Abstract
Thyroid transcription factor 1 (TTF-1 or NKX2-1) is an essential fetal lung developmental factor, which can be recurrently activated by gene amplification in adult lung cancer. We have discovered the first microRNA (i.e., miR-365) that directly regulates TTF-1 by interacting with its 3’-untranslated region. By gene expression profiling, we identified other putative targets of miR-365 and miR-365*. In line with the microRNA/target relationship, the expression patterns of miR-365 and TTF-1 were in an inverse relationship in human lung cancer. Exploration of human lung cancer genomics data uncovered that TTF-1 gene amplification was significantly associated with DNA copy number loss at one of the two genomic loci encoding the precursor RNA of mature miR-365 (i.e., mir-365-1). This implies the existence of genetic selection pressure to lose the repressive miR-365 that would otherwise suppress amplified TTF-1. We detected a signaling loop between transforming growth factor beta (TGFb) and miR-365 and this loop reinforced suppression of TTF-1 via miR-365. Mir-365 also targeted an epithelial mesenchymal transition (EMT)-promoting gene HMGA2. In summary, these data connect the lung transcriptional program to the microRNA network.
Introduction
Thyroid transcription factor 1 (TTF-1 or NKX2-1) plays a pivotal role in regulating lung development and morphogenesis.Citation1 Due to the unique expression pattern of TTF-1, which is largely restricted to lung and thyroid, it is routinely used by pathologists as an immunohistochemical marker for diagnosis of tumors of lung or thyroid origin.Citation2 TTF-1 contains a homeodomain which mediates its DNA binding activity.Citation3 Animals homozygous for the disruption of the Ttf-1 gene are born dead and lack lung parenchyma.Citation4 Genetically engineered mice expressing a mutant form of TTF-1 resistant to phosphorylation at seven serine residues died immediately after birth. These animals had defects in lung morphogenesis later in development.Citation5 Evidently, TTF-1 is a critical transcription factor in orchestrating lung development.
In 2007, four laboratories, including ours, independently reported that TTF-1 was activated via gene amplification in lung cancer.Citation6–Citation9 These findings imply that TTF-1 is a gain-of-function oncogene selected for gene amplification due to an undefined biological pressure in lung cancer cells. The studies related to the TTF-1-containing amplification at 14q13 were largely focused on lung adenocarcinomas (ACs). Intriguingly, the 14q13 amplicon also occurred in pulmonary squamous cell carcinomas (SqCCs)Citation6,Citation10 which were, by and large, negative for TTF-1 expression.Citation2 Consequently, it is less likely for TTF-1 to be the target gene of the 14q13 amplification in lung SqCCs. Indeed, our laboratory showed that there were two additional genes (NKX2–8 and PAX9) coamplified with TTF-1, and these two coamplified genes were important for tumor maintenance in the lung SqCC cells containing 14q13 amplicon.Citation6,Citation11 In contrast to the concept of TTF-1 being oncogenic, it was recently reported that Ttf-1 suppressed lung cancer progression in a mouse model system.Citation12 Despite the complexity surrounding TTF-1 in lung cancer, characterization of TTF-1 gene regulatory mechanisms is warranted, as the lung cancer subtype (AC) frequently amplified for TTF-1 presents the most common histologic type of lung cancer. To this end, we aimed at shedding light on TTF-1 signaling network from the angle of microRNAs (miRNAs). Specifically, we wished to identify the miRNAs directly regulating TTF-1, connecting TTF-1-based transcriptional program to the world of small non-protein-coding RNAs.
MiRNAs are small non-protein-coding RNAs that regulate gene expression primarily by binding to the 3′-untranslated region (3′UTR) of the target mRNAs.Citation13–Citation15 MiRNA genes reside in regions of the genome as distinct transcriptional units as well as in clusters of polycistronic units carrying the information of several microRNAs.Citation16–Citation18 Biogenesis of miRNA starts with the synthesis of a long transcript known as the pri-miRNA. In the nuclei, pri-miRNAs are processed to yield the next intermediates (pre-miRNAs) which are exported to cytoplasm for further processing/shortening to produce the final mature miRNAs (18∼22 mer). It is important to note that a single mature miRNA may be derived from multiple pri-miRNAs transcribed from distinct genomic loci. Although there is documented evidence that miRNAs influence lung biologyCitation19,Citation20 and cancer,Citation21–Citation23 to date how the miRNA network is intertwined with the pulmonary transcriptional program responsible for lung development and malignancy remains poorly defined.Citation24 To fill this void, we set out to uncover the miRNAs that directly regulate TTF-1. Initially, we used the in silico tool, TargetScan,Citation25 to select candidate miRNAs predicted to bind to TTF-1 3′UTR. Using a luciferase-based reporter system in which the luciferase reporter gene was fused to the 3′UTR of TTF-1, we discovered that a relatively uncharacterized miRNA, miR-365, directly repressed TTF-1. Based on the miRbase annotation,Citation26 there are two longer stem-loop RNA precursors, hsa-mir-365-1 and hsa-mir-365-2, that would give rise to the same mature miR-365 species. We showed that, indeed, both stem loops yielded mature miR-365 when expressed in human lung cells, which, in turn, repressed endogenous TTF-1 protein expression. Analysis of human lung cancer DNA copy number alteration data revealed co-occurrence of TTF-1 gene amplification and gene loss at either or both of the two genomic loci of miR-365 precursors. This suggests a tendency for cancer cells to lose repressive microRNAs of amplified oncogenes. Analysis of human lung cancer tissues indicated an inverse relationship between TTF-1 RNA level and miR-365 RNA expression. Saito et al. reported an unexpected activity of TTF-1: inhibition of TGFβ-induced EMT by TTF-1.Citation27 They further demonstrated that enhancement of autocrine TGFβ signaling would decrease TTF-1 expression, thus blunting the anti-EMT effect derived from TTF-1.Citation27 However, the molecular mechanism accounting for how TGFβ modulates TTF-1 expression was not elucidated in the study. We postulated that miR-365 mediates the TGFβ-dependent suppression of TTF-1. Indeed, we detected a positive feedback loop between miR-365 and TGFβ signaling. Detection of a mesenchymal phenotype in lung epithelial cells with enforced expression of miR-365 was made difficult by the unexpected finding that miR-365 also repressed a pro-EMT factor downstream to TTF-1 (i.e., HMGA2).Citation12 Gene expression profiling identified multiple candidate genes that were putative targets of miR-365 and miR-365* beyond TTF-1. As far as we know, miR-365 is the first miRNA discovered to directly regulate TTF-1. Future studies will be dedicated to elucidating the interconnections between miRNAs and the lung transcriptional network.
