Abstract
Histone gene expression is tightly controlled during cell cycle. The epigenetic mechanisms underlying this regulation remain to be fully elucidated. Pygopus 2 (Pygo2) is a context-dependent co-activator of Wnt/beta-catenin signaling and a chromatin effector that participates in histone modification. In this study, we show that Pygo2 is required for the optimal expression of multiple classes of histone genes in cultured human mammary epithelial cells. Using chromatin immunoprecipitation assay, we demonstrate that Pygo2 directly occupies the promoters of multiple histone genes and enhances the acetylation of lysine 56 in histone H3 (H3K56Ac), previously shown to facilitate yeast histone gene transcription, at these promoters. Moreover, we report reduced global levels of H3K56Ac in Pygo2-depleted cells that occur in a cell cycle-independent manner. Together, our data uncover a novel regulator of mammalian histone gene expression that may act in part via modifying H3K56Ac.
Introduction
Histones are major protein constituents of the nucleosome, structural unit of eukaryotic chromatin.Citation1 Two copies of histone H2A, H2B, H3 and H4 constitute the core of the nucleosome, whereas histone H1 associates with the linker DNA between nucleosomes. Through regulating DNA accessibility, histone proteins play important roles in diverse chromatin transactions, including DNA replication, repair, recombination and transcription.Citation2 The expression of histone genes is tightly controlled and usually coupled to DNA synthesis during S phase of the cell cycle.Citation3,Citation4 Characteristically, the five classes of replication-dependent histone genes are clustered together in the genome in metazoans,Citation5 a unique organization that allows highly coordinated co-regulation. To date, studies have uncovered transcriptional factors that regulate either a specific subtype (e.g., octamer binding protein 1, histone nuclear factor P) or multiple subtypes [e.g., yin yang 1, FADD-like IL-1β-converting enzyme associated huge protein and nuclear protein ataxia-telangiectasia locus (NPAT)] of replication-dependent histone genes.Citation6–Citation10 Clearly, additional regulators of histone gene transcription, particularly during cell cycle progression, remain to be discovered.
Post-translational modification of histones has emerged as a key epigenetic mechanism for transcriptional regulation.Citation11 Such modifications, including acetylation and methylation, can lead to altered chromatin configuration that allows or inhibits loading of the transcriptional machinery responsible for transcription initiation or elongation. It is well-established that acetylation of lysine (K) residues in the N-terminal tails of histone H3 (e.g., H3K9Ac and H3K14Ac) and H4 participates in gene activation.Citation12,Citation13 A role in transcription has also been strongly implicated for acetylation of lysine 56 in the globular domain of histone H3 (H3K56Ac).Citation14–Citation17 Originally identified and extensively studied in yeast, H3K56Ac has been well-recognized for its importance in regulating nucleosomal assembly following DNA replication and repair, whereas its specific role in transcription is less defined but seems to also involve nucleosomal dynamics.Citation16,Citation18–Citation21 H3K56Ac is highly enriched in yeast histone genes. and the loss of this modification as well as the histone acetyltransferase(s) (HAT) that is responsible for it results in compromised histone gene transcription.Citation15 H3K56Ac is also enriched at nearly all canonical histone genes in human embryonic stem cells.Citation14 However, the functional involvement of H3K56Ac in transcriptional control, e.g., of histone genes, in mammalian cells remains unclear.
Mammalian Pygopus 2 (Pygo2) is a member of the Pygopus family of proteins that are evolutionarily conserved across species.Citation22 Initially identified in Drosophila, Pygopus functions as a transcriptional co-activator of the Wnt (Wg)/β-catenin signaling pathway.Citation23–Citation26 In mammals, the involvement of Pygopus proteins in Wnt/β-catenin signaling is context-dependent. For example, while acting in a β-catenin-independent manner in lens development, Pygo2 regulates mammary gland development and stem/progenitor cell expansion at least in part by regulating Wnt/β-catenin-signaling.Citation27,Citation28 The conserved plant homeo domain at the C-termini of Pygopus proteins directly binds to histone H3, which is di- or tri-methylated at lysine 4 (H3K4me2/3), histone marks associated with active transcription.Citation28,Citation29 In addition to its ability to bind nuclear β-catenin via adaptor protein BCL9, Pygo2 also associates with histone-modifying enzymes, such as histone methyltransferase (e.g., myeloid/lymphoid or mixed-lineage leukemia protein 2) and HAT [e.g., CREB binding protein (CBP)/E1A binding protein p300 (p300) and general control of amino-acid synthesis 5-like 2 (GCN5)] complexes, and recruits them to target chromatin loci.Citation28,Citation30,Citation31 These molecular interactions enable Pygo2 to act as a chromatin effector that assists with both “reading” and “writing” of the histone code. The full spectrum of downstream targets of this important chromatin effector, whether Wnt/β-catenin-dependent or -independent and in different biological contexts, remains to be elucidated.
In the current work, we studied the impact of RNAi-mediated Pygo2 knockdown on histone gene expression in human mammary epithelial MCF10A cells. We found that Pygo2 is required for the expression of a majority of histone genes and for the acetylation of histone H3 at K56 both at specific histone gene promoters and globally in the cells.
Results
Pygo2 is required for histone gene expression.
To better understand the cellular and molecular function of human Pygo2, we sought to identify novel Pygo2-responsive genes by DNA microarray analysis. cDNA was prepared from MCF10A cells at 24 h after transfection of control or a Pygo2-specific siRNA. At this time point, both mRNA and protein levels of Pygo2 were reduced by ∼2-fold ( and C). In keeping with a co-activator function of Pygo2 in the Wnt signaling pathway, several known Wnt targets, including c-Myc and cyclin D1, were downregulated in Pygo2 siRNA-treated cells (data not shown). Interestingly, a large number of histone genes (50 of 66 total non-overlap probes ≥ 75% of the gene family, p < 0.001) showed decreased expression in Pygo2-knockdown cells ( and Table S1). Results of reverse transcription and semi-quantitative PCR (RT-qPCR) analysis confirmed the decreased mRNA levels of representative members of histone H2A, H2B, H3 and H4 gene families (). These results are consistent with our previous finding of reduced histone H3 protein levels in Pygo2-deficient MCF10A cells and mammary glands.Citation28
The activation of most histone genes is DNA replication-dependent.Citation3,Citation4 Moreover, our previous study showed that Pygo2 is necessary for efficient G1-S cell cycle transition, as S-phase population was significantly reduced 3 d after Pygo2 siRNA treatment.Citation28 It is therefore possible that the observed reduction in histone expression arises as an indirect consequence of the cell cycle arrest. To address this, we compared the cell cycle profiles of control and Pygo2-knockdown cells at 24 h after siRNA transfection, when changes in histone gene expression already occurred. Cells were pulse-labeled with BrdU for 30 min, subjected to staining with FITC-labeled anti-BrdU antibody and PI and analyzed by flow cytometry. As shown in , the fraction of BrdU-positive cells was not significantly affected by Pygo2 knockdown at this early time point (20.2 ± 4.6% with siPygo2 vs. 24.9 ± 1.7% with siControl, p = 0.17). Thus, the reduction in histone gene expression occurred prior to the cell cycle arrest.
To rule out potential off-target effects of Pygo2 siRNA, we generated high-titer lentiviruses expressing two shRNAs targeting different regions of Pygo2 (sh Pygo2-A and sh Pygo2-B, as indicated in ) and two control shRNAs containing a scrambled sequence (sh Scramble) or a sequence against green fluorescence protein (GFP) (sh GFP). To explore the kinetics of histone reduction after Pygo2 knockdown, we performed time-course analysis. At 48 h after infection, Pygo2 shRNA-expressing viruses, but not control viruses (sh Scramble or sh GFP), decreased Pygo2 (, left) and histone gene expression (, left). At this early time, the impact of the shRNAs on cell cycle progression was unremarkable (, top). By 72 h post-infection, histone gene expression was further downregulated in Pygo2-depleted cells, along with a concomitant partial arrest of cells at the G1 and/or S phase (, bottom and , right). Again, the observable reduction in histone gene expression preceded the apparent defect in cell cycle progression, in accordance with data from the Pygo2 siRNA experiments above. These results suggest that Pygo2 has a primary effect on the optimal expression of multiple histone genes in MCF10A cells.
Pygo2 binds to multiple histone promoters and is required for the maximal acetylation of histone H3 K56 at these promoters.
The effect of Pygo2 depletion on the expression of a large number of histone genes led us to wonder whether Pygo2 is involved in the normal organization of histone gene clusters in the nucleus. As a unique marker for the histone gene clusters, NPAT specifically localizes to large nuclear foci or histone locus bodies (HLBs) where these gene clusters reside, and activates the transcription of histone genes of different subtypes.Citation7 During the cell cycle, there are usually two NPAT-associated HLBs on chromosome 6p21 in G1 and G2 phases, four HLBs on 6p21 and 1q21 in S phase and none in M phase. We examined the effect of Pygo2 knockdown on HLBs in MCF10A cells. In contrast to the diffused presence of endogenous Pygo2 within the cell nucleus, as evident by immunofluorescence analysis using two different Pygo2-specific antibodies ( and ); NPAT was concentrated only at discrete HLBs as expected (). At 24 h after transfection with Pygo2 siRNA, when histone gene expression was already reduced (see ), the number of HLBs was not significantly affected (). Similar results were obtained at 72 h after siRNA transfection (data not shown). Therefore, Pygo2 is not required for nuclear foci formation of NPAT on HLBs in MCF10A cells.
We next asked whether Pygo2 protein itself physically occupies the histone gene promoters. In chromatin immunoprecipitation (ChIP) assays, Pygo2 was specifically detected on several histone gene promoters examined, including those of H2B/r, H3/c and H4/e (). In contrast, no binding to the Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) promoter was observed. Given the demonstrated role of Pygo2 in regulating histone modification and the importance of H3K56Ac in histone gene activation in yeast,Citation15,Citation28,Citation30 we tested whether Pygo2 occupancy at the histone promoters co-exists with H3K56Ac, and, if so, whether Pygo2 depletion causes reduction in this histone modification. MCF10A cells were infected with recombinant lentiviruses expressing sh GFP or sh Pygo2-A and subjected to ChIP analysis. Indeed, appreciable levels of H3K56Ac were detected at the histone promoters in the same regions bound by Pygo2 in control cells, and a 2–3-fold reduction was seen in sh Pygo2-A-treated cells ( and D). We also examined H3K4me3 at these histone promoters, but signals were weak even in control cells (data not shown). However, the levels of H3K9/K14Ac were high at the histone promoters, but they were minimally affected by depletion of Pygo2 ( and F). These results demonstrate that Pygo2 is recruited to the histone promoters and is required for their maximal H3K56 acetylation in MCF10A cells.
Pygo2 is important for global H3K56Ac throughout the cell cycle.
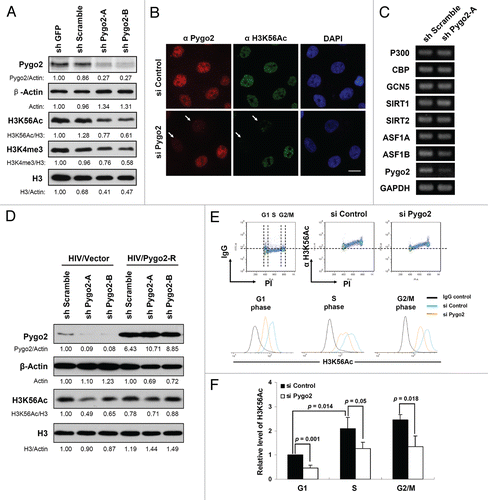
We next wondered whether Pygo2 regulates H3K56Ac more broadly than at the histone promoters, as we have found for H3K4me3 previously in reference Citation28. To address this, we measured the levels of global H3K56Ac by protein gel blot analysis of whole-cell extracts prepared from control and Pygo2-depleted MCF10A cells. As with H3K4me3, depletion of Pygo2 using two different shRNAs led to significantly reduced levels of H3K56Ac, after normalizing against total histone H3 protein levels (). In keeping with the changes in transcript levels (, C and ), total histone H3 protein levels were markedly reduced in Pygo2-knockdown cells compared with control cells. Immunofluorescence analysis of single cells confirmed that cells with efficient Pygo2 knockdown displayed a reduced H3K56Ac staining signal in their nuclei (). Of note, a similar effect was observed when an anti-H3K9Ac antibody was used (data not shown). Overall, the reduced histone H3 acetylation was not accompanied by significantly altered levels of expression of the following enzymes or chaperones: CBP, p300 and GCN5; histone deacetylases sirtuin1 (SIRT1) and sirtuin2 (SIRT2) and histone chaperones anti-silencing function 1A (ASF1A) and anti-silencing function 1B (ASF1B) ().Citation32.Citation35 To further substantiate the observation of Pygo2-dependent H3K56Ac, we performed RNAi-rescue experiments using recombinant lentiviruses that express an RNAi-resistant form of Pygo2. Expression of exogenous Pygo2 was able to restore global H3K56Ac in Pygo2-knockdown cells, as evident by the elevated levels of H3K56Ac after normalization against total H3 levels, which were also elevated ().
H3K56Ac is an S phase-enriched histone mark in yeast,Citation20,Citation36,Citation37 but its cell cycle distribution in mammals is controversial.Citation32,Citation38,Citation39 Since the observed reduction in global H3K56Ac occurred after Pygo2 knockdown at an early time point (24 h), when the G1-S cell cycle arrest was not yet significant, we surmise that it is unlikely that this reduction is simply a consequence of defective G1-S progression. To further corroborate this notion, we performed flow cytometry analysis of both H3K56Ac level and DNA content in control or Pygo2 siRNA-treated cells. H3K56Ac signals were observed in all stages of the cell cycle in control siRNA-treated cells but displayed an apparent rise during S phase (, top middle and ). The knockdown of Pygo2 resulted in significantly reduced levels of H3K56Ac in G1-, S- and G2/M-phase cells (peaks shifting to the left in , bottom parts and ). Therefore, Pygo2 appears to regulate the global level of H3K56Ac at all stages of the cell cycle in MCF10A cells.
Discussion
Despite emerging evidence that Pygo2 is a chromatin effector that regulates histone post-translational modifications at target loci, so far known downstream targets of Pygo2 have been limited to the several genes that are also regulated by the Wnt/β-catenin pathway, such as c-Myc and Lef1.Citation28,Citation30,Citation31,Citation40,Citation41 In this study, we identified genes in three of the core histone classes as novel direct targets of Pygo2 in MCF10A cells. Hence, Pygo2 is likely to exert multiple effects on the cell cycle, including regulating G1-S transition through c-Myc and cyclin D1 and S-phase progression through the histone genes. Histone genes have not been previously reported to be Wnt targets. Examination of the sequences within the PCR regions (∼−150 to +50) for the three core histone promoters that Pygo2 occupies in our ChIP analysis revealed no potential LEF/TCF binding sites (data not shown). However, consensus LEF/TCF sites were found in sequences further upstream (up to −5 kb; 4, 4 and 2 such sites in H2B/r, H3/c and H4/e, respectively). It remains possible that Pygo2 converges with Wnt signaling to regulate some downstream targets such as c-Myc, Cyclin D1 or Lef1,Citation27,Citation28 but regulates additional targets, such as the histone genes, in a Wnt-independent manner. Future studies outside the scope of this work will examine whether Pygo2 binding to the histone promoters occurs via non-canonical LEF/TCF sites or LEF/TCF-independent mechanisms.
Our work adds Pygo2 to the growing list of epigenetic factors that regulate the expression of histone genes beyond a specific subtype. Moreover, we showed that Pygo2 is required for maximal acetylation of histone H3K56 at the responsive histone promoters. The correlative changes in histone gene expression and H3K56Ac at histone promoters in response to Pygo2 depletion imply, but do not prove, the functional importance of H3K56Ac in histone gene activation in mammalian cells. One technical note worthy of consideration is potential off-target effects of siRNA/shRNAs. Although we tested multiple siRNA/shRNAs and they all led to a reduction in histone gene expression and H3K56Ac, we did not always observe a strict correlation between knockdown efficiency and the extent of reduction in histone gene expression/H3K56Ac (e.g., compare and D). Thus, some off-target effects may still exist. Alternatively, Pygo2 function may be very sensitive to the dosage of itself as well as of the proteins that it associates with. With all that said, the rescue of Pygo2 depletion-induced molecular changes by reintroduction of Pygo2 via a siRNA-resistant cDNA provides corroborative evidence that Pygo2 has a specific involvement in histone gene expression and H3K56Ac in MCF10A cells.
Although deemed specific in previous studies in references Citation32 and Citation42, it is important to note that the anti-H3K56Ac antibody used in our study may still cross-react with other forms of histone acetylation, such as H3K9/K14Ac. We note that our ChIP analysis using H3K9/K14Ac antibody did not detect a similar Pygo2-dependent response at the histone gene promoters as using H3K56Ac antibody. Based on this demonstrated specificity, we surmise that the bulk of the global signals detected by protein gel blotting are likely due to H3K56Ac. The global regulation of H3K56Ac by Pygo2 makes it intriguing to speculate a broader role for Pygo2 in gene transcription. It is also possible that Pygo2 is involved in additional DNA metabolic processes, such as DNA replication and repair. In support of this idea, we found Pygo2 knockdown to render cells more sensitive to DNA damage-inducing treatments, including irradiation and UV (Gu B and Dai X, unpublished results).
In yeast, H3K56Ac occurs in a cell cycle-specific manner, peaking earlier than histone gene expression in S phase.Citation15 Whether H3K56Ac is S phase-enriched in mammalian cells is a controversial issue.Citation32,Citation38,Citation39 We found, through flow cytometry analysis in human MCF10A cells, that H3K56Ac is present throughout the cell cycle, but its level is elevated upon entry into S phase. Importantly, Pygo2 knockdown reduced global H3K56Ac levels in cells at all stages of the cell cycle, arguing for a cell cycle-independent role of Pygo2 in enhancing this histone mark. Further studies are necessary to determine whether Pygo2 controls the cell cycle directly through regulation of H3K56Ac and histone gene expression.
How does Pygo2 affect H3K56Ac? This does not seem to occur indirectly via regulation of expression of the enzymes/chaperones that are responsible for H3K56Ac. Pygo2 physically associates with CBP/p300 and GCN5 HAT complexes,Citation30,Citation31 both of which have been shown to possess the ability to acetylate H3K56 in mammalian cells.Citation32,Citation33,Citation43 Thus, it is plausible to propose that Pygo2 facilitates H3K56Ac via recruiting these HATs onto the histone promoters and to the chromatin. Future work will be needed to probe into the underlying mechanism.
Materials and Methods
Cell culture, siRNA and lentiviral shRNA.
Human mammary epithelial MCF10A cells and embryonic kidney 293T cells were cultured as previously described in reference Citation28. siRNA (Applied Biosystems) for negative control (AM4635) and Pygo2 (AM16706, ID# 123812, 5′-GGA CGU AUC UUC UUC AUA CTT-3′) (at a final concentration of 60 nM) were transfected into MCF10A cells using Lipofectamine 2000 (Invitrogen, 11668-019,) according to the manufacturer's instructions. Recombinant lentiviruses expressing Pygo2 or shRNAs were generated following the instruction provided by Addgene. Briefly, 293T cells were cotransfected with packaging plasmid psPAX.2 (12260), envelope plasmid pMD2.G (12259) and viral vector pLKO.1 containing the shRNA expression cassette for scrambled (1864, 5′-CCT AAG GTT AAG TCG CCC TCG-3′), GFP (12273, 5′-GCA AGC TGA CCC TGA AGT TCA-3′), Pygo2-A (TRCN0000021860, Open Biosystems, 5′-CCT TCT CTG TCC CAA CGA TTT-3′), or Pygo2-B (TRCN0000021861, Open Biosystems, 5′-AGA AGC GAA GGA AGT CAA ATA-3′). Viral infection was performed with 4 µg/ml Polybrene (H9268, Sigma-Aldrich) overnight, and samples were harvested for analysis 48 or 72 h after infection as indicated. For rescue experiments, lentiviruses carrying the vector control [HIV/Vector; plasmid 21373, Addgene, contributed by Z. Werb's laboratory,Citation44 or RNAi-resistant Pygo2 cDNA (HIV/Pygo2-R)] were used to infect cells at 72 h after infection with the shRNA-expressing lentiviruses. Cell lysates were then prepared 24 h later for protein gel blot analysis. The following primers were used to make the Pygo2-R construct: Pygo2 start-EcoRI, 5′-GAA TTC ACC ATG GCC GCC TCG GCG CCG CC-3′, Pygo2 stop-XbaI, 5′-TCT AGA CTT CAC CCA TCG TTA GCA GCC A-3′; shPygo2-A mt, forward, 5′-CCG TCG CTT TCA CAG CGG TTT GCT CAG CCA GGG GCT CC-3′, reverse, 5′-CCG CTG TGA AAG CGA CGG TGG GCC CAG CTC TGC TCT GG-3′; shPygo2-B mt, forward, 5′-AAA AAA CGG AGA AAA TCC AAC ACT CAG GGG CCC TGC ATA CTC-3′, reverse, 5′-GTT GGA TTT TCT CCG TTT TTT TTC TGG ACT CTT CAT TTG CA-3′.
RNA isolation and microarray analysis.
Total RNA was isolated from cells using TRIzol Reagent according to manufacturer's protocol (15596-018, Invitrogen). For microarray analysis, RNA was collected at 24 h after transfection with control or Pygo2 siRN, and was further purified using RNeasy mini kit (74104, QIAGEN). Hybridization of arrays (GeneChip Human Gene 1.0 ST, 901086, Affymetrix) was performed in duplicate. Genes with normalized expression levels over detection threshold were called and analyzed for differential expression using the Cyber-T program (http://cybert.ics.uci.edu/).Citation45 The cutoff value for differential histone gene expression was arbitrarily set as a > 1.35-fold change in transcript level with a t-test p-value < 0.001.
Reverse transcription and semi-quantitative PCR.
