Abstract
We report that simultaneous expression of Akt and angiopoietin-1 (Ang-1) transgenes supported mitogenesis in stem cells with a critical role for microRNA-143 (miR-143) downstream of FoxO1 transcription factor. Mesenchymal stem cells (MSC) from young male rats were transduced with Ad-vectors encoding for Akt (AktMSC) and Ang-1 (Ang-1MSC) transgenes for their individual or simultaneous overexpression (AAMSC; > 5-fold gene level and > 4-fold Akt and Ang-1 protein expression in AAMSC vs. Ad-Empty transduced MSC; EmpMSC). AAMSC had higher phosphorylation of FoxO1, which activated Erk5, a distinct mitogen-induced MAPK that drove transcriptional activation of cyclin D1 and Cdk4. Flow cytometry showed > 10% higher S-phase cell population that was confirmed by BrdU assay (15%) and immunohistology for Ki67 (11%) in AAMSC using EmpMSC as controls. miR array supported by real-time PCR showed induction of miR-143 in AAMSC (4.73-fold vs.. EmpMSC). Luciferase assay indicated a dependent relationship between miR-143 and Erk5 in AAMSC. FoxO1-specific siRNA upregulated miR-143, whereas inhibition of miR-143 did not change FoxO1 activation. However, miR-143 inhibition repressed phosphorylation of Erk5 and abrogated cyclin D1 with concomitant reduction in cells entering cell cycle. During in vivo studies, male GFP+ AAMSC transplanted into wild-type female infarcted rat hearts showed significantly higher number of Ki67 expressing cells (p < 0.05 vs. EmpMSC) 7 days after engraftment (n = 4 animals/group). In conclusion, co-overexpression of Akt and Ang-1 in MSC activated cell cycle progression by upregulation of miR-143 and stimulation of FoxO1 and Erk5 signaling.
Introduction
Stem cell therapy for the infarcted heart is confronted with the problem of massive death of the donor stem cells post-transplantation, which results in poor prognosis. The problem is generally addressed by either priming of the cells by activation of survival signaling for improved survival or by increasing the number of donor cells for transplantation to compensate for the cell loss in the cytokine-rich microenvironment of the ischemic heart. With regards to the former approach, we have previously reported that preconditioning of stem cells by pharmacological manipulation,Citation1 growth factor treatmentCitation2 or by exposure to intermittent repeated cycles of anoxia/reperfusionCitation3 effectively promoted donor stem cell survival in the ischemic heart. As a part of the latter strategy, up to 1 billion cells have been transplanted in the experimental animal models, and dose-escalating clinical studies have shown that increasing the number of donor cells supported better therapeutic outcome.Citation4 We hypothesized that manipulation of donor stem cells for sustenance of their inherent property of self-renewal during the acute phase after transplantation would compensate for the massive cell loss. Given that stem cells are excellent carriers of transgenes, the strategy of genetic reprogramming of stem cells can be exploited to improve their post-transplantation characteristics, including survival and proliferation, for effective participation in myocardial repair process. Moreover, additive actions associated with genetic manipulation of stem cells may also include their angiomyogenic differentiation and altered paracrine activity in the heart.Citation5,Citation6 We have already shown that simultaneous overexpression of Akt and angiopoietin-1 (Ang-1) in bone marrow-derived mesenchymal stem cells (MSC) led to coordinated interaction between the two transgenes and gave better stem cell survival, promoted their angiomyogenic differentiation and stably improved global heart function.Citation7,Citation8 The present study was designed to determine the downstream signaling involved during coordinated interaction between Akt and Ang-1 transgenes in MSC and their effect on cell proliferation. We observed that MSC with Akt and Ang-1 co-expression (AAMSC) phosphorylated forkhead box O1 (FoxO1) transcription factor to abrogate its activity in the cells. FoxO proteins are known tumor suppressors, cause cell cycle arrest and induce apoptosis besides their vital involvement in maintenance of cellular homeostasis.Citation9 We observed that Akt-dependent phosphorylation of FoxO1 in AAMSCs was associated with activation of non-classical Erk5 and the expression of nuclear factor cyclin D1, an essential regulator of cell cycle progression via G1/S-phase transition. We have also shown mechanistic involvement of micro RNA-143 (miR-143) in AAMSC as a critical regulator of cell cycle signaling.
Results
Purity of MSC culture and transgene expression.
Bone marrow MSC isolated from male donor rats were expanded in vitro for 3–4 passages before use in further experimentation. Analysis of their surface marker expression showed that purified cells were 94.3 ± 1.4%, 91.5 ± 2.1% and 4.1 ± 0.3% pure for CD29, CD90 and CD45 expression, respectively (). Simultaneous expression of Akt and Ang-1 transgenes in MSC was determined by double fluorescence immunostaining and RT-PCR for the respective transgene expression in the transduced cells ( and C). These data were confirmed by western blot, which showed significant increase in total Akt and Ang-1 protein expression in AktMSC and Ang-1MSC at 48 h after transduction (). However, Akt and Ang-1 protein expression in AAMSC was significantly higher as compared with EmpMSC as well as single gene transduced AktMSC and Ang-1MSC (). Interestingly, pAkt was also increased significantly higher in AAMSC as compared with any other transduced cells.
FoxO1 expression in AAMSC.