Results
Discovery of miR-365.
lists the top 10 candidate miRNAs predicted to interact with the 3′UTR of TTF-1 using TargetScan release 5.1 Citation25,Citation28,Citation29 in a manner focusing on the context scores without emphasis on evolutionary conservation. These 10 candidate miRNAs included all of the four human miRNAs regarded by TargetScan as conserved sites in either of these two categories: broadly conserved among vertebrates (hsa-miR-23a and hsa-miR-23b) or conserved only among mammals (hsamiR-599 and hsa-miR-186). To functionally screen each of the top 10 candidate miRNAs, we co-transfected a luciferase-based reporter plasmid containing the full-length TTF-1 3′UTR with individual miRNA mimic oligonucleotides into three types of host cells: HeLa, NCI-H2170 (H2170), or NCI-H441 (H441) cells. The luciferase activities were read 48 h after transfection and the compilation of all the data are shown in , with all data points normalized to the reporter plasmid plus negative control (a short RNA derived from C. elegans without known human targets). Only miR-365 and a small interfering RNA (siTTF-1, targeting TTF-1 3′UTR outside the miR-365 binding site) reproducibly repressed the Renilla luciferase activity which reports TTF-1 3′UTR. Since complementarity in the seed sequence between a miRNA and its target RNA is crucial, the reporter assay was repeated with two mutant reporters. Four of the seven seed-sequence nucleotides were altered in Mutant A, whereas the entire 7-nt seed sequence was removed in Mutant B (). The resultant reporter luciferase activities indicated complete de-repression by miR-365 on both mutant reporters but the de-repression was not detected with siTTF-1 ().
Suppression of endogenous TTF-1 protein by miR-365.
The reporter assay suggested that miR-365 would repress endogenous TTF-1 protein expression. To test this hypothesis, we transfected miR-365 oligonucleotide into a human lung cancer cell line (H441) that expresses TTF-1 endogenously. As shown in , immunoblotting analysis showed that miR-365 knocked down the expression of TTF-1. Based on the miRbase annotation,Citation26 the mature 22-mer RNA of miR-365 is derived from two longer RNA precursors transcribed from two separate intergenic genomic loci, hsa-mir-365-1 (16p13.12, MI0000767) and hsa-mir-365-2 (17q11.2, MI0000769). These precursor RNA species are referred to as mir-365-1 and mir-365-2 hereafter. We cloned the genomic sequences (∼700 bp) encompassing mir-365-1 or mir-365-2 into a retroviral expression vector and transfected the vector into H441 cells for stable expression of individual precursor RNAs. We reasoned that the enforced expression of the precursor RNAs would result in intracellular accumulation of miR-365 mature species. As a control, a mutant form of mir-365-1 in which the 7-nt miR-365 seed sequence was deleted (denoted as mir-365-1m) was also transfected into H441 genome alongside with the empty vector control. Using a locked nucleic acid (LNA)-based quantitative PCR (QPCR) probe specific to the miR-365 22-mer, we detected accumulation of mature miR-365 species in the stable recombinant cells with enforced expression of either mir-365-1 or mir-365-2 (Fig. S1). However, the miR-365 buildup in the mir-365-1m transfectants was invisible to the LNA probe due to the deletion of the miR-365 seed sequence which was also critical for the LNA probe binding (Fig. S1). QPCR probes unique to mir-365-1 or mir-365-2 detected increased presence of respective RNA species without any cross-reactivity (Fig. S2A). Immunoblotting was performed with the cell lysates prepared from the H441-based transfectant cells for quantification of the endogenous TTF-1 protein levels. Both miR-365 precursors repressed the level of endogenous TTF-1 protein, and the mir-365-2 transfectant cells experienced a more severe suppression of TTF-1 (62.5% vs. 34.9% in mir-365-1 transfectants, ). It is intriguing to note that endogenous TTF-1 protein was reduced to a lesser degree in the mir-365-1m-expressing cells (). The more severe TTF-1 knockdown in mir-365-2 transfected cells may be explained by the higher accumulation of mature miR-365 species seen in the mir-365-2-expressing cells (Fig. S1).
MiR-365 expression in normal human tissues and the miR-365* species.
To further characterize miR-365, we determined the RNA expression of miR-365 and TTF-1 in 20 normal human tissues. As expected, TTF-1 RNA was only detectable in pulmonary and thyroid tissues (). Interestingly, miR-365 mature species was detected in all 20 tissues, with muscle (cardiac and skeletal) and adipose having the highest level of miR-365 (). This observation suggests that there are likely other targets to miR-365 beyond TTF-1. Comparison of the two panels of revealed that the two TTF-1-positive tissues (lung and thyroid) were among the tissues with low abundance of miR-365, consistent with the repressive function of miR-365. Eight normal human tissues were selected for further RNA expression analysis to differentiate the level of mir-365-1 vs. mir-365-2. The results demonstrate that mir-365-1 was generally more abundant than mir-365-2 (Fig. S2B), indicating a more robust transcriptional activity at the mir-365-1 locus. As mir-365* has been reported to derive from mir-365-2 by deep sequencing,Citation30 we profiled genome-wide mRNA expression alterations conferred by enforced expression of mir-365-2. Using the class comparison algorithm of BRB array tool (significance cutoff of univariate test at 0.001),Citation31 we detected nine downregulated and two upregulated RNAs in mir-365-2-expressing H441 cells relative to the empty vector control H441 cells (). Subsequently we employed TargetScan to explore these 11 RNAs for miR-365 binding sites and only scored KLRK1 and PFKFB4 as putative targets of miR-365. To determine if the remaining genes were targets of miR-365*, we ran the miRanda algorithmCitation32 locally to search for miR-365* binding sites in the differentially expressed RNAs. Unexpectedly, six of the nine downregulated RNAs were predicted to be targeted by miR-365*. These data provide a short list of putative targets of miR-365* for further study.
MiR-365 expression in human lung adenocarcinomas.
It is known that TTF-1 immunopositivity in lung adenocarcinoma tumors correlates with better survival outcome.Citation10,Citation33,Citation34 In light of our findings, we evaluated 67 fresh frozen lung adenocarcinoma tissues procured via the Penn State Cancer Institute Tumor Bank. Pathological evaluation of these tissues was performed by a board-certified pathologist. RNA levels of TTF-1 and miR-365 were quantified in each tissue (Fig. S3). Linear regression analysis of all 67 samples did not show a statistically significant negative correlation between TTF-1 RNA and miR-365 expression. To further dissect the data, we compared TTF-1 RNA expression in ten samples with the highest miR-365 level and ten samples with the lowest miR-365 level. The box plot shown in revealed a statistically significant inverse relationship for this microRNA/target connection.
Co-occurrence of mir-365-1/2 deletion and TTF-1 amplification.
Gene amplification leads to an increased dosage of genes residing inside the amplicon. Since TTF-1 is part of a recurrent lung cancer amplicon,Citation6–Citation9 we surmised that there might be concomitant DNA copy number loss events to reduce miR-365 expression in a fraction of lung tumors containing TTF-1 amplification. Previously, we profiled the genomic DNA copy number landscape in 211 human lung cancer genomes.Citation6 Interrogation of this data set using a less stringent setting to detect gene amplification [i.e., segmented DNA copy number median (segmed) of 1.2 as a cutoff to define gene amplification] identified 42 samples with TTF-1 gene amplification. Among these 42 samples, five and four samples had DNA copy number decrease at chromosomal regions covering mir-365-1 and mir-365-2, respectively (using the cutoff of segmed < 1/1.2, Fig. S4). None of the 42 TTF-1-amplified lung cancer genomes had DNA loss at both loci of miR-365 precursors. In the 169 samples with segmed of TTF-1 <1.2, there were four samples that also showed DNA copy number loss at mir-365-1 and mir-365-2, respectively (with one sample containing both mir-365-1 and mir-365-2 loss). By Fisher's exact test (Fig. S4), a statistically significant association (p ∼ 0.0169) was detected only between TTF-1 gene amplification (segmed > 1.2) and mir-365-1 deletion (segmed < 1/1.2). These observations suggest that DNA copy number loss is a factor accounting for the low miR-365 expression in lung cancer (), and that there is a selection pressure to reduce gene dosage at the mir-365-1 locus. This observation is in line with our expression analysis reported herein that the mir-356-1 locus is transcriptionally more active than the mir-356-2 locus.
MiR-365 and TGFβ form a feedback signaling loop.
Saito et al. recently reported that TTF-1 inhibits TGFβ-mediated EMT in lung cancer cells,Citation27 suggesting that TTF-1 is an anti-EMT factor. In their study, TGFβ treatment led to downregulation of TTF-1, blunting the anti-EMT phenotype conferred by the TTF-1 protein. However, the molecular mechanism of this process was unknown. In light of our discovery of the connection between miR-365 and TTF-1, we hypothesized that miR-365 mediates the TGFβ-induced suppression of TTF-1 expression. To test this hypothesis, we measured the miR-365 levels of a premalignant lung epithelial cell line (BEAS-2BCitation35) before and after TGFβ treatment. Following a 24-h incubation with TGFβ, miR-365 expression went up 4-fold (Fig. S5). To determine the generality of this induction, we treated a panel of three lung adenocarcinoma cell lines with TGFβ and followed the miR-365 levels over a period of 3 d. Interestingly, all three lung adenocarcinoma cell strains (NCI-H1944, NCI-H441 and A549) responded to TGFβ in terms of miR-365 induction, albeit to different extent (). Concomitantly, the TTF-1 RNA and protein expression decreased in the presence of TGFβ (data of H441 cells shown in Fig. S6). A chemical inhibitor of TGFβ receptor I kinase, LY364947, reversed the TGFβ-dependent repression of TTF1 RNA and protein expression (Fig. S6). However, miR-365 was not induced by TGFβ in the TTF-1-negative colon cancer cell line (HCT-15) (). The lack of TGFβ-induced miR-365 upregulation was also seen in breast (BT-20 and MDA-MB-453) and cervical (HeLa) cancer cells (Fig. S7). The lack of TTF-1 expression may not predict the absence of the TGFβ/miR-365 connection, as the TTF-1-negative lung cancer cell line A549 was positive for it. At this juncture, it is tempting to assume that the TGFβ-dependent upregulation of miR-365 can only be operant in pulmonary cells. The association between mir-365-1 deletion and TTF-1 amplification in human lung cancer samples implicates the biological importance of mir-365-1. This triggered us to hypothesize that mir-365-1, rather than mir-365-2, may be the primary respondent to TGFβ. The relative RNA levels of mir-365-1 and mir-365-2 were determined along the 3-d course of TGFβ incubation. Remarkably, the miR-365 precursor, mir-365-1, was quantitatively activated by TGFβ, but mir-365-2 was not (). The inhibitor of TGFβ receptor I kinase (LY364947) completely blocked the upregulation of miR-365 and mir-365-1 induced by TGFβ (Fig. S8). These data, together with the RNA expression survey data in normal human tissues presented above, suggest that mir-365-1 may be the primary source of mature miR-365 in human lung cells. An observation made by Saito et al. was that TTF-1 would blunt TGFβ signaling by suppressing TGFβ2 expression.Citation27 We were thus curious if miR-365 would induce TGFβ expression as a means to further counter the anti-TGFβ effect of TTF-1. The RNA expression of TGFβ1∼3 was determined using RT-QPCR in the H441-based stable transfectant cells. The results showed that the expression of TGFβ2, but not TGFβ1 and TGFβ3, was increased in the miR-365-expressing cells (). To extend this observation to a different background, we transfected miR-365 into A549 cells stably expressing TTF-1 cDNA with the full-length 3′UTR (TTF-1 + 3′UTR). Culture media were analyzed by ELISA assay to quantify TGFβ2 protein level. The data shown in revealed again that miR-365 induced TGFβ2 expression and a siTTF-1 phenocopied miR-365. These data clearly implicate the existence of a signaling loop between miR-365 and TGFβ.
MiR-365 also targets HMGA2.
Since we were unable to detect altered proliferation or wound healing/migration in lung cancer cells (H441) stably expressing miR-365 (data not shown), we chose to conduct a profiling experiment to monitor the RNA expression perturbation of ∼84 EMT genes in the stable miR-365-expressing H441 cells following TGFβ treatment using a commercial QPCR array (SA Biosciences). Surprisingly, few EMT-related genes exhibited altered expression between control and miR-365-expressing cells in response to TGFβ (Fig. S9). In reviewing the putative targets of miR-365 predicted by TargetScan, we noted that the 3′UTR of a strong pro-EMT gene, HMGA2 (high mobility group AT hook 2),Citation36 also harbors a broadly conserved miR-365 binding site among vertebrates. Suppression of HMGA2 by miR-365, if validated, may explain the lack of miR-365-dependent EMT phenotypes in our hands, i.e., simultaneous repression of TTF-1 (anti-EMT) and HMGA2 (pro-EMT) by miR-365 resulting in a net neutral effect. Through a luciferase assay reporting full-length 3′UTR of HMGA2 () and a RT-QPCR analysis of endogenous HMGA2 RNA level (), we concluded that HMGA2 was indeed a target of miR-365.
Discussion
MiRNAs modulate many if not nearly all biological pathways and signaling cascades.Citation13,Citation40 The first miRNA, lin-4, was discovered as a result of an investigation of mutant nematodes with defects in developmental stages.Citation41 Since then, numerous miRNAs have been discovered to influence development and maturation of tissues and organs. However, few genes critical to lung development have been recognized to be under miRNA regulation. Given this backdrop, we believed that it would be fruitful to search for the miRNAs that supervise TTF-1, a master regulator of lung development.Citation1 We first used an in silico method (i.e., TargetScan) to scan TTF-1 3′UTR to predict candidate miRNAs. We then selected the top ten candidate miRNAs for experimental validation using a cell system in which the luciferase activity reports the 3′UTR regulation of human TTF-1. The result was surprising in that only one miRNA, miR-365, could be experimentally validated as a bona fide miRNA that represses TTF-1. Although none of the available miRNA/target prediction algorithms is perfect,Citation42 the high false prediction rate of TargetScan seen in this study was somewhat surprising to us. During the course of our study, we had looked up the miRanda-based prediction for potential miRNA binders of TTF-1 3′UTR.Citation32 The data are listed at a website maintained by the Enright group at the EMBL-EBICitation26 (www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5). Interestingly, miR-365 was the top ranked hit by miRanda to bind TTF-1 3′UTR. Besides miR-365, there were only two overlapping hits (miR-323-5p and miR-599) among the top 10 predicted miRNAs between miRanda-based and our TargetScan-derived analyses. This discrepancy likely reflects the algorithmic difference of these tools: miRanda calculates sequence complementarity score with a weighting scheme,Citation32 whereas TargetScan mainly considers the complementarity of the seed sequence.Citation29 Clearly while the number of miRNA/target prediction algorithms continues to rise in the literature, the prediction accuracy remains to be improved.
Genes encoding miRNAs have been found to be frequently located at genomic regions exhibiting instability in cancers.Citation43 In our study we detected co-occurrence of deletion of either of the two genes encoding miR-365 precursors and the cognate target gene TTF-1 amplification in human lung cancers. This suggests that there is a selection force to lose the negative regulator of a gene undergoing DNA copy number amplification in order to maximize the functional advantage conferred by the amplified genes. As many oncogenes and tumor suppressor genes are under miRNA regulation,Citation44,Citation45 variation of DNA copy number of miRNA genes will inevitably alter the expression of the target genes. In cancers, the microRNA family let-7, miR17-92, miR-15a and miR-16-1 all undergo DNA copy number alterations and the expression of their cognate target genes vary accordingly.Citation44,Citation45 Since in the human genome, there are two non-protein-coding genes at two separate genomic loci that are predicted to produce the mature miR-365 RNA, we investigated both genes in cell-based systems and showed that, indeed, both genes, when under enforced expression, would lead to an accumulation of miR-365 sequence. But these two genes do not appear to be regulated equally. QPCR probes specific to the individual precursor species in general detected higher expression of mir-365-1 in normal human tissues, implicating a higher transcriptional activity at the mir-365-1 locus (). Molecular basis of this phenomenon is a focus of our future study.
Gene expression profiling of cells expressing miR-365 identified multiple putative targets of miR-365 and miR-356*. It is curious that TTF-1 and HMGA2 were not among identified putative targets. An explanation is that in our system, translational repression was the primary action mechanism of miR-365. The genetic connection of TTF-1 to lung cancer was first realized by the finding that TTF-1 undergoes a DNA copy number increase in human lung cancer.Citation6–Citation9 The concept of TTF-1 being oncogenic was further substantiated by Homminga et al., who detected ectopic expression of TTF-1 as a fusion gene in T-cell acute lymphoblastic leukemia.Citation46 However, in a murine mouse model of lung cancer Winslow et al. uncovered that Ttf-1 suppressed lung cancer progression by repressing HMGA2.Citation12 In an earlier study, Ttf-1 expression was observed to decrease progressively in a different mouse model of lung cancer.Citation47 To complicate the matter, the chromosomal area at 14q13 harboring the TTF-1 amplicon was discovered to also undergo allelic loss in lung cancer.Citation48 At this juncture, we do not have a unifying theory that accommodates all the reported observations regarding TTF-1 and cancer. Clearly, TTF-1 plays both pro- and anti-tumorigenic roles depending on the biological context. Nevertheless, our discovery of the first miRNA regulator of TTF-1 opens a new window for subsequent studies of lung tissue development and cancer. We believe that there is a significant crosstalk between the transcriptional program and miRNA network in the lung (diseased or normal), and miR-365 is merely “a tip of an iceberg.” In the intriguing signaling loop reported by Saito et al., TGFβ induced TTF-1 downregulation, thus blunting the anti-EMT activity associated with TTF-1,Citation27 but the molecules mediating this activity were unidentified. In this study, we present evidence that miR-365 mediates TTF-1 repression by TGFβ. This suggests that TGFβ may utilize multiple means to antagonize TTF-1 in view of the SMAD3-directed interference of TTF-1 DNA binding function.Citation49 Cells overexpressing miR-365 did not readily exhibit a detectable EMT phenotype. This may have been attributed to the incomplete reduction of TTF-1 protein level by miR-365. Alternatively, we suggest that simultaneous repression of an anti-EMT gene (TTF-1) and a pro-EMT gene (HMGA2) may have contributed to the lack of EMT phenotypes associated with miR-365 overexpression. Because of the well-documented importance of TTF-1 in fetal lung development, we predict that miR-365 plays a critical role in fetal lung development. In summary, the findings reported in this study present an entry point to explore how the pulmonary transcriptional program is interwoven with the miRNA network.
Materials and Methods
Reagents.
DMEM, RPMI-1640 medium, Penicillin/Streptomycin, 0.25% Trypsin/EDTA and Fetal Bovine Serum (FBS) were purchased from Mediatech. Keratinocyte-Serum Free Medium (K-SFM) was purchased from Invitrogen. Cell lines used in this study were from American Type Culture Collection (ATCC). TGFβ (i.e., TGFβ1) was purchased from R&D Systems. TGFβ receptor I inhibitor LY364947 was from Cayman Chemical.
Cell-based assays.
To functionally screen each of the top 10 candidate miRNAs, we modified a commercial luciferase reporter plasmid (psiCHECK-2) in which the full-length TTF-1 3′UTR was placed downstream to the Rellina luciferase. This reporter plasmid, with a built-in Firefly luciferase gene as a gauge of transfection efficiency, was cotransfected with individual miRNA mimic oligonucleotides into three types of host cells: HeLa, NCI-H2170 (H2170), or NCI-H441 (H441) cells. TTF-1 3′ UTR was amplified from human genomic DNA by PCR with an added XhoI cleavage site (Forward primer: ATC CCT CGA GCT ATA CGG TCG GAC CTG GTG; Reverse primer: ATC CCT CGA GTC AAA GCC ATT TAA AGC CAA A) and cloned into psi-CHECK-2 vector (Promega). As controls, two mutant constructs were also made by PCR-based mutagenesis using these primers: forward primer for Mutant A, CCT TCA CAT TTT TTA TAG ATC TTG ACA AAT CTG TGT ATA; reverse primer for Mutant A, TAT ACA CAG ATT TGT CAA GAT CTA TAA AAA ATG TGA AGG; forward primer for Mutant B, CCT TCA CAT TTT TTA TGA CAA ATC TGT GTA TA; reverse primer for Mutant B, TAT ACA CAG ATT TGT CAT AAA AAA TGT GAA GG. psiCHECK-HMGA2 3′ UTR reporter is a kind gift from Dr. Marcus Peter. For TTF-1 luciferase reporter assay, HeLa, H441 or H2170 cells were seeded at 1–2 × 104 cells/well in white 96-well plate (BD-falcon). After 12∼16 h, 200–300 ng wild-type or mutant TTF-1 3′UTR luciferase reporter constructs were co-transfected with miRNA mimics or negative control oligo (NC) or TTF-1 siRNA (positive control, Dharmacon) utilizing Dharmafect Duo reagent (Dharmacon) according to the manufacturer's instruction. For HMGA2 lufciferase assay, A549 or H1299 cells were seeded at 1–2 × 105/well in 12-well plate. After 12–16 h, 2 µg ofHMGA2 3′ UTR reporter construct was co-transfected with miR-365 mimic, negative control (NC) or let-7i mimic (Positive control, Dharmacon) utilizing Dharmafect Duo reagent (Dharma on). Before lucifease assay, cells were lysed, and lysates were tranferred into 96-well white plates for reading. Hsa-mir-365-1 and hsa-mir-365-2 sequences with 300 bps upstream and downstream each were amplified from genomic DNA by PCR with an added XhoI cleavage site (hsa-mir-365-1 forward primer, ATC CCT CGA GCA TCT GAA GTT TGG GGG AAA; reverse primer, ATC CCT CGA GCT TTC CGC TTT CCC TAC CTC; hsa-mir-365-2 forward primer, ATC CCT CGA GCT GCT CTC CAC ATT GGG ATT; reverse primer, ATC CCT CGA GCT TGT CCA GCC CAA CTC TGT). A commercial retroviral expression vector (pMSCV-hyg, Clontech) was used to mediate the stable expression of hsa-mir-365-1/2. As a control, the miR-365 seed binding sequence (AAT GCC C) was deleted in mir-365-1 by mutation-containing PCR primers. The primers used for introducing the deletion were: forward, TCC ATT CCA CTA TCA TCT AAA AAT CCT TAT TG; reverse, CAA TAA GGA TTT TTA GAT GAT AGT GGA ATG GA. To test the effect of miR-365 mimic on endogenous TTF-1 expression, H441 cells which have a high level of endogenous TTF-1 expression were seeded in 6-well plate at 4 × 105 cells/well. After a 12∼16-h incubation, 50 nmol, 100 nmol or 200 nmol of miR-365 mimic, NC or TTF-1 siRNA were transfected with Dharmafect Duo reagent. For luciferase reporter assay, we read the Firefly and Renilla luciferase signals with Dual-Glo luciferase assay System (Promega). For data analysis, Renilla luciferase signals were used to adjust for well-to-well transfection efficiencies. To make stable retroviral transfectants overexpressing mir-365-1 or mir-365-2, we used the Phoenix-ampho cells as the viral packaging cells (Allele Biotech) and followed the vendor's protocol.
RT-QPCR and protein analysis.
First Choice Human total RNA survey panel (Ambion) was used to represent normal tissue RNA. Total tumor and normal adjacent tissue RNA was isolated from approximately 100 mg of frozen tissues using Trizol (Invitrogen) and FastPrep instrument (Qbiogene, Inc.), with matrix D for 45 sec at a speed setting of 6. RNA from cell strains was prepared using Trizol. Genomic DNA of Trizol-derived RNA was removed by treatment with the TURBO DNA-free kit (Ambion). For RT-QPCR measurements of TTF-1, miR-365 precursors, TGFβs and HMGA2, pre-designed TaqMan probes were purchased from Applied Biosystems. High Capacity cDNA Reverse Transcription Kit and TaqMan universal PCR master mix (Applied Biosystems) were used to perform the reverse transcription step and QPCR step respectively. Relative quantity method (ΔΔCT) was used for data analysis. Human β-actin or GAPDH levels employed as the normalization basis. For RT-QPCR measurements of mature miR-365 expression, probes were purchased from Exiqon. Universal cDNA synthesis Kit and SYBR Green Master Mix Kit (Exiqon) were used to perform the reverse transcription step and QPCR step, respectively. Relative quantity method (ΔΔCT) was used for analysis, and SNORD38B (Exiqon) was used as the reference small RNA. To collect cell lysates for immunoblotting, cells grown in monolayer were lysed directly with RIPA buffer (150 mM NaCl, 50 nM Tris, 1% NP-40, 0.1% SDS) containing complete protease inhibitor cocktail (Roche). The volume of RIPA buffer used was 100 µL/well for 6-well plate or 500 µL/plate for 100 mm culture dish. Protein concentrations were then measured with BCA Protein Assay Kit (Thermo Fisher Pierce) according to the manufacturer's instruction. TTF-1 antibody was from DAKO (M3575, clone 8G7G3/1); β-tubulin antibody was from Cell Signaling. The densities of the blots on the scanned film image were quantified with UN-SCAN-IT gel6.1 software.
ELISA assay.
The cell culture supernatants were acidified with 1 N HCL for 10 min, neutralized with 1.2 N NaOH/0.5 M HEPES for 10 min, and subjected to TGFβ2 ELISA with Human TGFβ2 Quantikine ELISA kit (R&D) according to manufacturer's protocol.
RNA profiling and miRanda analysis.
Total RNA samples from H441 cells stably expressing mir-365-2 together with the empty vector control were collected using mirVana miRNA Isolation Kit (Ambion). The resultant RNAs were analyzed for quality using a Bioanalyzer (Agilent). RNA samples with a RNA Integrity Number (RIN) > 7 and 260/280 ratio between 1.8–2.0 were hybridized in duplicates to Affymetrix GeneChip Human Gene 1.0 ST arrays by Expression Analysis. Data were analyzed using the BRB array tool software (4.1.0 β 3 release).Citation31 Profiling data were deposited at GEO under accession number GSE33672. miRanda 3.0 was run locally using these parameters: Gap Open Penalty, −8.000000; Gap Extend, −2.000000; Score Threshold, 50.000000; Energy Threshold, −20.000000 kcal/mol; Scaling Parameter, 4.000000.
EMT PCR array.
H441 cells stably expressing mir-365-1, mir-365-1m, mir-365-2, or the empty vector control were incubated with TGFβ1 for 24 h. Subsequently, the mRNAs were collected using Qiagen RNeasy Mini Kit (Qiagen) combined with an on-column DNase treatment step. These samples were subjected to quality control analysis in the core facility using a Bioanalyzer. RNA samples with a RNA Integrity Number (RIN) > 7 and 260/280 ratio between 1.8–2.0 were used for RT-qPCR analysis on EMT PCR array plates with RTCitation2 first Strand Kit and SABiosciences qPCR master mix following manufacturer's protocol. Data were analyzed utilizing a web-based analysis tool from SABiosciences.
Disclsoure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Figures and Tables
Figure 1 Discovery of miR-365 regulating TTF-1. (A) The top 10 miRNAs predicted by TargetScan (release 5.1) to bind to the 3′UTR of TTF-1 RNA. The list is prioritized based on the context score defined by TargetScan. (B) Luciferase reporter-based screen of candidate miRNAs predicted to bind TTF-1 3′UTR. The psiCHECK-2 (Promega) vector-based luciferase reporter plasmid was cotransfected with individual miRNA oligonucleotides. The negative control (NC) RNA oligonucleotide bears no sequence identity with miRNAs in human, mouse and rat (Dharmacon, CN-001000-01). A validated small interfering RNA against TTF-1 3′UTR (siTTF-1) was included as a positive control (Dharmacon, D-019105-17-0005). The data columns labeled by an asterisk are statistically different from the data of Reporter + NC (p < 0.05, t-test). Y-axis shows the Renilla luciferase signal normalized to (Reporter + NC). Data shown were compiled from three host cells (H441, HeLa and H2170). (C) Seed region of miR-365-binding site and the mutant reporter plasmids. Mutant A includes a 4-bp alteration in the seed sequence (altered bases are underlined), whereas the seed sequence is removed in Mutant B. (D) The seed sequence in the TTF-1 3′UTR critical to miR-365 binding. Wild-type, Mutant A or B reporter plasmid was cotransfected into H441 cells to read out the impact of seed-region alterations in the reporter plasmid. siTTF-1 and wt-reporter were included as controls. An asterisk indicates p-value < 0.05 (t-test) in comparison with the data column of wtReporter + NC.
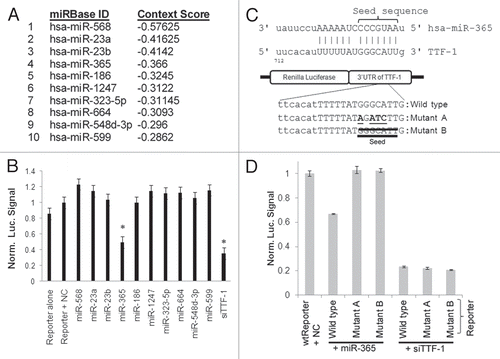
Figure 2 miR-365 suppresses endogenous TTF-1 expression and expression patterns of miR-365 in human tissues. (A) Immunoblotting analysis of H441 cells after transfection of varying amounts of miR-365 RNA oligonucleotide. siTTF-1 RNA oligonucleotide (targeting TTF-1 3′UTR) was included as a positive control. β-tubulin was used as a loading reference. TTF-1 protein levels were quantified and shown below the immunoblot. (B) Immunoblotting analysis of stable retroviral transfectant cells overexpressing mir-365-1, mir-365-2 or mir-365-1m. TTF-1 protein levels were quantified and shown below the immunoblot. (C) RT-QPCR-based RNA expression profiling of miR-365 in 20 normal human tissues. Skel, skeletal muscle; Small, small intestine. (D) Box plot of TTF-1 RNA expression in lung AC samples with highest or lowest miR-365 level. Each category contains 10 samples.
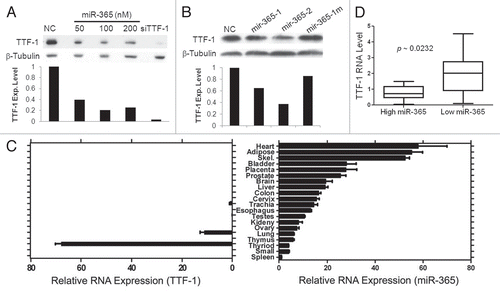
Figure 3 The signaling loop between miR-365 and TGFβ. (A) A time-course study of miR-365 induction by TGFβ. Three lung cancer cell lines and a colon cancer cell strain were subjected to a TGFβ treatment for 3 d. MiR-365 levels were quantified daily as indicated. All data were normalized to the time “0” data point. (B) The RNAs collected from the three lung cell strains treated with TGFβ were analyzed by RT-QPCR for the relative abundance of mir-365-1 and mir-365-2. An asterisk indicates p-value < 0.05 (t-test) in comparison with the respective data column of time 0. (C) TGFβ2 induced by miR-365. Stable transfectant cells (H441) were profiled for TGFβ1∼3 RNA expression by RT-QPCR as indicated. (D) A549 cells carrying empty vector or TTF-1 + 3′UTR were transfected with negative control (NC), miR-365 or siTTF-1. Two days after transfection, the culture media were analyzed using an ELISA assay specific for TGFβ2 (R&D Systems).