Total cDNA was synthesized from RNA using high capacity cDNA reverse transcription (RT) kit (4368814, Applied Biosystems) according to manufacturer's instructions. Semi-quantitative PCR was performed using titrated cycle numbers for each target. The level within linear range was quantified by ImageJ 1.43u (National Institutes of Health). GAPDH level serves as an internal control. Primers used for gene expression analysis are human Pygo2 (NM_138300), forward 5′-CCA GAA AAG AAG CGA AGG AAG TC-3′, reverse 5′-GTT GAA AGC AGG GCC CAT AGG ATT-3′; histone H2A/p (NM_021064), forward 5′-ATG TCT GGA CGT GGC CAA GCA-3′, reverse 5′-AGC TTG TTG AGC TCC TCG TG-3′;Citation6 histone H2B/j (AF531291), forward 5′-CCG AAG AAG GGC TCC AAG AA-3′, reverse 5′-TTA TTT GGA GCT GGT GTA CTT G-3′;Citation6 histone H3/c (AF531276), 5′-AGC TCG CAA GTC TAC CGG CG-3′, reverse 5′-CGT TTA GCG TGA ATA GCG CA-3′;Citation6 histone H4/k (NM_003546), forward 5′-CAA AGT TCT GCG CGA CAA CA-3′, reverse 5′-GCC GCC AAA GCC ATA CAG GG-3′;Citation6 P300 (NM_001429), forward, 5′-ATA TGC CAC CAT GGA GAA GC-3′, reverse 5′-TCC CGA CCA TCC ATC AGA TC-3′; CBP (NM_004380), forward, 5′-TCA CCA GTG CCA AGG AAC TG-3′, reverse 5′-TCT TGG CAT TCT TGC TGT CG-3′; GCN5 (NM_021078), forward, 5′-GGA GAT TGT CTT CTG TGC TG-3′, reverse 5′-TGA AGT AGC CGA TGG CGT AC-3′; SIRT1 (NM_012238), forward, 5′-CCA GAA CAG TTT CAT AGA GCC-3′, reverse 5′-TGC AGA TGA GGC AAA GGT TC-3′; SIRT2 (NM_012237), forward, 5′-TCT GTC ACT ACT TCA TGC GC-3′, reverse 5′-ATC CAG CTT AGC GGG TAT TC-3′; ASF1A (NM_014034) forward 5′-TAT GTG GGC TCT GCA GAA AG-3′, reverse 5′-CTG CAT CTG CAT CTG GAA TG-3′; ASF1B (NM_018154) forward 5′-ATC AGA TCC TAG ACT CGG TG-3′, reverse 5′-TGA ACT CCT GTC CAT GGT AG-3′; GAPDH (NM_002046), forward 5′-GGA CCT GAC CTG CCG TCT AGA A-3′, reverse 5′-GGT GTC GCT GTT GAA GTC AGA G-3′.
Protein gel blotting and immunofluorescence analysis.
Cells were lysed and sonicated in high-salt extraction buffer (20 mM Hepes, pH 7.9, 420 mM NaCl, 10% Glycerol, 1.5 mM MgCl2, 0.1% NP-40) containing protease inhibitors, and the amount of total protein in lysates was quantified by Bradford assay (500–0006, Biorad) for normalization purpose. Lysates were subjected to 12% sodium dodecyl sulfate (SDS)-PAGE. After transfer onto nitrocellulose membrane (09-301-128, Whatman), proteins were identified using antibodies including anti-Pygo2,Citation46 anti-β-Actin (ab6276, Abcam), anti-H3K56Ac (2134-1, Epitomics), anti-H3 (06–755, Millipore) and anti-H3K4me3 (05–745, Millipore). Indirect immunofluorescence analysis was performed as previously described in reference Citation47. Antibodies used for the analysis include anti-Pygo2 rabbit polyclonalCitation46 and mouse monoclonal (sc-81363, Santa Cruz Biotechnology), anti-NPAT (611344, BD Biosciences), and anti-H3K56Ac (2134-1, Epitomics).
Chromatin immunoprecipitation assay.
Chromatin immunoprecipitation (ChIP) was performed according to the protocol from Millipore. Briefly, treated cells were cross-linked in 1% formaldehyde at 37°C for 10 min, and chromatin was sheared into 500–1,500 bp fragments on ice using a Sonicator 3000 (Misonix). Lysates were precleared with Salmon sperm DNA/Protein A agarose slurry (16.157, Millipore) for one hour at 4°C, and the recovered supernatant was then incubated at 4°C overnight with control IgG (sc-2027, Santa Cruz Biotechnology), anti-Pygo2 (GTX119726, Genetex), anti-H3K56Ac (2134-1, Epitomics) or anti-H3K9/K14Ac (06–599, Millipore). Immunocomplexes were precipitated by Protein A beads, washed extensively and eluted. Cross-linking was reversed by 200 mM NaCl at 65°C for 5 h, and immunoprecipitated DNA was recovered using Phenol/Chloroform followed by QIAquick PCR purification kit (Qiagen, 28104). Primers for histone promoter regions were as described previously in references 6 and 7, including H2B/r (5′-GGA TTT GCG AAT CCT GAT TGG GCA-3′, 5′-GCA CTG TGT AGC TAT AAA GCG CC-3′), H3/c (5′-GAG TCT GAA CGT TTC TGG TG-3′, 5′-CCG CCG GTA GAC TTG CGA GCT-3′), and H4/e (5′-GCG GGA CTT CCC GCC GAC TTC TTC-3′, 5′-GCA GTA CTT TAC GGT GGC GCT TAG C-3′). Primers for the GAPDH promoterCitation48 (5′-CTG AGC AGA CCG GTG TCA CAT C-3′, 5′-GAG GAC TTT GGG AAC GAC TGA G-3′) were used as a negative control. The semi-quantitative PCR results were quantified by ImageJ 1.43u.
Flow cytometry analysis.
Cells were pulse labeled with BrdU for 30 min and immunostained with anti-BrdU-FITC according to manufacturer's instructions (559619, BD Biosciences). Immunostaining with control IgG (sc-2027, Santa Cruz Biotechnology), or anti-H3K56Ac (2134-1, Epitomics) was performed according to a previous report in reference Citation32. PI staining for DNA content was performed as previously reported in reference Citation47. Samples were analyzed by FACS Caliber (BD Biosciences) and the data analysis was performed by Flowjo 7.6.1.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Abbreviations
| Pygo2 | = | Pygopus 2 |
| H3K56Ac | = | histone H3 lysine 56 acetylation |
| H3K4me3 | = | histone H3 lysine 4 tri-methylation |
| H3K9/K14Ac | = | histone H3 lysine 9/lysine 14 acetylation |
| NPAT | = | nuclear protein ataxia-telangiectasia locus |
| HAT | = | histone acetyltransferase |
| GAPDH | = | glyceraldehyde-3-phosphate dehydrogenase |
| PI | = | propidium iodide |
| GFP | = | green fluorescence protein |
| HLBs | = | histone locus bodies |
| ChIP | = | chromatin immunoprecipitation |
| CBP | = | CREB binding protein |
| p300 | = | E1A binding protein p300 |
| GCN5 | = | general control of amino-acid synthesis 5-like 2 |
| SIRT1 | = | sirtuin1 |
| SIRT2 | = | sirtuin2 |
| ASF1A | = | anti-silencing function 1A |
| ASF1B | = | anti-silencing function 1B |
Figures and Tables
Figure 1 Altered histone gene expression in Pygo2-knockdown MCF10A cells. (A) Protein gel blot analysis showing effective depletion of Pygo2 protein at 24 h after siRNA treatment. (B) Pie diagram showing microarray data on histone gene expression upon Pygo2 knockdown. (C) RT-qPCR analysis of select histone genes. Shown are average values from two independent experiments with standard deviations. (D) Flow cytometry analysis of DNA content (propidium iodide, or PI) and DNA synthesis (BrdU) in siRNA-treated cells (24 h after treatment). Shown are profiles from a representative experiment as well as average values from three independent experiments with standard deviations. *p = 0.17 using two-tailed t-tests assuming equal variance.
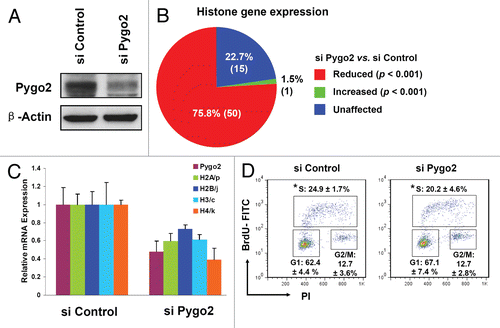
Figure 2 Histone gene expression change precedes cell cycle arrest in Pygo2-depleted MCF10A cells. (A) Schematic diagram showing the design of siRNA/shRNAs for human Pygo2. (B) Protein gel blot analysis of MCF10A whole-cell lysates showing knockdown of Pygo2 at 48 and 72 h post-infection. Signals are quantified by ImageJ 1.43u and normalized as indicated. (C) Flow cytometry analysis of cell cycle progression of infected cells as in (B). Shown are profiles from a representative experiment as well as average values from three independent experiments with standard deviations. (D) RT-qPCR analysis of select histone genes after knockdown of Pygo2. Similar results are obtained from two independent experiments. Expression levels are quantified and normalized against GAPDH control.
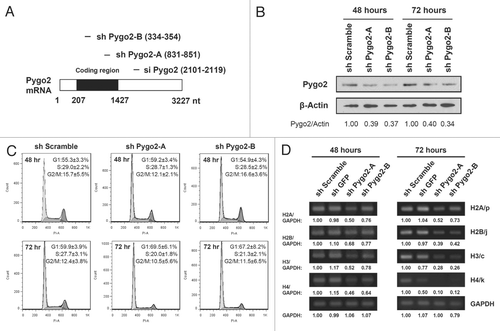
Figure 3 Pygo2 occupancy and H3K56 acetylation at select histone promoters. (A) Immunofluorescence analysis of Pygo2 (green) and NPAT-associated HLBs (red) 24 h after siRNA treatment. White arrows indicate Pygo2-depleted cells. Cell nuclei are stained blue with DAPI. Scale bar: 10 µm. Quantification of HLBs in control (n = 103) and Pygo2-knockdown (n = 91) cells is shown on the right. Error bars indicate standard deviations of two independent experiments. (B) ChIP analysis of Pygo2 occupancy on select histone gene promoters. GAPDH promoter was used as a negative control. (C–F) ChIP analysis of H3K56Ac (C and D) and H3K9/K14Ac (E and F) at select histone promoters after treatment with the indicated shRNAs. (D and F) are quantifications of ChIP experiments in (C and E), respectively. Error bars are standard deviations of two independent experiments.
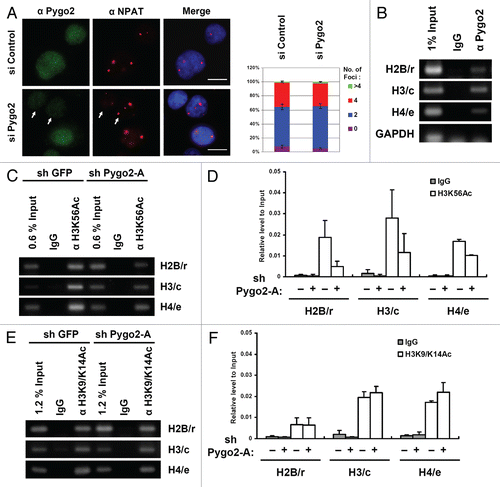
Figure 4 Reduced global H3K56Ac in Pygo2-knockdown cells. (A) Protein gel blot analysis of cellular H3K56Ac levels in shRNA-treated cells. MCF10A cells were infected with the indicated shRNA-expressing lentivirus for 3 d. Signals are quantified and normalized as indicated. (B) Immunofluorescence analysis of H3K56Ac in siRNA-treated cells (24 h). (C) RT/semi-quantitative PCR analysis of known H3K56Ac-regulating proteins 3 d after shRNA-viral infection. (D) Protein gel blot analysis of H3K56Ac in cells infected with lentiviruses that express the indicated shRNAs plus either lentiviruses that express a RNAi-resistant form of Pygo2 or vector control. Signals are quantified and normalized as indicated. (E) Flow cytometry analysis of H3K56Ac during cell cycle progression. Shown at the top are representative profiles of H3K56Ac/DNA content (PI) from siRNA-treated MCF10A cells (24 h). Bottom, altered H3K56Ac levels at the indicated cell cycle stages. (F) Quantification of results from (E). Error bars are standard deviations of three independent experiments. p-values are calculated using two-tailed t-tests assuming equal variance.
Additional material
Download Zip (78.9 KB)Acknowledgments
We thank the UCI Genomics High Throughput Facility (GHTF) and Sue and Bill Gross Stem Cell Research Center Core Facility for expert service. We also thank Julie Wells for technique assistance. This work was supported by NIH Grant R01-GM083089 (to X.D.). B.G. was supported by a California Breast Cancer Research Program (CBCRP) Postdoctoral Fellowship (14FB-0129). K.W. was supported by a US Department of Defense Breast Cancer Research Program (DOD BCRP) Postdoctoral Fellowship (W81XWH-10-1-0383).