Following transduction of MSC for Akt and Ang-1 overexpression, western blot studies revealed that phosphorylation of FoxO1 insignificantly changed in AktMSC and Ang-1MSC as compared with EmpMSC. However, simultaneous expression of Akt and Ang-1 transgenes significantly increased FoxO1 phosphorylation in AAMSC () besides abrogation of total FoxO1 in AAMSC (). Phosphorylation of FoxO1 was observed at both Thr24 and Ser256 sites. While Thr24-FoxO1 was mainly expressed in the cytoplasmic fraction, Ser256-FoxO1 was predominantly localized in the nuclear fraction of AAMSC (). Actin and CREB were used as internal controls for the cytoplasmic and nuclear fractions respectively. Immunostaining of AAMSC with an antibody specific for Thr24-FoxO1 confirmed these data, indicating exportation of Thr24-FoxO1 from the nucleus into the cytoplasm (red fluorescence; ). The nuclei were visualized by DAPI staining (blue fluorescence; ). Western blotting also showed significantly higher phosphorylation of Erk5 in AAMSC as compared with the other treatment groups of cells (). However, we observed a decrease in total Erk5 expression in AAMSC (). We also observed a concomitant increase in cyclin D1 expression in AAMSC (). Fluorescence immunostaining of AAMSC with antibody specific for cyclin D1 was consistent with western blot data and showed predominant nuclear localization of cyclin D1 (red fluorescence indicated by white arrows; ). The increase in cyclin D1 expression led to increased expression of downstream Cyclin dependent kinase 4 (Cdk4) (). Cyclin D1 and cyclin-dependent kinase expression have been widely regarded as regulatory promoters of cell cycle progression as well as indicators of cells entering S phase of the cell cycle.Citation10
Role of miR-143 in cell cycle of AAMSC.
Microarray profiling for miRs showed nearly 2-fold elevation of miR-143 in AAMSC (). These findings were confirmed by real-time PCR, which showed significant expression of miR-143 in AAMSC as compared with EmpMSC as control (). Transfection of native MSC (without genetic modification) with FoxO1-specific small integrated RNA (siFoxO1) simulated the effects of co-expression of Akt and Ang-1 in terms of miR-143 expression (). Treatment with siFoxO1 significantly increased miR-143 in native MSC as compared with scramble siRNA (Sc) treated native MSC, thus suggesting that FoxO1 activity was responsible for miR-143 induction () On the contrary, loss-of-function studies by abrogation of miR-143 using specific antagomir failed to recover FoxO1 expression, which was abrogated by simultaneous expression of Akt and Ang-1 in AAMSC (). These results confirmed that FoxO1 was upstream of miR-143.
In agreement with these observations, ectopic expression of pre-miR-143 in native MSC led to significant reduction in Erk5 expression (). However, a concomitant increase in Erk5 phosphorylation in MSC with ectopic expression of miR-143 was also observed as compared with the scramble-treated control cells (). Ectopic expression of miR-143 also increased cyclin D1 in the native MSC as compared with scramble transfected cells (). On the contrary, pre-treatment of AAMSC with miR-143 specific antagomir significantly increased total Erk5 (). However, we also observed concomitant reduction in pErk5 and abolished cyclin D1 expression (). These data showed that miR-143 regulated Erk5 and cyclin D1 signaling in AAMSC via its critical regulatory role in FoxO1 activity.
MiR-143 and Erk5 expression in AAMSC.
We examined three databases (miRANDA, Sanger MirBase and Targetscan algorithms) for in silico prediction of the potential targets of miR-143. Computational analysis showed a putative consensus site in the 3′UTR of Erk5 mRNA for miR-143 binding (). To determine a relationship between miR-143 and Erk5, a vector for Luc-pair miR-143 target clone for Luciferase assay was constructed specific for the 3′UTR region of Erk5 (). Using luciferase activity assay, co-transfection of a precursor miR-143 expression vector (pEZX-miR-143) with the vector containing 3′UTR of the Erk5 gene led to reduced luciferase activity in comparison to co-transfection with the miR-scramble vector (pEZX-miR-scramble), indicating that forced expression of miR-143 downregulated Erk5 by targeting 3′UTR of Erk5 gene ().
Functional aspects of miR-143 induction in AAMSC.
To demonstrate the functional aspect of miR-143 expression changes in response to co-overexpression of Akt and Ang-1 in AAMSC, BrdU assay was performed and showed that cell proliferation was increased in AAMSC (p < 0.01 vs. EmpMSC; and B). We observed 23.72 ± 3.4% BrdU positivity in AAMSC as compared with 9.71 ± 1.24% in EmpMSC. On the other hand, pretreatment of AAMSC with miR-143 antagomir significantly reduced BrdU positivity in AAMSC, unlike pre-treatment with scramble, which did not alter the proliferative activity of AAMSC. FACS analysis of AAMSC after propidium iodide labeling showed that co-overexpression of Akt and Ang-1 increased the number of AAMSC in S phase of the cell cycle to 15.24% ± 1.62% as compared with 4.220.49% in EmpMSC, used as a control. The proliferative effect of Akt and Ang-1 co-expression was significantly abolished by pre-treatment of AAMSC with miR-143 antagomir ( and D). These data was confirmed by fluorescence immunostaining specific for Ki67, a protein expressed in the nucleus of a cycling cell ( and F). Ki67-specific immunostaining showed up to 11.9 ± 1.5% increase in Ki67 positivity in AAMSC, which was reduced to 3.05 ± 0.23% in AAMSC with miR-143 antagomir pretreatment (p < 0.01; and F).
Proliferation of AAMSC post transplantation in the infracted myocardium.
Fluorescence immunostaining of histological sections for Ki67 expression was performed 7 d after transplantation of the cells with their respective treatment into the infarcted heart. The results showed AAMSC transplanted hearts had significantly higher total number of Ki67+ cells as compared with the EmpMSC- and DMEM-treated animal groups (). Double fluorescence immunostaining for Ki67 and GFP antigens was performed to visualize transplanted GFP+ donor cells that were positive for Ki67 expression ( and D). The number of GFP+Ki67+ cells was significantly higher in AAMSC- ( and D) transplanted animal hearts as compared with EmpMSC ( and D).