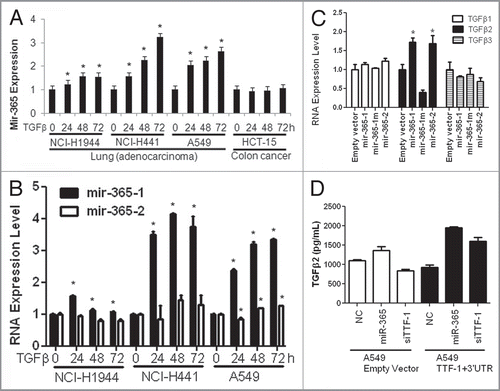
Figure 4 HMGA2 is a target of miR-365. (A) A luciferase reporter assay connected to HMGA2 3′UTR was used in two different host cells to detect the repressive effect by miR-365. Let-7i, a positive control, is known to target HMGA2.Citation37–Citation39 NC, negative control. (B) Endogenous HMGA2 RNA expression was reduced by miR-365. (C) A schematic summarizes the findings of this study. The signaling pathways (a,Citation12 bCitation36 and cCitation27) are known in the literature.
Table 1 RNAs with altered expression in miR-365-expressing H441 cells
Additional material
Download Zip (314.4 KB)Acknowledgments
We thank Scott Powers for access to the software tool (myGAD), which rapidly analyzes the human cancer DNA copy number profiles collected at the Cold Spring Harbor Laboratory; Richard Bruggeman and Kun Jiang for technical assistance; Daniel Beard and Molly Pells (Penn State Cancer Institute Tumor Bank) for human lung cancer tissues, and Marcus Peter for the HMGA2 3′UTR reporter construct. A.C.S. was supported in part by a NIH CTSA grant (UL1RR033184). D.M. was supported in part by NCI (CA127547) and the Uniting Against Lung Cancer foundation, New York.
References
- Maeda Y, Dave V, Whitsett JA. Transcriptional control of lung morphogenesis. Physiol Rev 2007; 87:219 - 244; PMID: 17237346; http://dx.doi.org/10.1152/physrev.00028.2006
- Ordóñez NG. Thyroid transcription factor-1 is a marker of lung and thyroid carcinomas. Adv Anat Pathol 2000; 7:123 - 127; PMID: 10721419; http://dx.doi.org/10.1097/00125480-200007020-00007
- Guazzi S, Price M, De Felice M, Damante G, Mattei MG, Di Lauro R. Thyroid nuclear factor 1 (TTF-1) contains a homeodomain and displays a novel DNA binding specificity. EMBO J 1990; 9:3631 - 3639; PMID: 1976511
- Kimura S, Hara Y, Pineau T, Fernandez-Salguero P, Fox CH, Ward JM, et al. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain and pituitary. Genes Dev 1996; 10:60 - 69; PMID: 8557195; http://dx.doi.org/10.1101/gad.10.1.60
- DeFelice M, Silberschmidt D, DiLauro R, Xu Y, Wert SE, Weaver TE, et al. TTF-1 phosphorylation is required for peripheral lung morphogenesis, perinatal survival and tissue-specific gene expression. J Biol Chem 2003; 278:35574 - 35583; PMID: 12829717; http://dx.doi.org/10.1074/jbc.M304885200
- Kendall J, Liu Q, Bakleh A, Krasnitz A, Nguyen KC, Lakshmi B, et al. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci USA 2007; 104:16663 - 16668; PMID: 17925434; http://dx.doi.org/10.1073/pnas.0708286104
- Kwei KA, Kim YH, Girard L, Kao J, Pacyna-Gengelbach M, Salari K, et al. Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene 2008; 27:3635 - 3640; PMID: 18212743; http://dx.doi.org/10.1038/sj.onc.1211012
- Tanaka H, Yanagisawa K, Shinjo K, Taguchi A, Maeno K, Tomida S, et al. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res 2007; 67:6007 - 6011; PMID: 17616654; http://dx.doi.org/10.1158/0008-5472.CAN-06-4774
- Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature 2007; 450:893 - 898; PMID: 17982442; http://dx.doi.org/10.1038/nature06358
- Perner S, Wagner PL, Soltermann A, LaFargue C, Tischler V, Weir BA, et al. TTF1 expression in non-small cell lung carcinoma: association with TTF1 gene amplification and improved survival. J Pathol 2009; 217:65 - 72; PMID: 18932182; http://dx.doi.org/10.1002/path.2443
- Hsu DS, Acharya CR, Balakumaran BS, Riedel RF, Kim MK, Stevenson M, et al. Characterizing the developmental pathways TTF-1, NKX2–8 and PAX9 in lung cancer. Proc Natl Acad Sci USA 2009; 106:5312 - 5317; PMID: 19279207; http://dx.doi.org/10.1073/pnas.0900827106
- Winslow MM, Dayton TL, Verhaak RGW, Kim-Kiselak C, Snyder EL, Feldser DM, et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature 2011; 473:101 - 104; PMID: 21471965; http://dx.doi.org/10.1038/nature09881
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136:215 - 233; PMID: 19167326; http://dx.doi.org/10.1016/j.cell.2009.01.002
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 2009; 10:704 - 714; PMID: 19763153; http://dx.doi.org/10.1038/nrg2634
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation 2004; 5:522 - 531
- Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism and Function. Cell 2004; 116:281 - 297; PMID: 14744438; http://dx.doi.org/10.1016/S0092-8674(04)00045-5
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science 2001; 294:853 - 858; PMID: 11679670; http://dx.doi.org/10.1126/science.1064921
- Melo SA, Esteller M. A precursor microRNA in a cancer cell nucleus: get me out of here!. Cell Cycle 2011; 10:922 - 925; PMID: 21346411; http://dx.doi.org/10.4161/cc.10.6.15119
- Bhaskaran M, Wang Y, Zhang H, Weng T, Baviskar P, Guo Y, et al. MicroRNA-127 modulates fetal lung development. Physiol Genomics 2009; 37:268 - 278; PMID: 19439715; http://dx.doi.org/10.1152/physiolgenomics.90268.2008
- Lizé M, Herr C, Klimke A, Bals R, Dobbelstein M. MicroRNA-449a levels increase by several orders of magnitude during mucociliary differentiation of airway epithelia. Cell Cycle 2010; 9:4579 - 4583; PMID: 21088493; http://dx.doi.org/10.4161/cc.9.22.13870
- Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA 2008; 105:3903 - 3908; PMID: 18308936; http://dx.doi.org/10.1073/pnas.0712321105
- Sozzi G, Pastorino U, Croce CM. MicroRNAs and lung cancer: from markers to targets. Cell Cycle 2011; 10:2045 - 2046; PMID: 21623159; http://dx.doi.org/10.4161/cc.10.13.15712
- Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell 2008; 13:48 - 57; PMID: 18167339; http://dx.doi.org/10.1016/j.ccr.2007.12.008
- Pacheco-Pinedo EC, Morrisey EE. Wnt and Kras signaling-dark siblings in lung cancer. Oncotarget 2011; 2:569 - 574; PMID: 21753228
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 2007; 27:91 - 105; PMID: 17612493; http://dx.doi.org/10.1016/j.molcel.2007.06.017
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res 2008; 36:154 - 158; PMID: 17991681; http://dx.doi.org/10.1093/nar/gkm952
- Saito RA, Watabe T, Horiguchi K, Kohyama T, Saitoh M, Nagase T, et al. Thyroid Transcription Factor-1 Inhibits Transforming Growth Factor-{beta}-Mediated Epithelial-to-Mesenchymal Transition in Lung Adenocarcinoma Cells. Cancer Res 2009; 69:2783 - 2791; PMID: 19293183; http://dx.doi.org/10.1158/0008-5472.CAN-08-3490
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009; 19:92 - 105; PMID: 18955434; http://dx.doi.org/10.1101/gr.082701.108
- Lewis BP, Burge CB, Bartel DP. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates that Thousands of Human Genes are MicroRNA Targets. Cell 2005; 120:15 - 20; PMID: 15652477; http://dx.doi.org/10.1016/j.cell.2004.12.035
- Bar M, Wyman SK, Fritz BR, Qi J, Garg KS, Parkin RK, et al. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells 2008; 26:2496 - 2505; PMID: 18583537; http://dx.doi.org/10.1634/stemcells.2008-0356
- Simon RM, Korn EL, McShane LM, Radmacher MD, Wright GW, Zhao Y. Design and analysis of DNA microarray investigations 2003; Springer
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol 2003; 5:1; PMID: 14709173; http://dx.doi.org/10.1186/gb-2003-5-1-r1
- Anagnostou VK, Syrigos KN, Bepler G, Homer RJ, Rimm DL. Thyroid transcription factor 1 is an independent prognostic factor for patients with stage I lung adenocarcinoma. J Clin Oncol 2009; 27:271 - 278; PMID: 19064983; http://dx.doi.org/10.1200/JCO.2008.17.0043
- Barletta JA, Perner S, Iafrate AJ, Yeap BY, Weir BA, Johnson LA, et al. Clinical significance of TTF-1 protein expression and TTF-1 gene amplification in lung adenocarcinoma. J Cell Mol Med 2009; 13:1977 - 1986; PMID: 19040416; http://dx.doi.org/10.1111/j.1582-4934.2008.00594.x
- Reddel RR, Ke Y, Gerwin BI, McMenamin MG, Lechner JF, Su RT, et al. Transformation of human bronchial epithelial cells by infection with SV40 or adenovirus-12 SV40 hybrid virus or transfection via strontium phosphate coprecipitation with a plasmid containing SV40 early region genes. Cancer Res 1988; 48:1904 - 1909; PMID: 2450641
- Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer 2007; 7:899 - 910; PMID: 18004397; http://dx.doi.org/10.1038/nrc2271
- Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev 2007; 21:1025 - 1030; PMID: 17437991; http://dx.doi.org/10.1101/gad.1540407
- Mayr C, Hemann MT, Bartel DP. Disrupting the Pairing Between let-7 and Hmga2 Enhances Oncogenic Transformation. Science 2007; 315:1576 - 1579; PMID: 17322030; http://dx.doi.org/10.1126/science.1137999
- Park SM, Shell S, Radjabi AR, Schickel R, Feig C, Boyerinas B, et al. Let-7 Prevents Early Cancer Progression by Suppressing Expression of the Embryonic Gene HMGA2. Cell Cycle 2007; 6:2585 - 2590; PMID: 17957144; http://dx.doi.org/10.4161/cc.6.21.4845
- Ambros V. The functions of animal microRNAs. Nature 2004; 431:350 - 355; PMID: 15372042; http://dx.doi.org/10.1038/nature02871
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75:843 - 854; PMID: 8252621; http://dx.doi.org/10.1016/0092-8674(93)90529-Y
- Zhang Y, Verbeek FJ. Comparison and integration of target prediction algorithms for microRNA studies. J Integr Bioinform 2010; 7
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 2004; 101:2999 - 3004; PMID: 14973191; http://dx.doi.org/10.1073/pnas.0307323101
- Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer 2006; 6:259 - 269; PMID: 16557279; http://dx.doi.org/10.1038/nrc1840
- Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol 2009; 4:199 - 227; PMID: 18817506; http://dx.doi.org/10.1146/annurev.pathol.4.110807.092222
- Homminga I, Pieters R, Langerak Anton W, de Rooi Johan J, Stubbs A, Verstegen M, et al. Integrated Transcript and Genome Analyses Reveal NKX2-1 and MEF2C as Potential Oncogenes in T Cell Acute Lymphoblastic Leukemia. Cancer Cell 2011; 19:484 - 497; PMID: 21481790; http://dx.doi.org/10.1016/j.ccr.2011.02.008
- Kang Y, Hebron H, Ozbun L, Mariano J, Minoo P, Jakowlew SB. Nkx2.1 transcription factor in lung cells and a transforming growth factor-β1 heterozygous mouse model of lung carcinogenesis. Mol Carcinog 2004; 40:212 - 231; PMID: 15264213; http://dx.doi.org/10.1002/mc.20034
- Harris T, Pan Q, Sironi J, Lutz D, Tian J, Sapkar J, et al. Both gene amplification and allelic loss occur at 14q13.3 in lung cancer. Clin Cancer Res 2011; In press PMID: 21148747; http://dx.doi.org/10.1158/1078-0432.CCR-10-1892
- Minoo P, Hu L, Zhu N, Borok Z, Bellusci S, Groffen J, et al. SMAD3 prevents binding of NKX2.1 and FOXA1 to the SpB promoter through its MH1 and MH2 domains. Nucleic Acids Res 2008; 36:179 - 188; PMID: 18003659; http://dx.doi.org/10.1093/nar/gkm871