References
- Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 1999; 98:285 - 294; PMID: 10458604; http://dx.doi.org/10.1016/S0092-8674(00)81958-3
- Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell 2004; 116:259 - 272; PMID: 14744436; http://dx.doi.org/10.1016/S0092-8674(04)00044-3
- Osley MA. The regulation of histone synthesis in the cell cycle. Annu Rev Biochem 1991; 60:827 - 861; PMID: 1883210; http://dx.doi.org/10.1146/annurev.bi.60.070191.004143
- Marzluff WF, Duronio RJ. Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr Opin Cell Biol 2002; 14:692 - 699; PMID: 12473341; http://dx.doi.org/10.1016/S0955-0674(02)00387-3
- Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ. The human and mouse replication-dependent histone genes. Genomics 2002; 80:487 - 498; PMID: 12408966; http://dx.doi.org/10.1006/geno.2002.6850
- Barcaroli D, Bongiorno-Borbone L, Terrinoni A, Hofmann TG, Rossi M, Knight RA, et al. FLASH is required for histone transcription and S-phase progression. Proc Natl Acad Sci USA 2006; 103:14808 - 14812; PMID: 17003125; http://dx.doi.org/10.1073/pnas.0604227103
- Zhao J, Kennedy BK, Lawrence BD, Barbie DA, Matera AG, Fletcher JA, et al. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev 2000; 14:2283 - 2297; PMID: 10995386; http://dx.doi.org/10.1101/gad.827700
- Eliassen KA, Baldwin A, Sikorski EM, Hurt MM. Role for a YY1-binding element in replication-dependent mouse histone gene expression. Mol Cell Biol 1998; 18:7106 - 7118; PMID: 9819397
- Fletcher C, Heintz N, Roeder RG. Purification and characterization of OTF-1, a transcription factor regulating cell cycle expression of a human histone H2b gene. Cell 1987; 51:773 - 781; PMID: 3677172; http://dx.doi.org/10.1016/0092-8674(87)90100-0
- Mitra P, Xie RL, Medina R, Hovhannisyan H, Zaidi SK, Wei Y, et al. Identification of HiNF-P, a key activator of cell cycle-controlled histone H4 genes at the onset of S phase. Mol Cell Biol 2003; 23:8110 - 8123; PMID: 14585971; http://dx.doi.org/10.1128/MCB.23.22.8110-23.2003
- Jenuwein T, Allis CD. Translating the histone code. Science 2001; 293:1074 - 1080; PMID: 11498575; http://dx.doi.org/10.1126/science.1063127
- Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev 2002; 12:142 - 148; PMID: 11893486; http://dx.doi.org/10.1016/S0959-437X(02)00279-4
- Berger SL. The complex language of chromatin regulation during transcription. Nature 2007; 447:407 - 412; PMID: 17522673; http://dx.doi.org/10.1038/nature05915
- Xie W, Song C, Young NL, Sperling AS, Xu F, Sridharan R, et al. Histone h3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Mol Cell 2009; 33:417 - 427; PMID: 19250903; http://dx.doi.org/10.1016/j.molcel.2009.02.004
- Xu F, Zhang K, Grunstein M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 2005; 121:375 - 385; PMID: 15882620; http://dx.doi.org/10.1016/j.cell.2005.03.011
- Williams SK, Truong D, Tyler JK. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc Natl Acad Sci USA 2008; 105:9000 - 9005; PMID: 18577595; http://dx.doi.org/10.1073/pnas.0800057105
- Värv S, Kristjuhan K, Peil K, Looke M, Mahlakoiv T, Paapsi K, et al. Acetylation of H3 K56 is required for RNA polymerase II transcript elongation through heterochromatin in yeast. Mol Cell Biol 2010; 30:1467 - 1477; PMID: 20065036; http://dx.doi.org/10.1128/MCB.01151-09
- Chen CC, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, et al. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell 2008; 134:231 - 243; PMID: 18662539; http://dx.doi.org/10.1016/j.cell.2008.06.035
- Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, et al. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 2008; 134:244 - 255; PMID: 18662540; http://dx.doi.org/10.1016/j.cell.2008.06.018
- Ozdemir A, Masumoto H, Fitzjohn P, Verreault A, Logie C. Histone H3 lysine 56 acetylation: a new twist in the chromosome cycle. Cell Cycle 2006; 5:2602 - 2608; PMID: 17172838; http://dx.doi.org/10.4161/cc.5.22.3473
- Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol Cell 2007; 27:393 - 405; PMID: 17679090; http://dx.doi.org/10.1016/j.molcel.2007.07.011
- Jessen S, Gu B, Dai X. Pygopus and the Wnt signaling pathway: a diverse set of connections. Bioessays 2008; 30:448 - 456; PMID: 18404694; http://dx.doi.org/10.1002/bies.20757
- Belenkaya TY, Han C, Standley HJ, Lin X, Houston DW, Heasman J. pygopus Encodes a nuclear protein essential for wingless/Wnt signaling. Development 2002; 129:4089 - 4101; PMID: 12163411
- Kramps T, Peter O, Brunner E, Nellen D, Froesch B, Chatterjee S, et al. Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear beta-catenin-TCF complex. Cell 2002; 109:47 - 60; PMID: 11955446; http://dx.doi.org/10.1016/S0092-8674(02)00679-7
- Parker DS, Jemison J, Cadigan KM. Pygopus, a nuclear PHD-finger protein required for Wingless signaling in Drosophila. Development 2002; 129:2565 - 2576; PMID: 12015286
- Thompson B, Townsley F, Rosin-Arbesfeld R, Musisi H, Bienz M. A new nuclear component of the Wnt signalling pathway. Nat Cell Biol 2002; 4:367 - 373; PMID: 11988739; http://dx.doi.org/10.1038/ncb786
- Song N, Schwab KR, Patterson LT, Yamaguchi T, Lin X, Potter SS, et al. pygopus 2 has a crucial, Wnt pathway-independent function in lens induction. Development 2007; 134:1873 - 1885; PMID: 17428831; http://dx.doi.org/10.1242/dev.001495
- Gu B, Sun P, Yuan Y, Moraes RC, Li A, Teng A, et al. Pygo2 expands mammary progenitor cells by facilitating histone H3 K4 methylation. J Cell Biol 2009; 185:811 - 826; PMID: 19487454; http://dx.doi.org/10.1083/jcb.200810133
- Fiedler M, Sanchez-Barrena MJ, Nekrasov M, Mieszczanek J, Rybin V, Muller J, et al. Decoding of methylated histone H3 tail by the Pygo-BCL9 Wnt signaling complex. Mol Cell 2008; 30:507 - 518; PMID: 18498752; http://dx.doi.org/10.1016/j.molcel.2008.03.011
- Chen J, Luo Q, Yuan Y, Huang X, Cai W, Li C, et al. Pygo2 associates with MLL2 histone methyltransferase and GCN5 histone acetyltransferase complexes to augment Wnt target gene expression and breast cancer stem-like cell expansion. Mol Cell Biol 2010; 30:5621 - 5635; PMID: 20937768; http://dx.doi.org/10.1128/MCB.00465-10
- Andrews PG, He Z, Popadiuk C, Kao KR. The transcriptional activity of Pygopus is enhanced by its interaction with cAMP-response-element-binding protein (CREB)-binding protein. Biochem J 2009; 422:493 - 501; PMID: 19555349; http://dx.doi.org/10.1042/BJ20090134
- Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 2009; 459:113 - 117; PMID: 19270680; http://dx.doi.org/10.1038/nature07861
- Tjeertes JV, Miller KM, Jackson SP. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J 2009; 28:1878 - 1889; PMID: 19407812; http://dx.doi.org/10.1038/emboj.2009.119
- Michishita E, McCord RA, Boxer LD, Barber MF, Hong T, Gozani O, et al. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle 2009; 8:2664 - 2666; PMID: 19625767; http://dx.doi.org/10.4161/cc.8.16.9367
- McCord RA, Michishita E, Hong T, Berber E, Boxer LD, Kusumoto R, et al. SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging (Albany NY) 2009; 1:109 - 121; PMID: 20157594
- Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 2005; 436:294 - 298; PMID: 16015338; http://dx.doi.org/10.1038/nature03714
- Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 2007; 315:649 - 652; PMID: 17272722; http://dx.doi.org/10.1126/science.1135862
- Vempati RK, Jayani RS, Notani D, Sengupta A, Galande S, Haldar D. p300-mediated acetylation of histone H3 lysine 56 functions in DNA damage response in mammals. J Biol Chem 2010; 285:28553 - 28564; PMID: 20587414; http://dx.doi.org/10.1074/jbc.M110.149393
- Yuan J, Pu M, Zhang Z, Lou Z. Histone H3-K56 acetylation is important for genomic stability in mammals. Cell Cycle 2009; 8:1747 - 1753; PMID: 19411844; http://dx.doi.org/10.4161/cc.8.11.8620
- Nair M, Nagamori I, Sun P, Mishra DP, Rheaume C, Li B, et al. Nuclear regulator Pygo2 controls spermiogenesis and histone H3 acetylation. Dev Biol 2008; 320:446 - 455; PMID: 18614164; http://dx.doi.org/10.1016/j.ydbio.2008.05.553
- Gu B, Watanabe K, Dai X. Epithelial stem cells: an epigenetic and Wnt-centric perspective. J Cell Biochem 2010; 110:1279 - 1287; PMID: 20564229; http://dx.doi.org/10.1002/jcb.22650
- Dutta D, Ray S, Home P, Saha B, Wang S, Sheibani N, et al. Regulation of angiogenesis by histone chaperone HIRA-mediated incorporation of lysine 56-acetylated histone H3.3 at chromatin domains of endothelial genes. J Biol Chem 2010; 285:41567 - 41577; PMID: 21041298; http://dx.doi.org/10.1074/jbc.M110.190025
- Kong S, Kim SJ, Sandal B, Lee SM, Gao B, Zhang DD, et al. The type III histone deacetylase Sirt1 suppresses p300-mediated histone H3 Lysine 56 acetylation at Bclaf1 promoter to inhibit T cell activation. J Biol Chem 2011; Epub Ahead of Print PMID: 21454709; http://dx.doi.org/10.1074/jbc.M111.218206
- Welm BE, Dijkgraaf GJ, Bledau AS, Welm AL, Werb Z. Lentiviral transduction of mammary stem cells for analysis of gene function during development and cancer. Cell Stem Cell 2008; 2:90 - 102
- Long AD, Mangalam HJ, Chan BY, Tolleri L, Hatfield GW, Baldi P. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J Biol Chem 2001; 276:19937 - 19944; PMID: 11259426; http://dx.doi.org/10.1074/jbc.M010192200
- Li B, Rheaume C, Teng A, Bilanchone V, Munguia JE, Hu M, et al. Developmental phenotypes and reduced Wnt signaling in mice deficient for pygopus 2. Genesis 2007; 45:318 - 325; PMID: 17458864; http://dx.doi.org/10.1002/dvg.20299
- Gu B, Chen PL. Expression of PCNA-binding domain of CtIP, a motif required for CtIP localization at DNA replication foci, causes DNA damage and activation of DNA damage checkpoint. Cell Cycle 2009; 8:1409 - 1420; PMID: 19342888; http://dx.doi.org/10.4161/cc.8.9.8322
- Watanabe K, Meyer MJ, Strizzi L, Lee JM, Gonzales M, Bianco C, et al. Cripto-1 is a cell surface marker for a tumorigenic, undifferentiated subpopulation in human embryonal carcinoma cells. Stem Cells 2010; 28:1303 - 1314; PMID: 20549704; http://dx.doi.org/10.1002/stem.463