Discussion
Self-renewal in stem cells involves an intricate mechanism regulated by various cyclin-dependent kinases (Cdks). The activity of Cdks gets modulated in response to different stimuli, and this differentially determines cell cycle progression in various cell types.Citation11 MiRs, in this regard, have emerged as new players that are critically involved in the signaling relevant to cell survival, differentiation and proliferation.Citation12–Citation14 For their critical participation in cell proliferation, miRs have been shown to post-transcriptionally regulate the expression of genes that encode for proteins critically involved in cell cycle progression.Citation15–Citation17 Whereas miR-34 transcriptionally controls p53 to downregulate a set of genes involved in cycling tumor cells,Citation18 oncogene Ras-mediated induction of miR-21 was attributed to Ras-dependent tumor cell proliferation by controlling gene expression of cell cycle checkpoint regulators.Citation19 More recent studies have determined the role of miRs in stem cell proliferation and reported that miRs relevant to cell cycle progression in embryonic stem cells included miR-290 cluster which suppressed many cell cycle regulating proteins.Citation15,Citation20 Our present study showed that miR-143 was significantly increased in stem cells genetically modified to co-overexpress Akt and Ang-1. Our data also implied Erk5 as its putative target gene to regulate cell cycle in AAMSC. The novel findings of our study were that (1) phosphorylation of FoxO1 significantly increased miR-143 in response to simultaneous expression of Akt and Ang-1 in stem cells; (2) ectopic expression of miR-143 in native MSC significantly reduced total Erk5 expression, and (3) miR-143 regulated G1-S-phase transition in AAMSC by targeting Erk5/cyclin D1 signaling. These molecular events led to cell cycle progression in AAMSC.
Our strategy of simultaneous expression of Ang-1 and Akt resulted in a significantly higher pAkt/Akt ratio in AAMSC as compared with either AktMSC or Ang-1MSC, using EmpMSC as a baseline control. Activation of Akt signaling is generally associated with cell growth, proliferation, cell energy metabolism as well as pro-survival effects.Citation21 The FoxO subfamily of forkhead transcription factors are known regulators of these cellular processes as well, and their activity is regulated by PI3K/Akt.Citation22,Citation23 Abrogation of PI3K/Akt signaling activates FoxO transcription factors, thus leading to cell cycle arrest and apoptosis.Citation22 On the contrary, phosphorylation of FoxO transcription factors by pAkt at Thr24, Ser256 and Ser319 curtails their ability to bind with DNA in the nucleus and, subsequent to complexation with 14-3-3 protein, they are exported out of the nucleus into the cytoplasm for degradation.Citation24 Therefore, shuttling of FoxO transcription factors between the nucleus and cytoplasm remains central to its transcriptional activity. FoxO contains both a nuclear export and a nuclear localization sequence; the latter gets masked subsequent to phosphorylation by Akt.Citation25 In relation to cell cycle progression, exportation of FoxO1 from nuclei into cytoplasm and phosphorylation by Akt allows its cytoplasmic localization, which promotes cyclin D1 and D2 expression to support cell cycle phase transition.Citation26,Citation27 Our strategy of co-overexpression of Akt and Ang-1 led to significant phosphorylation of FoxO1 with concomitant elevation in miR-143. Loss-of-function studies showed that inhibition of FoxO1 repressed miR-143 in AAMSC, whereas abrogation of miR-143 in AAMSC did not change FoxO1 activity. miR-143 is one of the most enriched miRs during embryonic development, with an essential role for myocardial proliferation, cardiac function and cardiogenesis.Citation28 Besides, miR-143 also showed reversed expression in some carcinomas and regenerative process of epithelial mucosa,Citation29–Citation31 thus suggesting that miR-143 may promote different cellular functions based upon lineage background as well as differences in the genetic make-up of the cells.
MiR-143 is a new entrant in cell cycle signaling. Although computational and experimental databases have identified multiple targets for miR-143, it remains undetermined how the variable targets influence the versatile biological activity of miR-143.Citation29,Citation30,Citation32,Citation33 One of the important direct target genes of miR-143 is Erk5, also known as big MAP kinase (Bmk1), a member of atypical mitogen-activated kinase family. Erk5 mostly gets activated in response to oxidative stress, hyperosmolarity and treatment of cells with serum and growth factors such as epidermal growth factor.Citation34 Although little information is available regarding substrates of Erk5, published data shows that activated Erk5 phosphorylates connexin-43 Citation35 and transcription factor MEF2c.Citation25 In relation to its role in cell proliferation, Erk5 interacts with cyclin D1 kinase to promote cell growth and proliferation in response to cytokine stimulation.Citation36 Cyclin D1 and its binding partner Cdk4 are known important regulators of cell migration, invasion as well as cell cycle progression from G1 to S phase in different cell types.Citation37,Citation38 Expression of cyclin D1 forms active complexes that promote cell cycle progression via phosphorylation and inhibition of the retinoblastoma protein and are linked to the development of several forms of carcinoma.Citation39 Cdk4 is a member of the Ser/Thr protein kinase family that serves as a catalytic subunit of the protein kinase complex involved in G1-phase progression of cell cycle.Citation32 The activity of Cdk4 is restricted to G1-S phase, which is controlled by the regulatory subunits D-type cyclins and Cdk inhibitor p16 (INK4a).Citation32 Our data provided clear evidence that cyclin D1 as well as Cdk4 were upregulated in AAMSC, and the functional outcome of these molecular events was evident from more than 15% increase in S-phase AAMSC.
In conclusion, we report for the first time that combined expression of Akt with angiocompetent Ang-1 enhances stem cell proliferation through upregulation of mir-143 and stimulation of signaling downstreams of FoxO1 and Erk5. Given that AAMSC have superior angiomyogenic capacity, the strategy will be important in addressing the problem of massive donor cell death that ensues during acute phase after transplantation.
