Abstract
Despite intense studies, questions still remain regarding the molecular mechanisms leading to the development of hereditary breast and ovarian cancers. Research focused on elucidating the role of the breast cancer susceptibility gene 1 (BRCA1) in the DNA damage response may be of the most critical importance to understanding these processes. The BRCA1 protein has an N-terminal RING domain possessing E3 ubiquitinligase activity and a C-terminal BRCT domain involved in binding specific phosphoproteins. These domains are involved directly or indirectly in DNA double-strand break (DSB) repair. As the two terminal domains of BRCA1 represent two separate entities, understanding how these domains communicate and are functionally altered in regards to DSB repair is critical for understanding the development of BRCA1-related breast and ovarian cancers and for developing novel therapeutics. Herein, we review recent findings of how altered functions of these domains might lead to cancer through a mechanism of increased aberrant homologous recombination and possible implications for the development of BRCA1 inhibitors.
Introduction
Breast cancer is an inherited disease in 5–10% of all cases, while the remaining are sporadic in nature.Citation1 Of the hereditary type, 40–45% of cases are linked to the breast cancer susceptibility gene 1 (BRCA1).Citation1,Citation2 BRCA1 is involved in a multitude of cellular functions, including homologous recombination (HR) and perhaps some forms of nonhomologous end joining (NHEJ) (reviewed in refs. Citation3 and Citation4). The N-terminal RING and C-terminal BRCT domains confer ubiquitin-ligase activity and specific phospho-protein binding to BRCA1, respectively. Inherited mutations in these two regions that alter ubiquitylation or binding functions may be the most deleterious in hereditary breast and ovarian cancers. Ubiquitylation is now known to regulate many of the processes involved in DNA repair and, in particular, those associated with BRCA1 (reviewed in ref. Citation5). BRCA1's RING domain acts in concert with BRCA1-associated RING domain 1 (BARD1) as an E3 ubiquitin-ligase. For example, BRCA1 directly ubiquitylates its BRCT binding partner CtIP,Citation6 while another partner, RAP80, binds ubiquitin-modified proteins at DNA double-strand breaks (DSBs).Citation7–Citation9 In addition, a recent study showed that BRCA1 maintains heterochromatin structure by ubiquitylating histone H2A, which modulates DSB repair and possibly suppresses genomic instability and tumorigenesis.Citation10 Therefore, it is somewhat surprising that the BRCA1 synthetic RING domain mutant I26A, which specifically abrogates BRCA1 ubiquitin-ligase activity without disrupting BARD1 binding, was reported not to affect tumorigenesis and DSB repair.Citation11–Citation13
The BRCA1 BRCT domain is known to bind at least three phosphoproteins [Abraxas-RAP80, BRIP1 (also known as BACH1 or FANCJ) and CtIP, referred to as the A, B and C complexes] that have unique S-X-X-F motifs phosphorylated on the S residue, with all involved in DSB repair.Citation7–Citation9,Citation14–Citation19 The B complex is involved in the process of lesion bypass during DNA replication, although its precise role is still unclear.Citation20,Citation21 The roles of the A and C complexes in G2/M checkpoint control and DNA end resection at DSBs, respectively, have been defined in more detail.Citation9,Citation16,Citation18,Citation19 In this perspective, we will focus on reviewing recent findings that have examined how the disruption of specific BRCA1 BRCT protein interactions increases HR. We will also discuss the implications of these findings for the development of synthetic molecules designed to bind BRCA1 to prevent specific protein interactions for therapeutic and diagnostic purposes.
Mechanism of BRCA1-Directed, Enhanced and Aberrant Homologous Recombination
Until recently, it was believed that mutations in the BRCA1 BRCT domain lead to reduced HR, resulting in genomic instability and, ultimately, the development of cancer. However, there is a new and growing literature suggesting that the disruption of specific BRCA1 protein complexes instead increases DNA resection, resulting in elevated HR,Citation22 which is likely to be aberrant and lead to increased genomic instability (reviewed in ref. Citation23). Two reports examined the effects of silencing BRCT-interacting proteins, while a third examined the effect of mutating the binding pocket of the BRCT domain to prevent these interactions, with all reaching similar conclusions.Citation24–Citation26 Silencing Abraxas, RAP80 or the BRCC36 deubiquitinase, all part of the A complex, resulted in increased or hyper-HR (HHR). Similarly, synthetic (K1702M) and naturally occurring (M1775R) mutants of BRCA1 that disrupt most, if not all, BRCT pS-X-X-F-binding interactions also resulted in aberrant HHR. On the other hand, silencing BRCA1, BRIP1 or CtIP resulted in decreased HR, suggesting that HHR is dependent on the presence of BRCA1, and that the disruption of interactions within the BRCA1-A complex is the critical step resulting in these effects.Citation24,Citation25 Together, these three reports concluded that disruption of BRCT interactions, whether resulting from mutations in the BRCT domain or from specifically preventing the A complex from binding to the BRCT domain, resulted in HHRCitation24–Citation26 ( and C). Other results from these reports are in line with HHR; both RAD51 and RPA were associated with repair foci at abnormally high and persistent levels, suggesting that DNA resection at DSBs was extensive. Direct measurements of DNA resection using ChIP and BrdU analyses were in line with this idea.Citation24,Citation26 In addition, overexpression of RAD51 and RPA was observed by immunohistochemistry in malignant regions of a breast cancer tissue sample with the M1775R mutation but not in other samples with different BRCA1 mutations, suggesting that HHR is also observed in patients harboring this mutation.Citation26 Interestingly, while normal HR was not affected by BRCA1 I26A, in line with recent reports in references Citation11–Citation13 ( and B), combining this mutation with K1702M led to the abrogation of HHR and the concomitant reduction in BRCA1 complex ubiquitylationCitation26 ( and D). On the other hand, NHEJ was not affected,Citation26 in agreementCitation24 or in disagreementCitation25 with results from silencing members of the A complex (). This surprising finding suggests that BRCA1-directed ubiquitylation is needed after the accumulation of RPA and RAD51 at resected DNA ends. Since the BRCA1 RING and BRCT domains are known to interact functionally,Citation6 it is possible that the prevention of specific BRCT interactions could redirect the activity of the RING domain. Therefore, the HHR seen with certain BRCT mutants may result from redirected, inappropriate ubiquitylation that is abrogated when both the BRCT and RING domains are inactivated. The underlying cause of this malfunctioning ubiquitylation process is likely linked to the inability of BRCA1 K1702M and M1775R to anchor and presumably direct early DNA resection via CtIP and MRE11/RAD50/NBS1 (MRN), giving nuclease complexes, such as Exo1-BLM-Dna2, uncontrolled access to DNA ends resulting in excessive resectionCitation27 ().
Despite earlier reports suggesting that Abraxas-RAP80 is critical early in the recruiting phase of BRCA1 to DSBs,Citation7–Citation9 all three recent studies suggested that HHR occurred because of abnormal processing late in the recombination process.Citation24–Citation26 Interestingly, expression of M1775R resulted in highly unusual, clustered RPA foci and peculiar juxtaposed promyelocytic leukemia (PML)-nuclear bodies (NBs).Citation26 PML-NBs are associated with post-translational protein processing such as (de)ubiquitylation, (de)sumoylation, and the (dis)assembly of repair complexes,Citation28–Citation30 and these post-translational modifications are critical for efficient DSB repair.Citation5,Citation31,Citation32 In fact, a recent report supports a causal link between PML-NBs and HR.Citation33 Thus, it is possible that such peculiar PML-NBs seen in cells expressing M1775R represent failed repair centers that are “stuck” late in recombination due to a malfunctioning ubiquitylation process (). Alternatively, these erroneous recombination intermediates may be sequestered in nuclear compartments to limit their adverse impact. An indication of this scenario taking place is the finding that I26A reduced K1702M-induced HHR to normal levels, suggesting that HHR depends on the BRCA1 ubiquitin-ligase.Citation26 Altogether, based on these findings, we suggest that the expression of M1775R or K1702M results in the loss of a critical deubiquitination step late in the repair process, leading to stalled repair in a nuclear compartment associated with PML-NBs resulting in HHR (). It is tempting to speculate that this putative deubiquitinase is BRCC36, which is associated with the A complex.Citation34 However, caution should be exercised in interpreting results from various systems and cell-based assays, as HHR with BRCT mutants has so far only been observed in human cells with correlative findings from limited breast cancer tissues.Citation26 Murine models did not show any effect of BRCA1 I26A on tumorigenesis, and HHR has so far not been shown to occur in this species.Citation11,Citation12 Subtle differences in protein structure related to how certain BRCT mutants function in human and mouse and how ubiquitylation is regulated in these species cannot be ruled out at this point.
These recent findings have direct implications for the design of therapeutic strategies for breast and ovarian cancers. On the one hand, they suggest that mechanistic studies should be performed to see if HHR plays any role in the enhanced sensitivity of BRCA1 BRCT mutant tumors to DNA damaging agents like cisplatin,Citation35–Citation37 vosaroxinCitation38 and PARP inhibitors.Citation39–Citation42 Perhaps new therapeutic strategies can be developed to take advantage of the HHR phenotype. Alternatively, these recent reports also provide important insights for developing molecules targeting BRCA1 itself for therapeutics or as research tools. Indeed, the expression of certain BRCA1 BRCT mutants along with RAP80 silencing have been shown to increase the radiosensitivity of cells in culture,Citation7–Citation9,Citation17,Citation26,Citation43 which underscores the notion that increased radiosensitivity can result from abnormal DSB repair that can be either underactive or overactive, both of which could lead to genomic instability and cancer. The remainder of this perspective focuses on strategies for targeting BRCA1 itself.
Development of Therapeutic Peptides and Small Molecule Inhibitors Targeting the BRCT and Other BRCA1 Domains
The radio- and chemo-sensitive phenotype associated with BRCA1 deficiency points to inhibition of BRCA1 as a potential therapeutic strategy. However, most of BRCA1's functions are mediated by protein-protein interactions (PPIs).Citation3 Historically, achieving PPI inhibition has been challenging due to the fact that the contact surface of PPIs is often little more than a flat, large surface void of suitable binding pockets for small molecules.Citation44 However, an increased interest in PPIs, and the development of intermediate-sized therapeutic agents capable of binding to large surfaces, has made the inhibition of some PPIs a therapeutically attractive strategy.Citation45 The complete structure of BRCA1 has yet to be determined, but the crystal structures of the N-terminal RING and C-terminal tandem BRCT domains are available to guide inhibitor development.Citation46–Citation49
The BRCA1 RING domain is composed of a Zn2+ binding region of 8 Cys and His residues that form two separate Zn2+ binding sites and an adjacent coiled-coil region.Citation49 This domain is known to interact primarily with BARD1, forming a heterodimer that possesses E3 ubiquitin-ligase activity (). Mutations that result in the loss of BRCA1 ubiquitin-ligase activity, mainly due to the disruption of BARD1 binding,Citation52 render cells sensitive to ionizing radiation.Citation43,Citation53 Until recently, the BRCA1-BARD1 complex was thought to be constitutive. However, it was recently demonstrated that when BRCA1-BARD1 binds to p53 in the nucleus, BARD1 dissociates, leading to the export of BRCA1 to the cytoplasm and concomitant sensitization of cells to DNA damage.Citation54 Therefore, inhibitors of BRCA1 and BARD1 interaction should lead to radio- and chemo-sensitization. The binding surface between BRCA1 and BARD1 is primarily composed of a 4-helical bundle, with two helicies contributed by each protein. The interface is quite large (2,200 Å2), and presents a formidable challenge for disruption. There is, however, some precedent for disruption of helical bundles. For example, the HIV protein gp41 assembles into a six-helical bundle that is disrupted effectively with peptides, including the HIV drug Fuzeon.Citation55
Interestingly, most cancer-predisposing mutations in the BRCA1 RING domain occur not in the interface between BRCA1 and BARD1, but in the Zn2+ binding sites of the RING domain.Citation53,Citation56 Platinum based anticancer drugs have previously been shown to preferentially bind to Zn2+ finger domains, replacing Zn2+ and thereby altering the protein tertiary structure.Citation57 Recently, it was shown in vitro that platinum agents are able to bind to the RING domain and inhibit its E3 ubiquitin-ligase activity by ejecting Zn2+.Citation56 The Zn-ligating residue H117 of BRCA1 was demonstrated to be the primary platinum binding residue.Citation58 Further work is required to develop specificity for BRCA1 prior to implementing this strategy in living cells.
The BRCA1 tandem BRCT domain is a member of a family of BRCT motifs known to bind phosphorylated proteins involved in DNA repairCitation14,Citation15 () as well as having other functions (reviewed in ref. Citation59). Sometimes these domains exist as a single motif, but often they are found in series, as is the case with BRCA1. Mutations in this domain are among the most common BRCA1 mutations in hereditary breast cancer.Citation46 The BRCT domain has potential for inhibitor development due to its well-defined, relatively small binding cleft known to interact with proteins having pS-X-X-F motifs.Citation15,Citation46,Citation48 Early work using SPOT peptide libraries identified the preferred binding sequence as phospho-Ser-aromatic β-branched/aromatic-Phe and confirmed that the phospho-Ser and Phe are the primary requirements for binding.Citation14,Citation15,Citation60 The highest affinity peptide from this screen had an affinity of 162 nM. Further optimization by Natarajan and coworkers has led to a tetrapeptide with a 40 nM binding affinityCitation51,Citation61,Citation62 (). However, the presence of phospho-Ser in these peptides limits their usefulness as therapeutic agents. Recently, a high-throughput assay based on fluorescence polarization to identify small molecules that bind to the BRCA1 BRCT domain was developed.Citation63,Citation64 An initial screen of the NCI diversity database led to a single hit with an IC50 of 10 µM. Later, a dual fluorescence screen of 75,000 compounds identified 16 inhibitors with the lowest IC50 values in the single digit micromolar range. However, some of these compounds have intrinsic fluorescence or act as fluorescence quenchers, suggesting, as the authors acknowledge, that they may be false positives. Results showing the inhibition of BRCA1 function in cells by any of these inhibitors have yet to appear in the literature. Thus, therapeutically useful BRCA1 BRCT inhibitors remain elusive. Perhaps more powerful in vitro selection techniques able to create extremely diverse (1013-member) peptide libraries composed of drug-like peptides will be able to overcome this barrier.Citation65,Citation66
At present, little is known about the BRCA1 structure outside of its two terminal domains. Much of the internal region of BRCA1 contains evolutionarily conserved sequences, but their function remains to be fully determined.Citation3 There is, however, the SQ cluster (amino acid residues 1,241–1,530) with a number of S-Q residues phosphorylated by ATM and ATRCitation67–Citation70 (). These regions constitute “non-druggable” BRCA1 targets except indirectly by inhibiting these kinases. Elucidating how the phosphorylation of this internal domain affects the activities of the N- and C-terminal domains is still an underexplored area, yet is critical to the design of BRCA1-based therapeutics targeting this region. In addition, the coiled-coil domain (amino acid residues 1,364–1,437) encompassed by the SQ cluster was shown to interact with PALB2 which, in turn, associates with BRCA2.Citation71–Citation73 Phosphorylation of S-Q residues within the SQ cluster was shown not to affect PALB2 binding.Citation72 However, the disruption of this interaction by cancer patient-derived BRCA1 mutations lead to decreased HR and mitomycin C hypersensitivity,Citation71 making it an interesting target from a therapeutic standpoint. Interestingly, the disruption of PALB2 binding to the chromatin regulator MRG15 resulted in HHR.Citation74 This phenotype is similar to the HHR phenotype observed when the BRCA1 A complex is silenced or by the expression of BRCA1 K1702M or M1775R, giving strength to the idea that HHR could also occur by upsetting the binding of proteins to the internal region of BRCA1 and not just via the BRCT domain. To block the interaction of BRCA1 and PALB2, one approach could be the use of hydrocarbon-stapled peptides, which have been shown to disrupt protein-protein interactions involving helical interfaces.Citation75 In addition, screens to identify new binding partners of the BRCA1 internal region involved in DNA repair may continue to provide more targets, such as the recently discovered interaction with the actin-binding protein Filamin A.Citation76
From the greater understanding of the large network of DSB repair has arisen a great interest in mutational analysis of patient tumors for tailored therapeutics. This has been beneficial in many cases of breast cancer, but genotyping can be slow and expensive. Improvements in sequencing technologies have lessened this hurdle, but the number of BRCA1 mutations is vast, and their significance is not always readily apparent.Citation77 Furthermore, DNA sequencing cannot detect epigenetic alterations and other regulatory aberrations, nor can it provide information about what each mutation means in terms of BRCA1 activity.Citation78 Efforts are being made to develop assays to determine BRCA1 activity closer to the physiological context rather than simply sequencing BRCA1 for mutations.Citation79,Citation80 If selective, high-affinity BRCA1 binding peptides are identified, they could perhaps be used to rapidly analyze BRCA1 activity from tumor isolates to more clearly guide therapy. Based on the HHR phenotype, it is possible to detect increased RAD51 and RPA immunoreactivity of human tumor biopsies expressing M1775R.Citation26 Such diagnosis could lead to more effective treatment by guiding appropriate inhibitor selection for patient tumors. However, the particular BRCT mutation being examined is a critical aspect that should be taken into consideration when developing diagnostics that do not directly target BRCA1.
Conclusionss
In this perspective, we have reviewed recent findings of BRCA1-associated HHR and how these new insights might guide the development of research tools and therapeutic BRCA1 inhibitors for cancer. The centrality of BRCA1 in the network of DSB repair is evident in the sensitization of cells to DNA damaging agents upon loss of function at any one of its currently characterized domains. Thus, to inhibit BRCA1, one could focus on several potential regions of the protein, including the RING and BRCT domains as well as the PALB2 binding domain and the SQ cluster. However, the BRCT domain might represent the most attractive known target because it would lead to simultaneous inhibition of multiple PPIs critical for proper DSB repair. While the goal of BRCA1 inhibition would be to sensitize cancer cells to DNA damaging agents, recent findings suggest the possibility that BRCT inhibitors might actually stimulate aberrant DSB repair through disruption of the BRCA1-A complex. Specific disruption of any of the BRCA1-A, -B or -C complexes seems challenging, as all three complexes bind in the same pocket of the BRCT domain. Thus, while BRCT inhibitors could possibly radiosensitize non-BRCT mutant cancer cells by promoting excessive and aberrant HHR, resulting in increased genomic instability, these effects would likely also be deleterious to normal cells. Therefore, the delivery and targeting of BRCA1-specific therapeutic inhibitors to cancer cells is as critical as their design. A case can also be made that BRCA1-mediated ubiquitylation might be a better target in BRCT mutant breast cancer cells to offset the possible deleterious effects associated with HHR, since BRCA1 ubiquitylation activity dominated over the effects of disrupting BRCT binding.Citation26 Since HR plays a key role in the response of cancer cells to DNA damaging agents, further investigation of HHR is critical to the understanding of breast and ovarian cancers and for developing treatment strategies. Although selective inhibition of PPIs remains a challenge, exploiting BRCA1 for the development of research tools and cancer therapeutics remains promising.
Note added in proof
While this perspective was under review, yet another report was published on the enigmatic role of the BRCA1 RING domain in HR (Drost R, Bouwman P, Rottenberg S, Boon U, Schut E, Klarenbeek S, et al. BRCA1 RING function is essential for tumor suppression but dispensable for therapy resistance. Cancer Cell 2011; 20:797–809). A hypomorphic activity of the naturally occurring BRCA1-C61G mutant, although unable to prevent tumor development, affected the response to chemotherapy. As opposed to BRCA1 I26A, C61G leads to the disruption of the BRCA1-BARD1 interaction in addition to the loss of E3 ubiquitin-ligase activity. Interestingly, the number of cells with RAD51 foci after irradiation was significantly higher in C61G cells compared with BRCA1-knockout cells derived from tumors, suggesting that increased HR might occur in this model when the BRCA1 RING ubiquitin-ligase is inactivated.
Figures and Tables
Figure 1 Effects of BRCA1 RING and BRCT domain mutations on homologous recombination. (A) The BRCA1 protein consists of an N-terminal RING domain possessing E3 ubiquitin-ligase activity and two tandem C-terminal BRCT repeats that bind phosphoproteins such as Abraxas-RAP80 (BRCA1-A complex). (B) Mutations in the RING domain that disrupt the ubiquitin-ligase activity of BRCA1, but not BARD1 binding, do not affect HR.Citation11,Citation26 (C) Certain mutations in the BRCT binding pocket that disrupt interactions with phosphoproteins, or silencing of the BRCA1 A complex, results in aberrant, hyper-HR (HHR).Citation24–Citation26 (D) Mutations in both the RING and BRCT domains diminish HHR, suggesting a dependence of ubiquitinylation specifically in HHR but not in HR.Citation26 ↑, increase; ↓, decrease; ↔, no change; ?, unknown; *, no effectCitation24,Citation26 or decreasedCitation25 NHEJ.
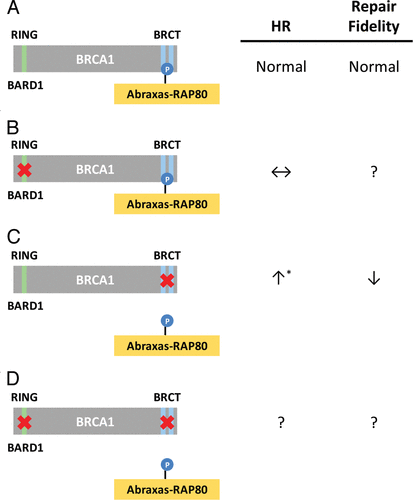
Figure 2 Possible involvement of BRCA1 and PML nuclear bodies in the temporal processing of DSBs during homologous recombination. Top portion (above the arrow) depicts the spatio-temporal process of normal HR whereas the bottom (below the arrow) depicts HHR. BRCA1 is likely playing a critical structural role in the temporal ‘handing-over’ process during HR in which the N- and C-terminal domains, along with the internal domain, communicate and coordinate the various steps and ensure a timely execution and conclusion of the repair process. The anchoring of BRCA1 to repair centers via phosphorylation, ubiquitinylation and sumoylation, and subsequent processing of these post-translational modifications, drives the DSB repair process from the time ionizing radiation (IR) damage occurs until it is repaired. PML-NBs are believed to be ‘factories’ for the (dis)assembly of DNA repair proteins such as BRCA1 and RAD51.Citation28 Silencing of the BRCA1 A complex, or the expression of specific BRCA1 BRCT mutants (e.g., K1702M or M1775R), results in extensive DNA resection at the DSB and ‘stalled’ repair late in the recombination process (with accumulated levels of RPA and RAD51Citation24–Citation26 associated with ssDNA) that seems to occur in close proximity to the PML-NBs.Citation26 Excessive resection might occur because of the inability of BRCA1 BRCT mutants to bind CtIP and recruit MRN in the initial stages of resection thereby allowing other nuclease complexes such as Exo1-BLM-Dna2 uncontrolled access to DNA ends.Citation27 Ub, ubiquitin; Dub, deubiquitinase; ![]()
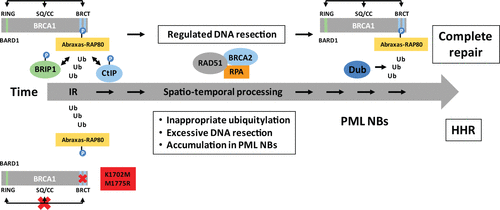
Figure 3 Potential BRCA1 therapeutic targets. BRCA1 with its RING, tandem BRCT, and overlapping SQ cluster and coiled-coil domains are indicated. Although the BRCT domain and, to a lesser extent, the RING domain, have been the focus of inhibitor design, others such as the coiled-coil domain may also be viable targets. The Zn2+ binding sites of the RING domain can be non-specifically inhibited by platinum compounds listed here in order of their affinity for the domain.Citation50 Extensive exploration of phosphopeptides that bind to the BRCT domain has resulted in the peptide shown which has a Ki of 40 nM.Citation51 Structural representations are of the BRCA1 RING and BRCT domains co-crystallized with the RING domain of BARD1 and a BACH1 phosphopeptide, respectively, and were adapted from PDB entry codes 1JM7Citation49 and 1T29,Citation46 using the PyMOL Molecular Graphics System (Schrödinger, LLC).
Acknowledgements
Supported in part by NIH R01NS064593 and R21ES016636 (K.V.), and the Concern Foundation (M.H.).
References
- Venkitaraman AR. Linking the cellular functions of BRCA genes to cancer pathogenesis and treatment. Annu Rev Pathol 2009; 4:461 - 487; PMID: 18954285; http://dx.doi.org/10.1146/annurev.pathol.3.121806.151422
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 1994; 266:66 - 71; PMID: 7545954; http://dx.doi.org/10.1126/science.7545954
- Huen MSY, Sy SMH, Chen J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol 2010; 11:138 - 148; PMID: 20029420; http://dx.doi.org/10.1038/nrm2831
- Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol 2010; 11:196 - 207; PMID: 20177395; http://dx.doi.org/10.1038/nrm2851
- Messick TE, Greenberg RA. The ubiquitin landscape at DNA double-strand breaks. J Cell Biol 2009; 187:319 - 326; PMID: 19948475; http://dx.doi.org/10.1083/jcb.200908074
- Yu X, Fu S, Lai M, Baer R, Chen J. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev 2006; 20:1721 - 1726; PMID: 16818604; http://dx.doi.org/10.1101/gad.1431006
- Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 2007; 316:1198 - 1202; PMID: 17525341; http://dx.doi.org/10.1126/science.1139516
- Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 2007; 316:1202 - 1205; PMID: 17525342; http://dx.doi.org/10.1126/science.1139621
- Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 2007; 316:1194 - 1198; PMID: 17525340; http://dx.doi.org/10.1126/science.1139476
- Zhu Q, Pao GM, Huynh AM, Suh H, Tonnu N, Nederlof PM, et al. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature 2011; 477:179 - 184; PMID: 21901007; http://dx.doi.org/10.1038/nature10371
- Reid LJ, Shakya R, Modi AP, Lokshin M, Cheng JT, Jasin M, et al. E3 ligase activity of BRCA1 is not essential for mammalian cell viability or homology-directed repair of double-strand DNA breaks. Proc Natl Acad Sci USA 2008; 105:20876 - 20881; PMID: 19088202; http://dx.doi.org/10.1073/pnas.0811203106
- Shakya R, Reid LJ, Reczek CR, Cole F, Egli D, Lin CS, et al. BRCA1 tumor suppression depends on BRCT phosphoprotein binding, but not its E3 ligase activity. Science 2011; 334:525 - 528; PMID: 22034435; http://dx.doi.org/10.1126/science.1209909
- Greenberg RA. Cancer. BRCA1, everything but the RING?. Science 2011; 334:459 - 460; PMID: 22034420; http://dx.doi.org/10.1126/science.1214057
- Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science 2003; 302:639 - 642; PMID: 14576433; http://dx.doi.org/10.1126/science.1088753
- Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science 2003; 302:636 - 639; PMID: 14576432; http://dx.doi.org/10.1126/science.1088877
- Liu Z, Wu J, Yu X. CCDC98 targets BRCA1 to DNA damage sites. Nat Struct Mol Biol 2007; 14:716 - 720; PMID: 17643121; http://dx.doi.org/10.1038/nsmb1279
- Yan J, Kim YS, Yang XP, Li LP, Liao G, Xia F, et al. The ubiquitin-interacting motif containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response. Cancer Res 2007; 67:6647 - 6656; PMID: 17621610; http://dx.doi.org/10.1158/0008-5472.CAN-07-0924
- Kim H, Huang J, Chen J. CCDC98 is a BRCA1-BRCT domain-binding protein involved in the DNA damage response. Nat Struct Mol Biol 2007; 14:710 - 715; PMID: 17643122; http://dx.doi.org/10.1038/nsmb1277
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, et al. Human CtIP promotes DNA end resection. Nature 2007; 450:509 - 514; PMID: 17965729; http://dx.doi.org/10.1038/nature06337
- Cantor S, Drapkin R, Zhang F, Lin Y, Han J, Pamidi S, et al. The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations. Proc Natl Acad Sci USA 2004; 101:2357 - 2362; PMID: 14983014; http://dx.doi.org/10.1073/pnas.0308717101
- Kumaraswamy E, Shiekhattar R. Activation of BRCA1/BRCA2-associated helicase BACH1 is required for timely progression through S phase. Mol Cell Biol 2007; 27:6733 - 6741; PMID: 17664283; http://dx.doi.org/10.1128/MCB.00961-07
- Harris JL, Khanna KK. BRCA1 A-complex fine tunes repair functions of BRCA1. Aging (Albany NY) 2011; 3:461 - 463; PMID: 21805697
- Guirouilh-Barbat JK, Wilhelm T, Lopez BS. AKT1/BRCA1 in the control of homologous recombination and genetic stability: the missing link between hereditary and sporadic breast cancers. Oncotarget 2010; 1:691 - 699; PMID: 21321378
- Hu Y, Scully R, Sobhian B, Xie A, Shestakova E, Livingston DM. RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes Dev 2011; 25:685 - 700; PMID: 21406551; http://dx.doi.org/10.1101/gad.2011011
- Coleman KA, Greenberg RA. The BRCA1-RAP80 complex regulates DNA repair mechanism utilization by restricting end resection. J Biol Chem 2011; 286:13669 - 13680; PMID: 21335604; http://dx.doi.org/10.1074/jbc.M110.213728
- Dever SM, Golding SE, Rosenberg E, Adams BR, Idowu MO, Quillin JM, et al. Mutations in the BRCT binding site of BRCA1 result in hyper-recombination. Aging (Albany NY) 2011; 3:515 - 532; PMID: 21666281
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, et al. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev 2011; 25:350 - 362; PMID: 21325134; http://dx.doi.org/10.1101/gad.2003811
- Dellaire G, Bazett-Jones DP. PML nuclear bodies: dynamic sensors of DNA damage and cellular stress. Bioessays 2004; 26:963 - 977; PMID: 15351967; http://dx.doi.org/10.1002/bies.20089
- Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol 2007; 8:1006 - 1016; PMID: 17928811; http://dx.doi.org/10.1038/nrm2277
- Praefcke GJ, Hofmann K, Dohmen RJ. SUMO playing tag with ubiquitin. Trends Biochem Sci 2012; PMID: 22018829; http://dx.doi.org/10.1016/j.tibs.2011.09.002
- Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 2009; 462:935 - 939; PMID: 20016603; http://dx.doi.org/10.1038/nature08657
- Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 2009; 462:886 - 890; PMID: 20016594; http://dx.doi.org/10.1038/nature08593
- Boichuk S, Hu L, Makielski K, Pandolfi PP, Gjoerup OV. Functional connection between Rad51 and PML in homology-directed repair. PLoS One 2011; 6:25814; PMID: 21998700; http://dx.doi.org/10.1371/journal.pone.0025814
- Shao G, Lilli DR, Patterson-Fortin J, Coleman KA, Morrissey DE, Greenberg RA. The Rap80-BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8-Ubc13-dependent ubiquitination events at DNA double strand breaks. Proc Natl Acad Sci USA 2009; 106:3166 - 3171; PMID: 19202061; http://dx.doi.org/10.1073/pnas.0807485106
- Cass I, Baldwin RL, Varkey T, Moslehi R, Narod SA, Karlan BY. Improved survival in women with BRCA-associated ovarian carcinoma. Cancer 2003; 97:2187 - 2195; PMID: 12712470; http://dx.doi.org/10.1002/cncr.11310
- Price M, Monteiro AN. Fine tuning chemotherapy to match BRCA1 status. Biochem Pharmacol 2010; 80:647 - 653; PMID: 20510205; http://dx.doi.org/10.1016/j.bcp.2010.05.015
- Tan DS, Rothermundt C, Thomas K, Bancroft E, Eeles R, Shanley S, et al. “BRCAness” syndrome in ovarian cancer: a case-control study describing the clinical features and outcome of patients with epithelial ovarian cancer associated with BRCA1 and BRCA2 mutations. J Clin Oncol 2008; 26:5530 - 5536; PMID: 18955455; http://dx.doi.org/10.1200/JCO.2008.16.1703
- Hawtin RE, Stockett DE, Wong OK, Lundin C, Helleday T, Fox JA. Homologous recombination repair is essential for repair of vosaroxin-induced DNA double-strand breaks. Oncotarget 2010; 1:606 - 619; PMID: 21317456
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005; 434:917 - 921; PMID: 15829967; http://dx.doi.org/10.1038/nature03445
- Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 2009; 361:123 - 134; PMID: 19553641; http://dx.doi.org/10.1056/NEJMoa0900212
- Saha T, Smulson M, Rosen EM. BRCA1 regulation of base excision repair pathway. Cell Cycle 2010; 9:2471 - 2472; PMID: 20581465
- Dedes KJ, Wilkerson PM, Wetterskog D, Weigelt B, Ashworth A, Reis-Filho JS. Synthetic lethality of PARP inhibition in cancers lacking BRCA1 and BRCA2 mutations. Cell Cycle 2011; 10:1192 - 1199; PMID: 21487248; http://dx.doi.org/10.4161/cc.10.8.15273
- Scully R, Ganesan S, Vlasakova K, Chen J, Socolovsky M, Livingston DM. Genetic analysis of BRCA1 function in a defined tumor cell line. Mol Cell 1999; 4:1093 - 1099; PMID: 10635334; http://dx.doi.org/10.1016/S1097-2765(00)80238-5
- Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov 2004; 3:301 - 317; PMID: 15060526; http://dx.doi.org/10.1038/nrd1343
- Verdine GL, Walensky LD. The challenge of drugging undruggable targets in cancer: lessons learned from targeting BCL-2 family members. Clin Cancer Res 2007; 13:7264 - 7270; PMID: 18094406; http://dx.doi.org/10.1158/1078-0432.CCR-07-2184
- Shiozaki EN, Gu L, Yan N, Shi Y. Structure of the BRCT repeats of BRCA1 bound to a BACH1 phosphopeptide: implications for signaling. Mol Cell 2004; 14:405 - 412; PMID: 15125843; http://dx.doi.org/10.1016/S1097-2765(04)00238-2
- Clapperton JA, Manke IA, Lowery DM, Ho T, Haire LF, Yaffe MB, et al. Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nat Struct Mol Biol 2004; 11:512 - 518; PMID: 15133502; http://dx.doi.org/10.1038/nsmb775
- Williams RS, Lee MS, Hau DD, Glover JN. Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1. Nat Struct Mol Biol 2004; 11:519 - 525; PMID: 15133503; http://dx.doi.org/10.1038/nsmb776
- Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat Struct Biol 2001; 8:833 - 837; PMID: 11573085; http://dx.doi.org/10.1038/nsb1001-833
- Atipairin A, Canyuk B, Ratanaphan A. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by the platinum-based anticancer drugs. Breast Cancer Res Treat 2011; 126:203 - 209; PMID: 20878461; http://dx.doi.org/10.1007/s10549-010-1182-7
- Yuan Z, Kumar EA, Campbell SJ, Palermo NY, Kizhake S, Mark Glover JN, et al. Exploiting the P-1 pocket of BRCT domains toward a structure guided inhibitor design. ACS Med Chem Lett 2011; 2:764 - 767; PMID: 22046493; http://dx.doi.org/10.1021/ml200147a
- Brzovic PS, Keeffe JR, Nishikawa H, Miyamoto K, Fox D 3rd, Fukuda M, et al. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc Natl Acad Sci USA 2003; 100:5646 - 5651; PMID: 12732733; http://dx.doi.org/10.1073/pnas.0836054100
- Ruffner H, Joazeiro CA, Hemmati D, Hunter T, Verma IM. Cancer-predisposing mutations within the RING domain of BRCA1: loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proc Natl Acad Sci USA 2001; 98:5134 - 5139; PMID: 11320250; http://dx.doi.org/10.1073/pnas.081068398
- Jiang J, Yang ES, Jiang G, Nowsheen S, Wang H, Wang T, et al. p53-dependent BRCA1 nuclear export controls cellular susceptibility to DNA damage. Cancer Res 2011; 71:5546 - 5557; PMID: 21742769; http://dx.doi.org/10.1158/0008-5472.CAN-10-3423
- Wild CT, Shugars DC, Greenwell TK, McDanal CB, Matthews TJ. Peptides corresponding to a predictive α-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc Natl Acad Sci USA 1994; 91:9770 - 974; PMID: 7937889; http://dx.doi.org/10.1073/pnas.91.21.9770
- Brzovic PS, Meza JE, King MC, Klevit RE. BRCA1 RING domain cancer-predisposing mutations. Structural consequences and effects on protein-protein interactions.. J Biol Chem 2001; 276:41399 - 41406; PMID: 11526114; http://dx.doi.org/10.1074/jbc.M106551200
- Almaraz E, de Paula QA, Liu Q, Reibenspies JH, Darensbourg MY, Farrell NP. Thiolate bridging and metal exchange in adducts of a zinc finger model and Pt(II) complexes: biomimetic studies of protein/Pt/DNA interactions. J Am Chem Soc 2008; 130:6272 - 6280; PMID: 18422317; http://dx.doi.org/10.1021/ja711254q
- Atipairin A, Canyuk B, Ratanaphan A. Cisplatin affects the conformation of apo form, not holo form, of BRCA1 RING finger domain and confers thermal stability. Chem Biodivers 2010; 7:1949 - 1967; PMID: 20730959; http://dx.doi.org/10.1002/cbdv.200900308
- Leung CC, Glover JN. BRCT domains: easy as one, two, three. Cell Cycle 2011; 10:2461 - 2470; PMID: 21734457; http://dx.doi.org/10.4161/cc.10.15.16312
- Rodriguez M, Yu X, Chen J, Songyang Z. Phosphopeptide binding specificities of BRCA1 COOH-terminal (BRCT) domains. J Biol Chem 2003; 278:52914 - 52918; PMID: 14578343; http://dx.doi.org/10.1074/jbc.C300407200
- Lokesh GL, Muralidhara BK, Negi SS, Natarajan A. Thermodynamics of phosphopeptide tethering to BRCT: the structural minima for inhibitor design. J Am Chem Soc 2007; 129:10658 - 10659; PMID: 17685618; http://dx.doi.org/10.1021/ja0739178
- Yuan Z, Kumar EA, Kizhake S, Natarajan A. Structure-activity relationship studies to probe the phosphoprotein binding site on the carboxy terminal domains of the breast cancer susceptibility gene 1. J Med Chem 2011; 54:4264 - 4268; PMID: 21574625; http://dx.doi.org/10.1021/jm1016413
- Lokesh GL, Rachamallu A, Kumar GDK, Natarajan A. High-throughput fluorescence polarization assay to identify small molecule inhibitors of BRCT domains of breast cancer gene 1. Anal Biochem 2006; 352:135 - 141; PMID: 16500609; http://dx.doi.org/10.1016/j.ab.2006.01.025
- Simeonov A, Yasgar A, Jadhav A, Lokesh GL, Klumpp C, Michael S, et al. Dual-fluorophore quantitative high-throughput screen for inhibitors of BRCT-phosphoprotein interaction. Anal Biochem 2008; 375:60 - 70; PMID: 18158907; http://dx.doi.org/10.1016/j.ab.2007.11.039
- Hartman MC, Josephson K, Szostak JW. Enzymatic aminoacylation of tRNA with unnatural amino acids. Proc Natl Acad Sci USA 2006; 103:4356 - 4361; PMID: 16537388; http://dx.doi.org/10.1073/pnas.0509219103
- Ma Z, Hartman MC. In vitro selection of unnatural cyclic peptide libraries via mRNA display. Methods Mol Biol 2012; 805:367 - 390; PMID: 22094817; http://dx.doi.org/10.1007/978-1-61779-379-0_21
- Gatei M, Scott SP, Filippovitch I, Soronika N, Lavin MF, Weber B, et al. Role for ATM in DNA damage-induced phosphorylation of BRCA1. Cancer Res 2000; 60:3299 - 3304; PMID: 10866324
- Gatei M, Zhou BB, Hobson K, Scott S, Young D, Khanna KK. Ataxia telangiectasia mutated (ATM) kinase and ATM and Rad3 related kinase mediate phosphorylation of Brca1 at distinct and overlapping sites. In vivo assessment using phospho-specific antibodies. J Biol Chem 2001; 276:17276 - 17280; PMID: 11278964; http://dx.doi.org/10.1074/jbc.M011681200
- Xu B, O'Donnell AH, Kim ST, Kastan MB. Phosphorylation of serine 1387 in Brca1 is specifically required for the Atm-mediated S-phase checkpoint after ionizing irradiation. Cancer Res 2002; 62:4588 - 4591; PMID: 12183412
- Cortez D, Wang Y, Qin J, Elledge SJ. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science 1999; 286:1162 - 1166; PMID: 10550055; http://dx.doi.org/10.1126/science.286.5442.1162
- Sy SM, Huen MS, Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci USA 2009; 106:7155 - 7160; PMID: 19369211; http://dx.doi.org/10.1073/pnas.0811159106
- Zhang F, Ma J, Wu J, Ye L, Cai H, Xia B, et al. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol 2009; 19:524 - 529; PMID: 19268590; http://dx.doi.org/10.1016/j.cub.2009.02.018
- Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell 2006; 22:719 - 729; PMID: 16793542; http://dx.doi.org/10.1016/j.molcel.2006.05.022
- Sy SM, Huen MS, Chen J. MRG15 is a novel PALB2-interacting factor involved in homologous recombination. J Biol Chem 2009; 284:21127 - 21131; PMID: 19553677; http://dx.doi.org/10.1074/jbc.C109.023937
- Kritzer JA. Stapled peptides: Magic bullets in nature's arsenal. Nat Chem Biol 2010; 6:566 - 567; PMID: 20644540; http://dx.doi.org/10.1038/nchembio.407
- Velkova A, Carvalho MA, Johnson JO, Tavtigian SV, Monteiro AN. Identification of Filamin A as a BRCA1-interacting protein required for efficient DNA repair. Cell Cycle 2010; 9:1421 - 1433; PMID: 20305393; http://dx.doi.org/10.4161/cc.9.7.11256
- Weaver JM, Edwards PA. Targeted next-generation sequencing for routine clinical screening of mutations. Genome Med 2011; 3:58; PMID: 21910929; http://dx.doi.org/10.1186/gm274
- Bellosillo B, Tusquets I. Pitfalls and caveats in BRCA sequencing. Ultrastruct Pathol 2006; 30:229 - 235; PMID: 16825125; http://dx.doi.org/10.1080/01913120500521281
- Rowling PJ, Cook R, Itzhaki LS. Toward classification of BRCA1 missense variants using a biophysical approach. J Biol Chem 2010; 285:20080 - 20087; PMID: 20378548; http://dx.doi.org/10.1074/jbc.M109.088922
- