling an epithelial to mesenchymal transition (EMT) in hESC.
Embryonic stem cells are derived from the inner cell mass (ICM) of the blastocyst and have the capacity for unlimited proliferation while retaining their potential to differentiate into a wide variety of cell types when cultured in vitro.Citation1,Citation2 Recently, it has been shown that a combination of three or four factors, such as, Oct4, Sox2, KLf4 and c-Myc can reprogram somatic cells to generate induced pluripotent stem cells or iPSC that have similar characteristics to ESC.Citation3-Citation7 For mouse ESC and to a lesser extent, primate and human ESC, the cell cycle profile has been well described characterized by a short G1 phase and a high proportion of cells in S-phase resulting in an average cell cycle of 9 h (for mouse ESC).Citation8-Citation10 Mouse ESC have been found to express very low levels of CDK inhibitory protein levels such as the Ink family (p16INK4a), Cip/Kip family (p21Cip1, p27Kip1) and are specifically dependent on cyclin A expression for pluripotency.Citation11,Citation12 When ESC differentiate, their cell cycle structure changes dramatically so as to incorporate a significantly longer G1 phase and their mechanism of cell cycle regulation changes to that typically seen in other mammalian somatic cells.Citation10
Recently, new roles for p27Kip1 independent from its function on cell cycle have been described in promoting differentiation suggesting that expression of p27Kip1 is a key protein for cell cycle to differentiation switches during development.Citation13-Citation20 Here we describe a gain and loss of function study to define the functional role of a single cell cycle gene, namely p27Kip1, in self-renewal and pluripotency of human embryonic stem cells.
Results
We evaluated the cell cycle structure of two hESC and two hiPSC lines in undifferentiated conditions, and after differentiation, by flow cytometry analysis of 4',6-dimidino-2-phenylindole (DAPI)-stained cells (). An analog of BrdU, namely EdU, was used to analyze the percentage of cells in S phase. In order to perform a cell cycle analysis of undifferentiated cells we selected pluripotent TRA-1–60 positive-stained cells. We observed a strong resemblance between the cell cycle profile of undifferentiated hESC and hiPSC, where 54–62% of the population was in S phase and a low proportion of cells in G0/1 phase of the cell cycle (17–33%) (). On differentiation, the cell cycle structure for all cell lines analyzed, similarly changed with a striking increase of cells in the G0/1 phase (87–93%) and a reduction of cells in the S phase (2–8%) (). We confirmed these results in mESC (Sup. Figure 1). Taken together, the results showed no differences in the cell cycle profile of hESC, hiPSC or mESC suggesting a conserved cell cycle structure for the pluripotent cells.
Figure 1. Pluripotent hESC and hiPSC share a conserved cell cycle structure that changes upon differentiation paralleled by an increase of p27Kip1. (A) Timeline of general differentiation protocol and representative phase contrast images of hESC in undifferentiated conditions and during general differentiation. Scale bars 500 μm. (B) Quantitative flow cytometry analysis of cell cycle in two different hESC lines and two different iPSC lines reveals a cell cycle structure change along differentiation with a high percentage of cells in S phase and low in G0/1 phase of cell cycle in undifferentiated cells, and a reduction of cells in S phase and increase of cells in G0/1 phase of cell cycle when cells are differentiated. Shown are the representative results obtained in two independent experiments. (C) western blot of main cell cycle proteins in undifferentiated conditions and during general differentiation in hESC reveals changes in the expression of some cell cycle proteins along differentiation. MEFs were used as a control. (D) western blot of human p27Kip1 in two different hESC lines and two different iPSC reveals low levels in undifferentiated conditions and high levels in differentiated conditions.
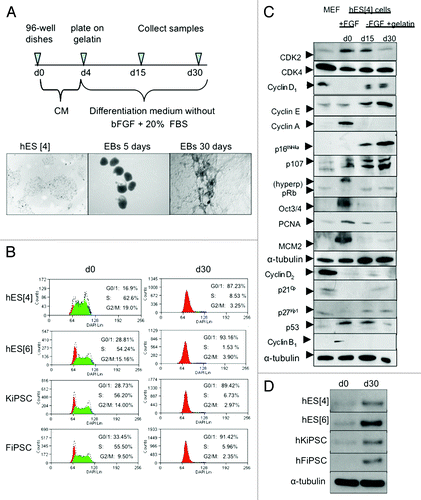
Next, we decided to study the cell cycle, in more detail, at the protein level because many of the cell cycle proteins are regulated at the post-transcriptional level.Citation23 We performed comprehensive series of western blot analysis for the cell cycle proteins regulating the G1 phase in mESC and hESC because the G1 phase is when the cell commits to go into cell cycle. We analyzed the cell cycle protein profile in undifferentiatiated conditions and compared with ESC under general differentiation conditions ( and Sup. Figure 1 for mouse ESC). Among the changes in protein levels during differentiation we found the cell cycle inhibitor p27Kip1. Interestingly, with differentiation, p27Kip1 protein levels increased over time, suggesting that low levels of p27Kip1 were important for the typical cell cycle structure of pluripotent cells ( and Sup. Figure 1 for mouse ESC). Reduction in Oct3/4 protein levels was used to monitor differentiation (). Given this important increase in both mESC, as well as, hESC and because, previously, it has been shown to have critical functions in determining cell fate we decided to investigate p27Kip1 in other hESC and hiPSC lines.Citation13-Citation20 In all human pluripotent cell lines analyzed, the expression of p27Kip1 was low in undifferentiated conditions and increased during differentiation suggesting that low levels of p27Kip1 were important for maintaining hESC self-renewal and pluripotency (). We also confirmed these results with another differentiation protocol toward neuronal cells using hESC (Sup. Figure 2).
Figure 2. Gain and loss of function of p27Kip1 in hESC. (A) Quantitative RT-PCR analysis of p27Kip1 expression in hESC infected with lentiviral tet-off inducible vector encoding for human p27Kip1 shows a gradual increase in the levels of p27Kip1 mRNA after removing doxycycline and a repression when doxycycline is added to the medium. Western blot analysis of the level of p27Kip1 protein in hESC infected with lentiviral tet-off inducible vector encoding for human p27Kip1 cultured in the presence or absence of doxycycline confirmed qRT-PCR results. Uninfected hESC were used for control of endogenous p27Kip1. (B) Quantitative RT-PCR analysis of p27Kip1 expression in hESC infected with 3 different lentivirus-derived GFP plasmids. These plasmids generated short hairpin RNA (shRNA) with different percentages of efficiency. The construct number 3 reached ~70% of knockdown. A Non-Target shRNA lentivirus GFP plasmid vector (NT shRNA) that does not target human genes was used as a control. Western blot analysis confirmed qRT-PCR results. (C) Immunofluorescence analysis of hESC after overexpression and knockdown of p27Kip1, in undifferentiated conditions, reveals an enlarged phenotype in cells overexpressing p27Kip1 and a fibroblastoid scatter-cell phenotype in knockdown cells. Scale bars 250 μm for lower magnification and 75 μm for higher magnification.