Materials and Methods
All experimental procedures were performed in accordance with the standard human care guidelines of the “Guide for the Care and Use of Laboratory Animals” and Institutional Animal Care and Use of Committee of University of Cincinnati, which conformed to National Institutes of Health guidelines.
Isolation, expansion and purity of MSC.
MSC were obtained from young male Fisher-344 rats by flushing and culturing the bone marrow harvested from the cavity of femurs and tibias as described previously in reference Citation7. For surface marker analysis of the MSC cell cultures, the cells were immunstained with fluorescently conjugated antibodies at concentration of 1 µg/ml and analyzed by flow cytometry (FACSCanto, BD Biosciences) for CD29, CD90 and CD45.Citation7
Viral vector propagation and transduction of MSC.
Replication-deficient adenoviral vectors (Ad) with any therapeutic gene or encoding for Akt and Ang-1 transgenes were gifts from Dr. Xu Meifeng (University of Cincinnati) and Dr. Ge Ruowen (National University of Singapore). The vectors were individually propagated in HEK-293 cells cultured in DMEM supplemented with 10% fetal bovine serum (FBS). The cells were transduced with respective viral vector for 2 cycles of 6 h each and a subsequent recovery period of 48 h before use in further experimentation.Citation7 Fluorescence immunostaining, RT-PCR and western blotting were performed to determine percentage transduction and expression efficiency of the transgenes.Citation7
RNA isolation and reverse transcription-PCR (RT-PCR).
For RT-PCR, RNeasy kit (Qiagen) was used for isolation of total mRNA from the cells per manufacturer's instructions and RT-PCR was run for 35 cycles using specific primers (Table S1).Citation7 Densitometry for quantification of mRNAs was performed using computer software (FluorChem SP, Alpha Innotech).
Western blotting.
Cells were harvested and homogenized at 4°C for 30 min using lysis buffer (HEPES 50 mM, EDTA 5 mM, NaCl 50 mM, Triton X-100 1%, NaF 50 mM, Na3VO4 1 mM, Na4P2O7 10 mM, aprotinin 10 µg/ml, leupeptin 10 µg/ml, phenylmethylsulfonyl fluoride 1 mM). The whole-cell lysate samples were centrifuged for 10 min at 4°C at 14,000 G. Cytoplasmic and nuclear protein fractions were isolated using commercial kits (NE-PER Nuclear and Cytoplasmic Extraction Reagents, Thermo Scientific) per supplier's protocol. Concentration of protein was measured using Bradford's method, and protein lysates were aliquoted and stored at −80°C until used. A total of 25–30 µg protein samples were electrophoresed in each well of SDS-polyacrylamide gels (Invitrogen) at 100 V using running buffer (MOPS buffer, Invitrogen). After electroblotting of the proteins from the gel on to PVDF membranes overnight (BioRad), membranes were then washed with TBST buffer, blocked with 5% non-fat milk, incubated with specific primary and secondary antibodies (Table S2) and developed onto X-ray films (Denville Scientific) using chemiluminescent reagents (ECL, GE Healthcare Biosciences or SuperSignal, West Femto Thermo Scientific). Densitometry was performed with FluorChem SP (Alpha Innotech) computer software.
MiRNA target prediction.
Computational prediction for best complementarity of the target genes was performed using available databases (target scan.org and microrna.org).
MiRNA isolation, miRNA array and real-time PCR.
Isolation of miRNA from various treatment groups of cells was performed using a commercially available kit (Ambion) following the instructions of the manufacturer.Citation3 For miRNA array studies, the samples were sent to LC Sciences. For confirmation and reproducibility assessment of the microarray data of the miRNAs of interest, qRT-PCR was performed using specific miRNA primers (Exiqon) following manufacturer's instruction. For loss-of-function studies, specific small interference RNA siRNA (Dharmacon Thermo Scientific), antagomirs and scramble siRNA (Sc siRNA) were purchased from Exiqon, and transfection of the chemicals was performed using tranfection reagent (DharmaFECT Duo, Thermo Scientific) per the instructions of the companies.
Luciferase reporter assay.
Precursor miR-143 expression clone was constructed in a feline immunodeficiency virus (FIV)-based lentiviral vector system (pEZX-miR-143), and luciferase reporter constructs containing the 3′UTR of Erk5 were designed to encompass the miR-143 binding sites (GeneCopoeia). For luciferase assay, MSCs were plated in triplicate into 24-well plates and co-transfected with 0.8 µg of pEZX-miR-143 (or pEZX-miR-scramble) and reporter construct by using the Lipofectamine 2000™ (Invitrogen, Carlsbad). Transfection efficiency was normalized on the basis of luciferase activity. The luciferase activity was measured at 48 h after transfection using Luciferase Reporter Assay System kit according to the manufacturer's instructions.
Cell cycle analysis.
The different treatment groups of cells were trypsinized 48 h after transduction, washed with cold PBS and fixed with 70% ethanol at 4°C. The cells were washed twice with cold PBS and subjected to RNase/propidium iodide (PI) staining using Cell Cycle kit (GenScript) per manufacturer's instructions. The cells were analyzed using flow cytometer (FACSCanto), and a minimum of 10,000 events were analyzed at 488 nm excitation wavelength. The data was analyzed using computational software Modfit LT30 (Verity Software House).
The cells entering S phase of cell cycle were also determined by immunostaining for the expression of endogenous Ki67 and by bromodeoxyuridine assay (BrdU, BD PharMingen).
Fluorescence immunocytochemistry.
Non-transfected and transfected MSC were cultured in chambered glass slides, fixed with 4% paraformaldehyde. After washing twice with PBS, the cells were permeabilized with 0.5% Triton-X100 (Sigma Aldrich) for 30 min, washed twice with PBS and incubated with Cas-Block for 1 h (Invitrogen). The cells were then incubated with primary antibodies specific for Akt, Ang-1, phospho-Th24 FoxO1, cyclin D1, BrdU and Ki67 for 2 h at 37°C. Following this, the cells were washed and incubated with corresponding fluorescently labeled secondary antibodies for 1 h at 37°C. Finally, the nuclei were visualized by DAPI staining at 0.5 µg ml for 5 min. The slides were then washed and mounted with Flouromount-G solution (Southern Biotech) and observed under fluorescence microsope (BX-41, Olympus). The list of antibodies used is provided in Table S1. For Ki67- and BrdU-positive cells, random microscopic fields of 300 cells were photographed and counted. The cells positive for BrdU and Ki67 were considered as cells in S phase of mitosis.
In vivo studies.
Male donor bone marrow derived MSC from young GFP transgenic rats were transduced with Ad-Emp vector or vectors encoding for Akt and Ang-1. For in vivo studies, experimental model of acute myocardial infarction was developed in female Fisher-344 rats by permanent ligation of left anterior descending (LAD) coronary artery.Citation7 Ten minutes after LAD ligation, the animals were grouped for intramyocardial injection of 60 µl DMEM without cells (group-1) or containing GFP expressing (green fluorescence) 1 × 106 male EmpMSC (group-2) or AAMSC (group-3). The chest was sutured and animals were allowed to recover. During post-operational care, Buprenex (0.1 mg/kg b.i.d) was administered for 24 h to alleviate pain. For post-mortem studies, animals were harvested on day 7 (n = 4/group) after their respective treatment using an overdose of sodium pentobarbitol, and the heart tissue was collected for immunohistological studies. The heart tissue samples were fixed with 4% paraformaldehyde and cryosectioned at 7–8 µm thickness. The histological sections were immunostained with primary antibodies specific for Ki67 and GFP (Table S2). The primary antibody-antigen reaction was visualized with specific fluorescently labeled secondary antibodies and analyzed using fluorescent microscope (BX-41, Olympus) for the expression of Ki67 in transplanted cells.
Statistical analysis.
Data were expressed as mean ± SEM, t-test or Anova were used with appropriate post hoc analysis. A p-value < 0.05 was considered to be statistical significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Abbreviation
| Ad | = | adenoviral |
| Ang-1 | = | angiopoietin |
| AAMSC | = | MSC transduce for coexpression of Akt and Ang-1 |
| AktMSC | = | Ad-Akt transduced MSC |
| Ang-1MSC | = | Ad-angiopoietin-1 transduced MSC |
| Cdk4 | = | cyclin dependent kinase 4 |
| EmpMSC | = | Ad-empty transduced MSC |
| MSC | = | mesenchymal stem cells |
| miR | = | microRNA |
Figures and Tables
Figure 1 In vitro characterization of MSC for transgenes expression. (A) Flow cytometric analysis of hematopoietic and mesenchymal specific membrane markers CD45, CD29 and CD90 in the purified MSC culture used during the present studies. (B) Double fluorescence immunostaining for Akt (green fluorescence) and Ang-1 (red fluorescence) expression in AAMSC. (C) RT-PCR for Akt and Ang-1 transgene expression in various treatment groups of cells. Densitometry showed significantly higher Akt and Ang-1 in AktMSC, Ang-1MSC and AAMSC as compared with EmpMSC at 48 h after transduction. Transgene expression was normalized to actin. (D) Western blots showing total Akt, pAkt and Ang-1 protein expression standardized to actin in various treatment groups of MSC. Highest level of Akt, pAkt and Ang-1 was observed in AAMSC as compared with Ang-1MSC, AktMSC and EmpMSC.
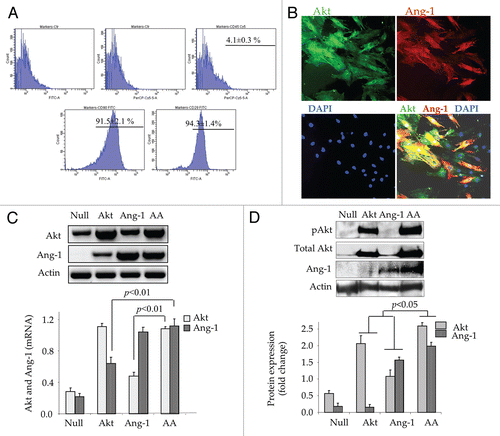
Figure 2 Phosphorylation of FoxO1 and its nuclear exportation in AAMSC. (A and B) Western blots showing significantly higher phosphorylation of FoxO1 at Ser256 with concomitant reduction in total FoxO1 in the whole cell lysate using actin for normalization during densitometry. (C) Representative western blots of FoxO1 and phosphorylated FoxO1 at Ser256 and Thr24 in the nuclear and cytoplasmic fractions of various groups of cells. Whereas Thr24-FoxO1 was mainly activated in cytoplasmic fraction and Ser256-FoxO1 was mainly activated in the nuclear fraction, actin was used as loading control for cytoplasmic fraction and CREB transcription factor was used for loading control in nuclear fraction. (D) Representative images of immunostained AAMSC showing cytoplasmic localization of Thr24-FoxO1 (red fluorescence). No Thr24-FoxO1 was observed in EmpMSC. Nuclei were visualized by DAPI staining (blue). (E and F) Proportion of pErk5 normalized to total Erk5 and total cyclin D1 normalized to actin in the whole cell lysate samples from after respective treatment. Highest level of pErk5 and Cyckin D1 were observed in AAMSC. (G) Representative images of AAMSC immunostained for cyclin D1 expression. Extensive nuclear localization of cyclin D1 (ref fluorescence) was observed in AAMSC. (H) Western blots showing significantly higher expression of Cyclin dependent kinase-4 expression in AAMSC as compared with other treatment groups of cells. Actin was used as an internal control.
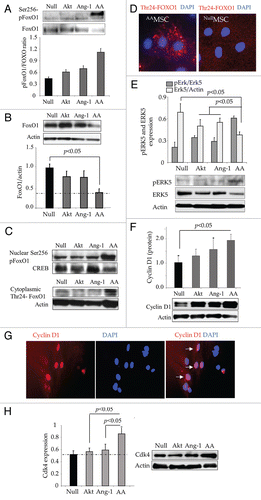
Figure 3 miR-143 expression in AAMSC regulated cell cycle signaling. (A) Heat map showing miR expression profiles in different treatment groups of MSC. (B) Validation of microarray data by Real-time PCR which confirmed significantly higher expression of miR-143 in AAMSC as compared with AktMSC, Ang-1MSC and EmpMSC as controls. (C) Expression of miR-143 was significantly increased in AAMSC. Similarly, when native MSC (without any viral vector transduction) were treated with siFoxO1, miR-143 expression was significantly increased (similar to AAMSC) whereas treatment of native MSC with scramble did not alter miR-143 expression, thus suggesting that miR-143 expression was FoxO1 dependent. (D) Western blots showing FoxO1 was abrogated by co-expression of Akt/Ang-1 in AAMSC as compared with EmpMSC. However, prior treatment of AAMSC with miR-143 specific antagomir did not rescue FoxO1 expression in AAMSC. (E) Western blots showing ectopic expression of miR-143 in native MSC led to abrogation of total Erk5 expression. However, we observed concomitant increase in phosphorylation of Erk5 in comparison with scramble transfected cells. (F) Western blots showing significantly higher expression of cyclin D1 in native MSC with ectopic expression of miR-143. Scramble transfection in AAMSC did not alter cyclin D1 expression. (G and H) Western blots showing significantly higher induction of Erk5 and abrogation of cyclin D1 in native MSC treated with miR-143 specific antagomir as compared with the scramble transfected native MSC as controls (p < 0.05). Although Erk5 was significantly increased after pretreatment with miR-143 specific antagomir, phosphorylation of Erk5 was significantly decreased in AAMSC as compared with scrambled treated AAMSC.
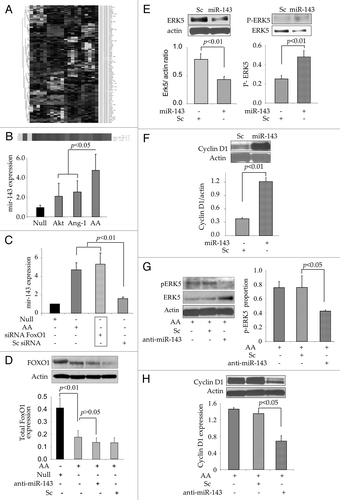
Figure 4 Erk5 is a direct target of miR-143. (A) Computational analysis predicting possible target site of miR-143 conserved in the 3′UTR of Erk5 mRNA. (B) The construction of pEZX-Luc-Erk5 3′UTR luciferase reporter plasmid and precursor miR-143 expression clone. Co-transfection of MSC with pEZX-Luc vector containing Erk5 3′ UTR together with a plasmid encoding miR-143 showed decreased luciferase activity (p < 0.01 vs. pEZX-miR-SC transfected cells). The ratio of luciferase activity was calculated either in the presence or absence of miR-143. (C) Luciferase assay measurement in AAMSC transfected with vectors carrying Luciferase and Erk5 genes.
Figure 5 Co-overexpression of Akt/Ang-1 transgenes enhanced cell proliferation. (A) Graph showing significantly higher BrdU uptake in AAMSC as compared with EmpMSC. However, prior treatment of AAMSC with miR-143 antagomir significantly reduced BrdU positivity in AAMSC as compared with scramble (Sc) treated AAMSC. (B) Representative merged fluorescence images of AAMSC immunostained for BrdU uptake (green fluorescence). The nuclei were visualized by DAPI staining (blue fluorescence) and showed nuclear localization of BrdU signals (green fluorescence). White boxed areas in AAMSC and EmpMSC images were magnified for clarity (original magnification = 40x). (C and D) Graph showing significantly higher percentage of AAMSC in G-S phase transition. The cells were stained with propidium iodide and analyzed by FACS at 488 nm. The native (non-transduced) MSC and EmpMSC were used as controls. Prior treatment of the AAMSC with miR-143 antagomir significantly reduced G-S phase transition of the cells. (D) Typical representative histograms from FACS analysis for G-S phase transition of native, AAMSC and AAMSC with miR-143 antagomir treatment or Sc treatment. (E and F) Fluorescence immunostaining of cells from various treatment groups for Ki67 expression (green fluorescence). The nuclei were visualized by DAPI staining which helped to determine nuclear specificity of Ki67 expression (original magnification = 40x). The percentage of Ki67+ cells was significantly increased in AAMSC which was abolished by pretreatment of the cells with miR-143 antagomir. Pretreatment with scramble (Sc) did not alter Ki67 positivity in AAMSC.
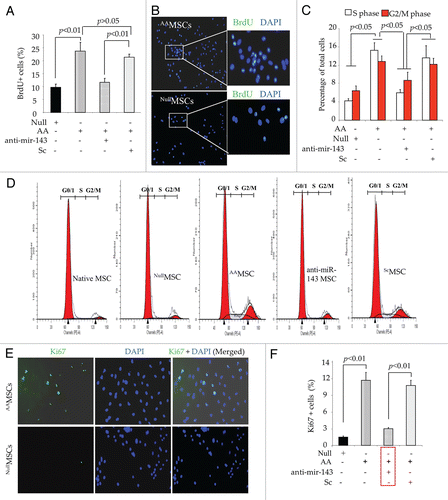
Figure 6 Proliferation of AAMSC post-transplantation in the infarct myocardium. (A) Quantification of Ki67+ positivity in the infarcted myocardium on day 7 after transplantation. The total number of Ki67+ cells was significantly higher in AAMSC transplanted animal hearts as compared with the other treatment groups. (B–D) Representative images of histological sections on day 7 after treatment with (B) EmpMSC and (C) AAMSC. The histological sections were immunostained for Ki67 (red) and GFP (green) antigens. (D) Graph showing higher number of Ki67+/GFP+ cells in AAMSCs group-3 animal hearts as compared with EmpMSC group-2 animal hearts (magnification = 20x).