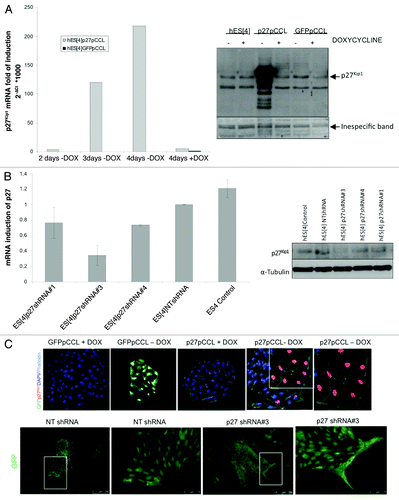
We sought to investigate whether overexpression of p27Kip1 could affect pluripotency or self-renewal properties. To this end, we overexpressed p27Kip1 in hESC using a doxycycline tet-off regulated lentiviral pCCL vector (kindly provided by the group of Dr. Naldini). We optimized the lentiviral transduction of hESC with a GFPpCCL vector to achieve over 76% of cells infected (Sup. Figure 3). Next, we used this infection conditions to transduce hESC with a p27pCCL vector. In this system, the expression of p27Kip1 was minimal in presence of doxycycline, and was induced by its withdrawal progressively after 4 d of infection, at both the mRNA and protein level (). Based on these results we performed all the experiments at 4 d post-infection. To perform loss of function assays, we used RNAi hairpins against p27Kip1 sequence cloned into a pLKO lentiviral vector and found that construct number three gave 70% knockdown (). With the aim to know the subcellular localization within the cell, we overexpressed p27Kip1 in hESC and performed immunofluorescence analysis after 4 d in the presence or absence of doxycycline ( and Sup. Figure 4). Surprisingly, we observed that p27Kip1 overexpressing hESC displayed a characteristic phenotype, with a large and spread cell morphology after staining with phalloidin (). Moreover, monitorization of p27Kip1 or GPF infected cells for 7 d, by time-lapse microscope photographs, demonstrated that large, spread p27Kip1 overexpressing cells could be observed after 4 d onwards (Sup. Figure 5). These results are consistent with previous work showing that overexpression of p27Kip1 in murine fibroblasts NIH-3T3 produces a flat phenotype and an augmented cell size.Citation24 Conversely, with loss of function of p27Kip1, we observed a elongated/scatter cell phenotype, whereby the cells elongated away from the hESC colony ( and Fig. S6). This data demonstrates that maintaining p27Kip1 at a certain level of expression is essential for normal hESC morphology.
Figure 3. Analysis of cell cycle and apoptosis in hESC. (A) Quantitative flow cytometry analysis of the cell cycle in p27Kip1 overexpressing and knockdown hESC. Overexpression of p27Kip1 induces an arrest in G1 phase of cell cycle in undifferentiated conditions. Representative results obtained in three independent experiments. (B) Quantitative flow cytometry analysis of apoptosis in p27Kip1 overexpressing and knockdown hESC in undifferentiated conditions.
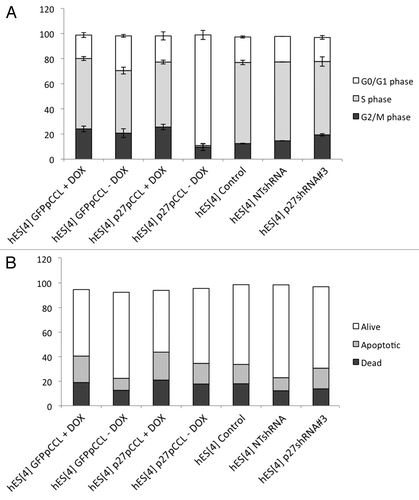
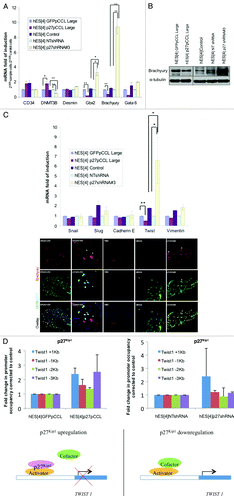
Figure 5. Manipulation of p27Kip1 expression in hESC affects pluripotency by regulating expression of Brachyury and Twist. (A) Quantitative RT-PCR validation of the markers presenting the most significant changes in p27Kip1 overexpressing hESC, p27Kip1 knockdown hESC and controls, reveals Brachyury repressed in p27Kip1 overexpressing cells and upregulated in p27Kip1 knockdown cells. Results represent the mean ± Standard Deviation of two independent experiments performed per duplicate. ANOVA test *p < 0.05, **p < 0.01. (B) western blot and immunofluorescence analysis for Brachyury expression in p27Kip1 overexpressing and knockdown hESC confirmed qRT-PCR results. Note the decrease of Brachyury expression in cells overexpressing p27Kip1 by immunofluorescence (denoted by an asterisk *) and the elongated morphology of p27Kip1 knockdown cells. These elongated cells were positive for Brachyury expression. Nuclei were counterstained with DAPI. Scale bars 75 μm. (C) qRT-PCR analysis for selected EMT markers in p27Kip1 overexpressing and knockdown hESC revealed a significant increase of Twist gene in p27Kip1 overexpressing hESC compared with controls in undifferentiated conditions. Results represent the mean ± Standard Deviation of two independent experiments performed per duplicate. ANOVA Test, p < 0.05. (D) ChIP assay for p27Kip1 in hESC treated with p27Kip1 RNAi (loss of function) or p27Kip1 cDNA (gain of function) on the Twist1 gene promoter (+1 Kb to -3 Kb). Schematic model suggesting that p27Kip1 binds the Twist1 gene promoter as part of a repressor complex in undifferentiated cell culture conditions of human embryonic stem cells.
In order to first explain the enlarged and flattened cell phenotype caused by p27Kip1 overexpression, we investigated a number of possible explanations: (1) cell cycle kinetics, (2) apoptosis, (3) induction of senescence and (4) differentiation. To analyze the cell cycle kinetics, we performed flow cytometry analysis of the cell cycle with DAPI and EdU substrate, for stain cells in S phase, of hESC immuno-stained for p27Kip1 expression. Control cells had low levels of p27Kip1 protein and had a normal hESC profile; however, with hESC overexpressing p27Kip1, 90% of the population arrested in G1 phase of the cell cycle (). On the contrary, with the p27Kip1 knockdown a slight increase in the proportion of cells in S phase and a decrease in the G1 phase could be observed. These results are proof of principle that a single cell cycle protein; namely, p27Kip1, inhibits self-renewal of hESC by arresting hESC in the G1 phase of the cell cycle.
Next, we checked if p27Kip1 overexpressing hESCs were apoptotic. We first, analyzed the nuclear morphology by DAPI staining of DNA. Results showed that the nuclei appeared round, clear-edged and uniformly stained further demonstrating that the cells were not apoptotic (Fig. S4 and ). We, also, determined the apoptosis by flow cytometry analysis of the mitochondrial membrane potential, by DiLC incorporation, using both, gain and loss of p27Kip1 function in hESC (). No significant changes were detected in apoptosis with either gain or loss of p27Kip1 function in undifferentiation conditions (). Next, we determined if p27Kip1 overexpressing cells could be senescent by performing ®-galactosidase activity at pH 6. We did not detect ®-galactosidase activity at pH 6 in p27Kip1 overexpressing hESC, suggesting that these cells were not senescent (data not shown).
To investigate in more detail the effect of p27Kip1on pluripotency of hESC, we compared the transcriptional profiles for 84 genes related to embryonic stem cell pluripotency and differentiation (Fig. 4 and Fig. S7). We had observed from flow cytometry analysis of the cell size that p27Kip1 overexpressing hESC were larger than controls and displayed an increase of the mean of intensity of the pulse width signal (). Based on these results and, with the aim to obtain a homogeneous population of p27Kip1 overexpressing cells, we FACS sorted cells in G1 phase by staining with Hoechst 33342 and displaying a higher mean of intensity of the pulse width signal (). We confirmed a 25-fold increase of p27Kip1 mRNA expression, by qRT-PCR analysis, in p27Kip1 large cells FACS sorted compared with GFP large cells as controls (). This was confirmed by western blot analysis (). We, then, performed the superarray for embryonic stem cell pluripotency and differentiation genes (Fig. S7). Values that were two-times upregulated or 50% downregulated were considered as significant cut offs (). We found that functional manipulation of p27Kip1 gene expression affected mainly genes regulating differentiation than pluripotency (). To confirm this we assessed the effect on pluripotency genes in undifferentiated conditions (). Gene expression profiling of this array values, showed that p27Kip1 overexpression did not result in a significant change of the master pluripotency factors such as Oct3/4, Nanog and Sox2 (). Moreover, with p27Kip1 loss of function we did not observe a significant change in any of these pluripotent genes ().
Figure 4A and B. SuperArray Real-Time PCR of pluripotency and differentiation genes in p27Kip1 overexpressing hESC. (A) Quantitative flow cytometry analysis of the cell size of the G1 phase residing population in p27Kip1 overexpressing cells and GFP expressing cells. Shown are the representative results obtained in three independent experiments. p27Kip1 overexpressing cells present a significative increase in the mean of the pulse width signal. Student’s t-test **p < 0.01. The cells presenting the highest pulse width signal were separated from the ones presenting the lowest pulse width signal. (B) Quantitative RT-PCR and western blot analysis of p27Kip1 expression confirmed that the large cells selected by FACS were overexpressing p27Kip1.
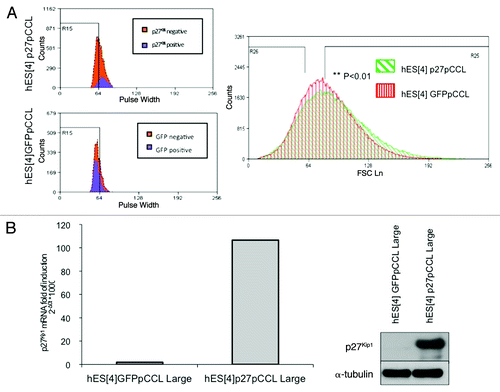
Figure 4C and D. SuperArray Real-Time PCR of pluripotency and differentiation genes in p27Kip1 overexpressing hESC. (C) Graph summarizing genes that were upregulated 2-fold or more or downregulated 50% or less were considered significant changes. (D) Quantitative RT-PCR of pluripotency markers in p27Kip1 overexpressing and knockdown hESC.
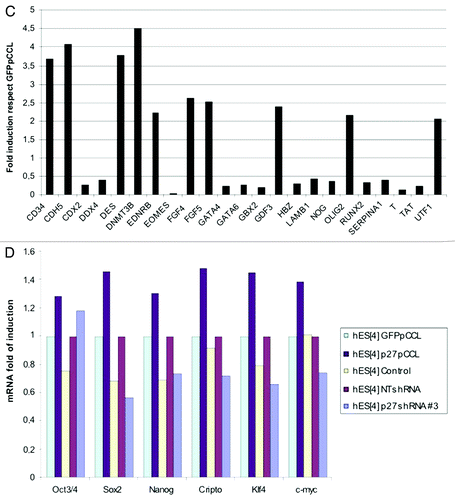
Next, we validated the expression of those genes that were more than two-times upregulated or 50% downregulated, by qRT-PCR, in p27Kip1 overexpressing, p27Kip1 depleted cells and respective controls (). These data revealed a decrease of early mesoderm differentiation marker Brachyury, Gbx2 and GATA6 with gain of function and, conversely, loss of function of p27Kip1 reverted this trend (). We decided to investigate Brachyury given that p27Kip1 had the largest effect on it compared with the other genes and due to its important role in differentiation. We confirmed by western blot that with loss of p27Kip1, Brachyury was induced, as seen with the real time PCR data, and conversely, with p27Kip1 gain of function, Brachyury was repressed (). The regulation of Brachyury was, also, seen by immunofluorescence analysis, in which, p27Kip1 overexpressing hESC had reduced levels of expression ( and see asterisk). Conversely, with p27Kip1 shRNA infected hESC, we observed that a high number of colonies, containing elongated shape cells, expressed Brachyury ().
Given that we had observed the major effect of p27Kip1 on a mesodermal marker, namely Brachyury, and that loss of p27Kip1 by shRNA lead to an elongated/scatter cell type resembling an EMT phenotype, we proceeded to analyze if other EMT markers could be regulated by p27Kip1. We analyzed a battery of EMT genes, by qRT-PCR, in both p27Kip1 overexpressing and shRNA infected hESC (). This analysis revealed an upregulation of Twist mRNA 6-fold compared with controls with loss of function. We did not detect any change in Vimentin, Snail1 or Snail2 genes, or E-Cadherin gene expression, suggesting that the effect of p27Kip1 was specific for Twist. Because we had detected p27Kip1 nuclear with overexpression in hESC, we performed ChIP analysis on the Twist1 promoter for p27Kip1 and methylation changes using antibodies against p27Kip1. We searched for p27Kip1 along 4,000 Kb of Twist1 promoter starting from +1.0 Kb to -3.0 Kb from the ATG. We found that p27Kip1 protein occupies the +1.0 Kb sites of the Twist1 promoter with overexpression of p27Kip1 in hESC but not seen in controls (based on 2-fold increases compared with control cells) (). This data suggests that p27Kip1 is a repressor of Twist1 gene expression in hESC by either binding directly or indirectly the promoter of Twist1.
Discussion
To date, most studies of the cell cycle in hESC have been descriptive, lacking functional studies that reveal the mechanisms of how the cell cycle maintains pluripotency and self-renewal of hESC.Citation7 In this study we sought to understand if overexpression or knockdown of a cell cycle gene, could regulate self-renewal or pluripotency of undifferentiated hESC. We focused on p27Kip1, given the low levels of expression in undifferentiated ESC and the critical role in cell fate decisions.Citation13-Citation20 We used a gain and loss of function strategy to define the functional role of the cell cycle in self-renewal or pluripotency by manipulating the expression of p27Kip1 in hESC.
The results demonstrate that a single cell cycle protein; namely, p27Kip1, inhibits self-renewal of hESC by arresting hESC in the G1 phase of the cell cycle. Moreover, we observe that p27Kip1 has an important role in the proper pluripotency of hESC (Fig. 4), supporting previous work in mESC showing an impairment of normal differentiation in p27Kip1 deficient cells after treatment with Retinoic Acid.Citation25 Under undifferentiating conditions the data demonstrates that p27Kip1 regulates proper pluripotency of hESC most likely via its effect in regulating Brachyury and Twist expression. We did not find that overexpression of p27Kip1 cuased senescence in hESC.
The idea that the cell cycle regulates pluripotency has been previously discussed joining a growing body of scientific data supporting a link between the cell cycle and pluripotency.Citation7,Citation26 Interestingly, loss of p27Kip1 initiates an elongated/scatter cell phenotype suggesting a possible regulation of the EMT phenotype. The discovery that loss of p27Kip1 initiates an elongated/scatter cell phenotype in hESC in undifferentiated cell conditions by regulating Twist and Brachyury has interesting implications in cell biology for a number of reasons. First, these results are in agreement with previous studies showing that overexpression of p27Kip1 promotes the maintenance of differentiation.Citation13-Citation20 Second, Twist induces an epithelial-mesenchymal transition to facilitate tumor metastasisCitation27,Citation28 and is upregulated in several types of epithelial cancers.Citation29 The novel discovery that p27Kip1 is present on the Twist1 gene promoter associated with gene expression levels suggests other functions of p27Kip1 in cell biology. A previous role for p27Kip1 in suppressing Twist in chicken muscle satellite cells (a type of stem cell) has been described, supporting further our own finding that p27Kip1 is an important and novel regulator of Twist in hESC.Citation30 A previous role for another similar cell cycle kinase inhibitor (CKI), p21waf1, has also been found to bind chromatin to regulate gene expression supporting the novel discovery that cell cycle inhibitor proteins may have additional roles to their kinase inhibitor functions by binding chromatin, most probably indirectly as a complex, to regulate gene expression.Citation31 If p27Kip1 regulates Twist1 as part of a repressor complex indirectly via other chromatic modeling proteins during gene regulation remains to be defined. Moreover, how p27Kip1 is regulating Brachyury at a gene level remains to be defined. These results are a proof of principle that proteins regulating the cell cycle play a critical role in self-renewal of hESC and also with making cell fate decisions that have far reaching implications for cell biology.
Previous work has shown that overexpression of cyclin D1, which we observed increases during differentiation, can induce senescence when the cell cycle is blocked.Citation32 Furthermore, pS6, a marker of mTOR activity is low in quiescent cells and this remains to be determined in hESC.Citation32
Materials and Methods
Cell culture
Human embryonic stem cells (hESC), human keratinocytes induced pluripotent stem cells (hKiPS) and human fibroblast induced pluripotent stem cells (hFiPS) were derived and characterized at the CMR[B].Citation4-Citation6 hESC, hKiPS and hFiPS were cultured on top of mitotically inactivated human fibroblasts and picked mechanically, or over Matrigel (BD Biosciences) coated plates with hESC conditioned medium from irradiated MEFs and passaged by trypsinization (0.05%).Citation21 hESC medium consisted of Knockout DMEM supplemented with 20% Knockout Serum Replacement, non-essential amino acids, 50 µM of 2-Mercaptoethanol, 100 U/mL penicillin, 100 µg/ mL streptomycin, 2 mM GlutaMAX (all Invitrogen) and 10 ng/ mL bFGF (Peprotech). Cultures were maintained at 37°C, 5% CO2 atmosphere, with medium changes daily. Feeder cells used were HFF-1: human foreskin fibroblasts, ATCC. MEFs were established from dissociated C57BL/6 mouse embryos (13.5 d gestation). Both HFF-1 and MEFs were mitotically inactivated by gamma irradiation (55 Gy) and grown in DMEM medium supplemented with 10% FBS, 100 U/mL penicillin, 100 µg/mL streptomycin and 2 mM GlutaMAX (all Invitrogen). Karyotypic analysis were performed routinely and in all cases normal karyotype was observed.
293T cells (CRL-12103 ATCC number, Rockville, MD) were cultured in DMEM medium supplemented with 10% FBS, 100 U/mL penicillin, 100 µg/mL streptomycin and 2 mM GlutaMAX (all Invitrogen). mESC were grown over MEFs feeder layer and the cell culture medium consisted of Knockout DMEM, 15% FBS (HyClone), 1% non-essential amino acids, 50 µM of 2-Mercaptoethanol, 100 U/mL penicillin, 100 µg/mL streptomycin, 2 mM GlutaMAX and 1,000 U/mL recombinant leukemia inhibitory factor (LIF) (ESGRO).
In vitro differentiation
For mechanically picked cells, colonies of undifferentiated cells were detached from the feeder layer using a modified glass pipette and transferred to low attachment plates 60 mm x 15 mm (Corning, NY) in hESC medium for 3–4 d. After 3–4 d the embryoid bodies were transferred to 0.1% gelatin (Chemicon) coated plates and cultured in general differentiation medium: Knockout DMEM (Invitrogen) supplemented with 20% Fetal Bovine Serum (HyClone), non-essential aminoacids, 100 U/mL penicillin, 100 µg/mL streptomycin, 2 mM GlutaMAX and 50 µM 2-Mercaptoethanol (all Invitrogen) for 30 d. Cell samples for western blot and FACs analysis were taken after 15 and 30 d. The medium was changed every other day.
For hESC cells passaged by trypsinization, 0.05% trypsin-EDTA (Invitrogen) was used for single cell suspension and 35,000–40,000 hESC were seeded in 200 µl of conditioned medium in each well of 96-well v-bottom low attachment plates (NUNC) and centrifuged at 950 g for 10 min to aggregate the cells. After 3–4 d embryoid bodies were transferred to 0.1% gelatin coated plates and cultured in general differentiation medium. Cell samples for western blot and FACs analysis were taken after 15 and 30 d. The medium was changed every other day.
For ectoderm differentiation: embryoid bodies were cultured in DMEM/F12 (Invitrogen), N2 and B27 supplements (Invitrogen), 1 mM l-glutamine, 1% nonessential aminoacids, 0.1 mM β-mercaptoethanol for 6 d, after which they were seeded onto matrigel chamber-slides with the same media plus 10−6 M Retinoic Acid (Sigma-Aldrich Quimica). Media was changed every two days. For mesoderm differentiation, embryoid bodies were replated onto gelatin-coated chambers-slide. Cultures were maintained in general differentiation medium supplemented with 100 µM Ascorbic Acid (Sigma-Aldrich Quimica). Media was changed every two days.
Flow cytometry analysis
For measuring apoptosis and proliferation we used the commercial kits “MitoProbe DilC1(5) Assay Kit” and “Click-iT EdU AlexaFluor647 Flow Cytometry Assay kit” respectively (both from Invitrogen) following the manufacturer’s instructions. Briefly, for the proliferation assay, 2 µl of 10 mM EdU solution was added to 2 mL of culture medium containing cells for 1 h, cells were then trypsinizated with 0.05% trypsin and resuspended in 100 µl of PBS supplemented with 1% BSA, 100 µl of Click-iT fixative solution was added to the cells, mixed for 5 sec and left at room temperature for 15 min. Cells were washed with 3 mL of PBS-1% BSA and collected by centrifugation (2,000 rpm, 3 min). 100 µl of 1x saponin-based permeabilization and wash buffer were added to the cell pellet and incubated with rabbit polyclonal antibodies against rabbit p27Kip1 (M-197, Santa Cruz Biotechnology) 1:20, and mouse Tra-1–60-FITC (BD) for 30 min at room temperature. Cells were then washed in PBS-1% BSA, collected by centrifugation and cells incubated with antibodies against p27Kip1, were incubated with alexa fluor 488-conjugated anti-rabbit IgG 1:50 for 15 min at room temperature. Cells were washed with 3 mL of the 1x saponin-based permeabilization and wash buffer and collected by centrifugation. 500 µl of Click-iT reaction cocktail containing 1x Click-iT EdU buffer additive, CuSO4 and Alexa Fluor 647 were added to the cells and left for 30 min at room temperature. After this time, cells were washed with saponin 1x buffer, collected by centrifugation and resuspended in 500 µl of saponin 1x buffer. Cells were stained by the addition of DAPI solution (0.1 M Tris Base pH 7.4; 0.9% or 150 mM NaCl; 1 mM CaCl2; 0.5 mM MgCl2; 0.2% BSA; 0.1% Nonidet P40; 10 mg/ mL DAPI) (Invitrogen) for 2 h at room temperature or overnight at 4°C. Finally cells were analyzed on a MoFlo cell sorter (DakoCytomation) with Summit software. A total of 10,000 events were collected for the cell cycle analysis.
For cell sorting of alive cells based on size, we selected the cells residing in G1 phase of the cell cycle by staining with 2 µg/mL of Hoechst 33342 for 30 min at 37°C. Hoechst 33342 was detected by using UV light excitation. Of this selected G1 phase residing cells, the biggest cells were separated from the smallest ones based on pulse-width signal at comparable cell numbers (58% population).
Western blot analysis
Cells were washed twice with cold PBS then harvested by scraping cells in ice-cold lysis buffer (150 µl for 10 cm Petri dishes) consisting of TRIS-HCl pH 7.4 (Roche), 250 mM NaCl, 1% Triton X100, 1 mM EDTA, 1 mM EGTA, 0.2 mM PMSF (all Sigma-Aldrich Quimica), 1x protease inhibitor cocktail (Roche) and 1x phosphatase inhibitor cocktail (Sigma-Aldrich Quimica). Lysates were passaged through a 1 mL syringe (Rubilabor) and a 25G needle (Terumo) and left for 30 min on ice. Lysates were clarified by centrifugation at 10,000 rpm for 10 min at 4°C and protein concentrations determined by Bradford assay (Bio-Rad Laboratories GmbH). Twenty µg of protein were mixed with 4x of a commercial loading buffer containing 20% 2-mercaptoethanol (Sigma-Aldrich Quimica), boiled at 100°C for 5 min and resolved on 8–12% SDS polyacrilamide electrophoresis gels. After electrophoresis, proteins were transferred to a nitrocellulose membrane using a submerged transfer apparatus (BioRad), filled with 25 mM Tris Base (Roche), 200 mM glycine (Sigma-Aldrich Quimica) and 20% methanol (Merck). Membranes were blocked in PBS containing 0.1% Tween-20 (PBST) (Sigma-Aldrich Quimica) and 5% milk powder (Sigma-Aldrich Quimica) for 1 h at room temperature. Membranes were incubated with primary antibody in PBST supplemented with 2% milk powder for 2 h at room temperature or overnight at 4°C. Primary antibodies were raised against p27Kip1 (M-197), E2F4 (A-20), p16 (H-156), p21Cip1 (F-5), p107 (C-18), Cdk2 (H-298), Cdk4 (H-22), Cyclin A (C-19), Cyclin D1 (H-295), Cyclin D2 (M-20), Cyclin E (M-20), PCNA (PC10), Rb (C-15), MCM2 (N-19), Cyclin B1 (GNS1), p53 (FL-393) (all Santa Cruz Biotechnology); Oct3/4 (ab19857) (Abcam); Cyclin D1 (Ab-2), Cyclin D2 (Ab-2) and Cyclin D3 (Ab-2) (all from Neomarkers) all primary antibodies were used as a 1: 1,000 dilution. Brachyury (AF2085, R&D Systems, Inc.) and E-cadherin (BD, Transduction Laboratories) were used as a 1:500 dilution. Alfa-tubulin (T6074, Sigma-Aldrich) was used as a 1:8,000. All antibodies were used according to the manufacturer’s specifications. Membranes were washed four times at room temperature for 5 min in PBST, then incubated for 1 h at room temperature with horseradish peroxidase (HRP)-conjugated secondary anti-rabbit, anti-mouse or anti-goat antibodies (Amersham Biosciences; 1:10,000) in PBST 2% milk powder. HRP activity was detected using ECL Plus detection kit (Pierce) following manufacturer’s instructions on Amersham ECL Hyperfilm (GE Healthcare, Limited).
Immunofluorescence
Cells were grown on plastic cover slide chambers (170920, NUNC), fixed with 4% paraformaldehyde (Sigma-Aldrich Quimica) for 20 min at room temperature or fixed with cold methanol for 5 min. After fixation cells were washed three times with TBS and blocked in TBS containing 0.5% Triton X-100 plus 6% donkey serum (S-30; Chemicon) for 30 min at room temperature. The following antibodies in TBS containing 0.1% Triton X-100 plus 6% donkey serum were used: p27Kip1 (M-197, Santa Cruz Biotechnology) 1:200, Brachyury (AF2085, R&D Systems, Inc.,) 1:25, E-Cadherin (BD, Transduction Laboratories) 1:100, GFP (GFP-1020, AVES) 1:250, Actin-〈 Smooth Muscle (A5228, Sigma) 1:400, ®-Tubulin III TUJ1 (MMS-435P, Covance) 1:500 and Nestin (AB5922, Chemicon) 1:100.
After overnight primary antibody incubation, samples were washed three times with TBS 0.1% Triton X-100 plus 6% donkey serum and incubated with CY-conjugated secondary anti-rabbit, anti-mouse, anti-goat or anti-chicken antibodies (Jackson) for 2 h at 37°C. Samples were washed three times in TBS buffer at room temperature for 5 min and were counterstained with DAPI (Invitrogen, 21490, 1:10,000) for 10 min at room temperature in the dark. Samples were covered with mounting medium. Images were taken using Leica SP5 confocal microscope.
Time-lapse microscopy analysis
Two x 105 hESC were infected with lenti-p27pCCL or GFPpCCL control vector for one hour and then plated in chamber slide flasks with a further 1 mL of hESC conditioned medium. Cells were left grow in a 5% CO2 atmosphere at 37°C and photographs of individual cells were taken after 4, 5, 6 and 7 d using Leica DMI4000B inverted microscope equipped with a Leica DFC320 digital fluorescence camera and Leica AF6000 software. After this time, cultures were fixed with 4% Paraformaldehyde for 20 min at room temperature and immunofluorescence analysis performed for detection of p27Kip1 expression. Cells were counterstained with DAPI 1:10,000 for 10 min to allow the identification of nuclei.
Isolation of RNA and quantitative RT-PCR analysis
Total RNA was isolated using TRIzol reagent (Invitrogen) as recommended by the manufacturer and 500 ng of mRNA were used to synthesize cDNA using Cloned AMV First-strand cDNA synthesis kit (Invitrogen). One µl of the reaction was mixed with SYBR GreenER qPCR superMix for ABI PRISM (Invitrogen) and corresponding primers. Each reaction was done per duplicate and run in an ABI Prism 7000 thermocycler (Applied Biosystems). Relative mRNA expression levels were determined using the comparative threshold cycle method. The graphics represent [2-CT (target)/2-CT (housekeeping)] cell line/[2-CT (target)/2-CT (housekeeping)] control cells. The sequences of the oligonucleotides used for the quantitative RT-PCR are shown in (Table S1).
For the expression analysis of stem cell related genes, we used the Human Stem Cell RT2 profiler PCR array (SuperArray Bioscences Corporation) with 500 ng of RNA isolated with Qiagen RNA easy mini kit following the manufacturer’s directions. Relative mRNA expression levels were determined using the comparative threshold cycle method. The graphics represent [2-CT (target)/2-CT (housekeeping)] cell line/[2-CT (target)/2-CT (housekeeping)] control cells. Values that were not two-times upregulated or 50% downregulated were not considered as significant changes.
Constructs and lentiviral production
Human p27 cDNA was amplified by PCR using a forward primer containing an Age I restriction enzyme site and a reverse primer containing a Cla I restriction enzyme site. Primer sequences were as follows:
Forward, 5′-GAT CAC CGG TGC CAT GGC TTA CCC ATA CGA TGT TCC AGA TTA CGC GTC AAA CGT GCG AGT GTC TAA CGG G-3′
Reverse, 5′-GAT CAT CGA TTT ACG TTT GAC GTC TTC TGA GGC CAG GC-3′ (amplified a fragment of 650 bp). PCR products were obtained after 20 cycles of amplification with an annealing temperature of 60°C. PCR products were cloned into pcRII (Invitrogen) to give pcRII-p27. The fragment was digested using Age I/Cla I restriction enzymes (NEB) and cloned into Age I and Cla I sites of pCCL TET OFF inducible lentiviral vector. GFP pCCL TET OFF inducible lentiviral vector was kindly provided by Dr. Antonella Consiglio.
p27 shRNA and non target shRNA constructs were cloned into pLK0.1 puromycin lentiviral vector and were purchased from Sigma-Aldrich. Puromycin sequence was removed by digestion with Bam H1 (New England Biolabs) and Asp 718 (Roche) restriction enzymes and a GFP sequence containing Bam H1 and KpnI cloning sites was cloned instead. GFP cDNA was amplified by PCR using a forward primer containing a Bam H1 restriction enzyme site and a reverse primer containing a KpnI restriction enzyme site. Primer sequences were as follows:
Forward
5′-GAT CGG ATC CAT GGT GAG CAA GGG C-3′
Reverse
5′-GAT CGG TAC CTT ACT TGT ACA GC-3′
PCR products were obtained after 30 cycles of amplification with an annealing temperature of 58°C. PCR products were cloned into pGEM-T easy vector (Promega Biotech Iberica) and digested with Bam H1 and Asp 718 restriction enzymes. The fragment was cloned into Bam H1 and Asp 718 sites of p27 shRNA and non target shRNA.
Lentiviruses were independently produced by transfecting the cell line 293T with packaging and envelope vectors: pMDLg-pRRE (6.5 µg), pMD2.VSV.G (4 µg), pRSV-Rev (2.5 µg) kindly provided by L. Naldini (San Raffaele Telethon Institute for Gene Therapy-‘Vita-Salute San Raffaele’ University Medical School, Milano, Italy) and lentiviral constructs (10 µg) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After 16 h medium was replaced with 6 mL of hESC conditioned medium, and cells incubated at 37°C in a 5% CO2 atmosphere. Viral supernatant was harvested after 24 and 48 h, filtered and stored at -80°C in mL aliquots. Virus titer was tested using a HIV-1 p24 ELISA kit (Perkin Elmer). 2,00,000 human ESC were infected in suspension with 1 mL of viral supernatant at 37°C in a 5% CO2 atmosphere for one hour and then plated in a well of 6-well plates with a further 1 mL of hESC conditioned medium. The next day the medium was changed.
Chromatin immunoprecipitation assays
ChIP assays were performed as described,Citation22 by using chromatin from the human embryonic stem cells cultured and treated as described above. The antibodies used for ChIPs assays are listed in Figures. Quantification of ChIP was performed by real-time PCR using Roche Lightcycler (Roche). The change in promoter occupancy in the immunoprecipitated (IP) compared with input (Ref) fractions was calculated using the comparative Ct (the number of cycles required to reach a threshold concentration) method with the equation 2Ct(IP) - Ct(Ref). Real Time PCR primers for the human Twist1 gene from +1.0 Kb to -3.0 Kb at 1,000 bp intervals were bought from SA biosciences. Oct4 primers were used as a positive technical control to ensure the ChIP had worked.
Statistical analysis
All data were analyzed applying ANOVA or Student’s t-test statistic analysis. Probability values that were less than 0.05 were considered statistically significant.
Conclusion
In conclusion, we have proven using functional genetic approach that a cell cycle inhibitor, p27Kip1, controls self-renewal and pluripotency of hESC in undifferentiated conditions, by imposing a G1 arrest and controlling the expression of the specific differentiation genes Brachyury and Twist. This suggests that p27Kip levels must be maintained at a critical level in undifferentiated hESC in order to maintain low levels of mesenchymal markers and that alteration of expression either way unbalances the pluripotent state toward differentiation and cell cycle slowing.
Author Contributions
Cristina Menchón: Collection and assembly of data, data analysis and interpretation, manuscript writing. Michael J. Edel: Conception and design, financial support, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript. Juan Carlos Izpisúa Belmonte: Financial support, administrative support, final approval of manuscript, provision of study material or patients.
Additional material
Download Zip (3.7 MB)Acknowledgments
The authors are indebted to José Miguel Andrés Vaquero for assistance with flow cytometry, Meritxell Carrió for expert assistance with cell culture techniques, Mercé Martí Gaudes, Lola Mulero Pérez and Esther Melo for bioimaging assistance. Dr. Naldini for providing pCCL vector. Federico González for helpful expertise with the cloning. Members of the laboratory for helpful discussion. María José Barrero for assistance with the ChIP assay. Cristina Menchón is partly supported by a pre-doctoral grant from the Spanish Ministry (AP-2004–3523). This work was partially supported by grants from the Fondo de Investigaciones Sanitarias (P1071209), MICINN, Fundacion Cellex and Sanofi-Aventis.
References
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981; 292:154 - 6; http://dx.doi.org/10.1038/292154a0; PMID: 7242681
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, et al. Embryonic stem cell lines derived from human blastocysts. Science 1998; 282:1145 - 7; http://dx.doi.org/10.1126/science.282.5391.1145; PMID: 9804556
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126:663 - 76; http://dx.doi.org/10.1016/j.cell.2006.07.024; PMID: 16904174
- Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F, et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol 2008; 26:1276 - 84; http://dx.doi.org/10.1038/nbt.1503; PMID: 18931654
- Gonzalez F, Barragan Monasterio M, Tiscornia G, Montserrat Pulido N, Vassena R, Batlle Morera L, et al. Generation of mouse-induced pluripotent stem cells by transient expression of a single nonviral polycistronic vector. Proc Natl Acad Sci U S A 2009; 106:8918 - 22; http://dx.doi.org/10.1073/pnas.0901471106; PMID: 19458047
- Raya A, Rodríguez-Pizà I, Guenechea G, Vassena R, Navarro S, Barrero MJ, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature 2009; 460:53 - 9; http://dx.doi.org/10.1038/nature08129; PMID: 19483674
- Edel MJ, Menchon C, Menendez S, Consiglio A, Raya A, Izpisua Belmonte JC. Rem2 GTPase maintains survival of human embryonic stem cells as well as enhancing reprogramming by regulating p53 and cyclin D1. Genes Dev 2010; 24:561 - 73; http://dx.doi.org/10.1101/gad.1876710; PMID: 20231315
- Becker KA, Ghule PN, Therrien JA, Lian JB, Stein JL, van Wijnen AJ, et al. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J Cell Physiol 2006; 209:883 - 93; http://dx.doi.org/10.1002/jcp.20776; PMID: 16972248
- Fluckiger AC, Marcy G, Marchand M, Négre D, Cosset FL, Mitalipov S, et al. Cell cycle features of primate embryonic stem cells. Stem Cells 2006; 24:547 - 56; http://dx.doi.org/10.1634/stemcells.2005-0194; PMID: 16239321
- White J, Dalton S. Cell cycle control of embryonic stem cells. Stem Cell Rev 2005; 1:131 - 8; http://dx.doi.org/10.1385/SCR:1:2:131; PMID: 17142847
- Kalaszczynska I, Geng Y, Iino T, Mizuno S, Choi Y, Kondratiuk I, et al. Cyclin A is redundant in fibroblasts but essential in hematopoietic and embryonic stem cells. Cell 2009; 138:352 - 65; http://dx.doi.org/10.1016/j.cell.2009.04.062; PMID: 19592082
- Savatier P, Lapillonne H, van Grunsven LA, Rudkin BB, Samarut J. Withdrawal of differentiation inhibitory activity/leukemia inhibitory factor up-regulates D-type cyclins and cyclin-dependent kinase inhibitors in mouse embryonic stem cells. Oncogene 1996; 12:309 - 22; PMID: 8570208
- Buttitta LA, Edgar BA. Mechanisms controlling cell cycle exit upon terminal differentiation. Curr Opin Cell Biol 2007; 19:697 - 704; http://dx.doi.org/10.1016/j.ceb.2007.10.004; PMID: 18035529
- Nguyen L, Besson A, Heng JI, Schuurmans C, Teboul L, Parras C, et al. p27kip1 independently promotes neuronal differentiation and migration in the cerebral cortex. Genes Dev 2006; 20:1511 - 24; http://dx.doi.org/10.1101/gad.377106; PMID: 16705040
- Vernon AE, Devine C, Philpott A. The cdk inhibitor p27Xic1 is required for differentiation of primary neurones in Xenopus. Development 2003; 130:85 - 92; http://dx.doi.org/10.1242/dev.00193; PMID: 12441293
- Vernon AE, Movassagh M, Horan I, Wise H, Ohnuma S, Philpott A. Notch targets the Cdk inhibitor Xic1 to regulate differentiation but not the cell cycle in neurons. EMBO Rep 2006; 7:643 - 8; PMID: 16648822
- Messina G, Blasi C, La Rocca SA, Pompili M, Calconi A, Grossi M. p27Kip1 acts downstream of N-cadherin-mediated cell adhesion to promote myogenesis beyond cell cycle regulation. Mol Biol Cell 2005; 16:1469 - 80; http://dx.doi.org/10.1091/mbc.E04-07-0612; PMID: 15647380
- Vernon AE, Philpott A. A single cdk inhibitor, p27Xic1, functions beyond cell cycle regulation to promote muscle differentiation in Xenopus. Development 2003; 130:71 - 83; http://dx.doi.org/10.1242/dev.00180; PMID: 12441292
- Movassagh M, Philpott A. Cardiac differentiation in Xenopus requires the cyclin-dependent kinase inhibitor, p27Xic1. Cardiovasc Res 2008; 79:436 - 47; http://dx.doi.org/10.1093/cvr/cvn105; PMID: 18442987
- Deschênes C, Vézina A, Beaulieu JF, Rivard N. Role of p27(Kip1) in human intestinal cell differentiation. Gastroenterology 2001; 120:423 - 38; http://dx.doi.org/10.1053/gast.2001.21199; PMID: 11159883
- Raya A, Rodríguez-Pizà I, Arán B, Consiglio A, Barri PN, Veiga A, et al. Generation of cardiomyocytes from new human embryonic stem cell lines derived from poor-quality blastocysts. Cold Spring Harb Symp Quant Biol 2008; 73:127 - 35; http://dx.doi.org/10.1101/sqb.2008.73.038; PMID: 19028986
- Strutt H, Paro R. Mapping DNA target sites of chromatin proteins in vivo by formaldehyde crosslinking. Methods Mol Biol 1999; 119:455 - 67; PMID: 10804532
- Sherr CJ, Roberts JM. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev 1995; 9:1149 - 63; http://dx.doi.org/10.1101/gad.9.10.1149; PMID: 7758941
- Díez-Juan A, Andrés V. Coordinate control of proliferation and migration by the p27Kip1/cyclin-dependent kinase/retinoblastoma pathway in vascular smooth muscle cells and fibroblasts. Circ Res 2003; 92:402 - 10; http://dx.doi.org/10.1161/01.RES.0000059306.71961.ED; PMID: 12600894
- Bryja V, Pacherník J, Soucek K, Horvath V, Dvorák P, Hampl A. Increased apoptosis in differentiating p27-deficient mouse embryonic stem cells. Cell Mol Life Sci 2004; 61:1384 - 400; http://dx.doi.org/10.1007/s00018-004-4081-4; PMID: 15170516
- Edel MJ, Izpisua Belmonte JC. The cell cycle and pluripotency: Is there a direct link?. Cell Cycle 2010; 9:2694 - 5; http://dx.doi.org/10.4161/cc.9.14.12456; PMID: 20581443
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 2004; 117:927 - 39; http://dx.doi.org/10.1016/j.cell.2004.06.006; PMID: 15210113
- Karreth F, Tuveson DA. Twist induces an epithelial-mesenchymal transition to facilitate tumor metastasis. Cancer Biol Ther 2004; 3:1058 - 9; http://dx.doi.org/10.4161/cbt.3.11.1302; PMID: 15640618
- Puisieux A, Valsesia-Wittmann S, Ansieau S. A twist for survival and cancer progression. Br J Cancer 2006; 94:13 - 7; http://dx.doi.org/10.1038/sj.bjc.6602876; PMID: 16306876
- Leshem Y, Spicer DB, Gal-Levi R, Halevy O. Hepatocyte growth factor (HGF) inhibits skeletal muscle cell differentiation: a role for the bHLH protein twist and the cdk inhibitor p27. J Cell Physiol 2000; 184:101 - 9; http://dx.doi.org/10.1002/(SICI)1097-4652(200007)184:1<101::AID-JCP11>3.0.CO;2-D; PMID: 10825239
- Vigneron A, Cherier J, Barré B, Gamelin E, Coqueret O. The cell cycle inhibitor p21waf1 binds to the myc and cdc25A promoters upon DNA damage and induces transcriptional repression. J Biol Chem 2006; 281:34742 - 50; http://dx.doi.org/10.1074/jbc.M602492200; PMID: 16923815
- Demidenko ZN, Blagosklonny MV. Growth stimulation leads to cellular senescence when the cell cycle is blocked. Cell Cycle 2008; 7:3355 - 61; http://dx.doi.org/10.4161/cc.7.21.6919; PMID: 18948731