Additional material
Download Zip (63.4 KB)Funding
This work was supported by National Institutes of Health (NIH) Grants # R37HL074272; HL-080686; HL-087246 (M.A.) and HL-087288; HL-089535; HL106190-01 (Kh.H.H.).
References
- Niagara MI, Haider HKh, Jiang S, Ashraf M. Pharmacologically preconditioned skeletal myoblasts are resistant to oxidative stress and promote angiomyogenesis via release of paracrine factors in the infarcted heart. Circ Res 2007; 100:545 - 555; PMID: 17234963; http://dx.doi.org/10.1161/01.RES.0000258460.41160.ef
- Lu G, Haider HK, Jiang S, Ashraf M. Sca-1+ stem cell survival and engraftment in the infarcted heart: dual role for preconditioning-induced connexin-43. Circulation 2009; 119:2587 - 2596; PMID: 19414636; http://dx.doi.org/10.1161/CIRCULATIONAHA.108.827691
- Kim HW, Haider HK, Jiang S, Ashraf M. Ischemic preconditioning augments survival of stem cells via miR-210 expression by targeting caspase-8-associated protein 2. J Biol Chem 2009; 284:33161 - 33168; PMID: 19721136; http://dx.doi.org/10.1074/jbc.M109.020925
- Martin-Rendon E, Brunskill SJ, Hyde CJ, Stanworth SJ, Mathur A, Watt SM. Autologous bone marrow stem cells to treat acute myocardial infarction: a systematic review. Eur Heart J 2008; 29:1807 - 1818; PMID: 18523058; http://dx.doi.org/10.1093/eurheartj/ehn220
- Yau TM, Kim C, Li G, Zhang Y, Weisel RD, Li RK. Maximizing ventricular function with multimodal cell-based gene therapy. Circulation 2005; 112:123 - 128; PMID: 16159803
- Haider HKh, Jiang S, Idris NM, Ashraf M. IGF-1-overexpressing mesenchymal stem cells accelerate bone marrow stem cell mobilization via paracrine activation of SDF-1alpha/CXCR4 signaling to promote myocardial repair. Circ Res 2008; 103:1300 - 1308; PMID: 18948617; http://dx.doi.org/10.1161/CIRCRESAHA.108.186742
- Jiang S, Haider HKh, Idris NM, Salim A, Ashraf M. Supportive interaction between cell survival signaling and angiocompetent factors enhances donor cell survival and promotes angiomyogenesis for cardiac repair. Circ Res 2006; 99:776 - 784; PMID: 16960098; http://dx.doi.org/10.1161/01.RES.0000244687.97719.4f
- Shujia J, Haider HK, Idris NM, Lu G, Ashraf M. Stable therapeutic effects of mesenchymal stem cell-based multiple gene delivery for cardiac repair. Cardiovasc Res 2008; 77:525 - 533; PMID: 18032392; http://dx.doi.org/10.1093/cvr/cvm077
- Salih DA, Brunet A. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr Opin Cell Biol 2008; 20:126 - 136; PMID: 18394876; http://dx.doi.org/10.1016/j.ceb.2008.02.005
- Resnitzky D, Gossen M, Bujard H, Reed SI. Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol Cell Biol 1994; 14:1669 - 1679; PMID: 8114703
- Ito T, Yagi S, Yamakuchi M. MicroRNA-34a regulation of endothelial senescence. Biochem Biophys Res Commun 2010; 398:735 - 740; PMID: 20627091; http://dx.doi.org/10.1016/j.bbrc.2010.07.012
- Saunders LR, Sharma AD, Tawney J, Nakagawa M, Okita K, Yamanaka S, et al. miRNAs regulate SIRT1 expression during mouse embryonic stem cell differentiation and in adult mouse tissues. Aging (Albany NY) 2010; 2:415 - 431; PMID: 20634564
- Brett JO, Renault VM, Rafalski VA, Webb AE, Brunet A. The microRNA cluster miR-106b∼25 regulates adult neural stem/progenitor cell proliferation and neuronal differentiation. Aging (Albany NY) 2011; 3:108 - 124; PMID: 21386132
- Sayed D, Abdellatif M. AKT-ing via microRNA. Cell Cycle 2010; 9:3213 - 3217; PMID: 20814244; http://dx.doi.org/10.4161/cc.9.16.12634
- Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet 2008; 40:1478 - 1483; PMID: 18978791; http://dx.doi.org/10.1038/ng.250
- Xia W, Li J, Chen L, Huang B, Li S, Yang G, et al. MicroRNA-200b regulates cyclin D1 expression and promotes S-phase entry by targeting RND3 in HeLa cells. Mol Cell Biochem 2010; 344:261 - 266; PMID: 20683643; http://dx.doi.org/10.1007/s11010-010-0550-2
- Peck B, Schulze A. A role for the cancer-associated miR-106b∼25 cluster in neuronal stem cells. Aging (Albany NY) 2011; 3:329 - 331; PMID: 21483041
- He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network—another piece in the tumour-suppression puzzle. Nat Rev Cancer 2007; 7:819 - 822; PMID: 17914404; http://dx.doi.org/10.1038/nrc2232
- Frezzetti D, De Menna M, Zoppoli P, Guerra C, Ferraro A, Bello AM, et al. Upregulation of miR-21 by Ras in vivo and its role in tumor growth. Oncogene 2011; 30:275 - 286; PMID: 20956945; http://dx.doi.org/10.1038/onc.2010.416
- Lichner Z, Páll E, Kerekes A, Pállinger E, Maraghechi P, Bosze Z, et al. The miR-290–295 cluster promotes pluripotency maintenance by regulating cell cycle phase distribution in mouse embryonic stem cells. Differentiation 2011; 81:11 - 24; PMID: 20864249; http://dx.doi.org/10.1016/j.diff.2010.08.002
- Khwaja A. Akt is more than just a Bad kinase. Nature 1999; 401:33 - 34; PMID: 10485701; http://dx.doi.org/10.1038/43354
- Roy SK, Srivastava RK, Shankar S. Inhibition of PI3K/AKT and MAPK/ERK pathways causes activation of FOXO transcription factor, leading to cell cycle arrest and apoptosis in pancreatic cancer. J Mol Signal 2010; 5:10; PMID: 20642839; http://dx.doi.org/10.1186/1750-2187-5-10
- Wang H, Cui J, Bauzon F, Zhu L. A comparison between Skp2 and FOXO1 for their cytoplasmic localization by Akt1. Cell Cycle 2010; 9:1021 - 1022; PMID: 20160512; http://dx.doi.org/10.4161/cc.9.5.10916
- Van Der Heide LP, Hoekman MF, Smidt MP. The ins and outs of FoxO shuttling: mechanisms of FoxO translocation and transcriptional regulation. Biochem J 2004; 380:297 - 309; PMID: 15005655; http://dx.doi.org/10.1042/BJ20040167
- Cameron SJ, Abe J, Malik S, Che W, Yang J. Differential role of MEK5alpha and MEK5beta in BMK1/ERK5 activation. J Biol Chem 2004; 279:1506 - 1512; PMID: 14583600; http://dx.doi.org/10.1074/jbc.M308755200
- Huang H, Tindall DJ. Dynamic FoxO transcription factors. J Cell Sci 2007; 120:2479 - 2487; PMID: 17646672; http://dx.doi.org/10.1242/jcs.001222
- Zhang H. Skip the nucleus, AKT drives Skp2 and FOXO1 to the same place?. Cell Cycle 2010; 9:868 - 869; PMID: 20348850; http://dx.doi.org/10.4161/cc.9.5.11153
- Deacon DC, Nevis KR, Cashman TJ, Zhou Y, Zhao L, Washko D, et al. The miR-143-adducin3 pathway is essential for cardiac chamber morphogenesis. Development 2010; 137:1887 - 1896; PMID: 20460367; http://dx.doi.org/10.1242/dev.050526
- Lee SK, Teng Y, Wong HK, Ng TK, Huang L, Lei P, et al. MicroRNA-145 regulates human corneal epithelial differentiation. PLoS One 2011; 6:21249; PMID: 21701675; http://dx.doi.org/10.1371/journal.pone.0021249
- Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 2009; 460:705 - 710; PMID: 19578358
- Dijckmeester WA, Wijnhoven BP, Watson DI, Leong MP, Michael MZ, Mayne GC, et al. MicroRNA-143 and -205 expression in neosquamous esophageal epithelium following Argon plasma ablation of Barrett's esophagus. J Gastrointest Surg 2009; 13:846 - 853; PMID: 19190970; http://dx.doi.org/10.1007/s11605-009-0799-5
- Liu SP, Fu RH, Yu HH, Li KW, Tsai CH, Shyu WC, et al. MicroRNAs regulation modulated self-renewal and lineage differentiation of stem cells. Cell Transplant 2009; 18:1039 - 1045; PMID: 19523330; http://dx.doi.org/10.3727/096368909X471224
- Wang X, Hu G, Zhou J. Repression of versican expression by microRNA-143. J Biol Chem 2010; 285:23241 - 23250; PMID: 20489207; http://dx.doi.org/10.1074/jbc.M109.084673
- Kato Y, Tapping RI, Huang S, Watson MH, Ulevitch RJ, Lee JD. Bmk1/Erk5 is required for cell proliferation induced by epidermal growth factor. Nature 1998; 395:713 - 716; PMID: 9790194; http://dx.doi.org/10.1038/27234
- Cameron SJ, Malik S, Akaike M, Lerner-Marmarosh N, Yan C, Lee JD, et al. Regulation of epidermal growth factor-induced connexin 43 gap junction communication by big mitogen-activated protein kinase1/ERK5 but not ERK1/2 kinase activation. J Biol Chem 2003; 278:18682 - 18688; PMID: 12637502; http://dx.doi.org/10.1074/jbc.M213283200
- Clapé C, Fritz V, Henriquet C, Apparailly F, Fernandez PL, Iborra F, et al. miR-143 interferes with ERK5 signaling, and abrogates prostate cancer progression in mice. PLoS One 2009; 4:7542; PMID: 19855844; http://dx.doi.org/10.1371/journal.pone.0007542
- Zhong Z, Yeow WS, Zou C, Wassell R, Wang C, Pestell RG, et al. Cyclin D1/cyclin-dependent kinase 4 interacts with filamin A and affects the migration and invasion potential of breast cancer cells. Cancer Res 2010; 70:2105 - 2114; PMID: 20179208; http://dx.doi.org/10.1158/0008-5472.CAN-08-1108
- Li Z, Wang C, Prendergast GC, Pestell RG. Cyclin D1 functions in cell migration. Cell Cycle 2006; 5:2440 - 2442; PMID: 17106256; http://dx.doi.org/10.4161/cc.5.21.3428
- Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer 2011; 11:558 - 572; PMID: 21734724; http://dx.doi.org/10.1038/nrc3090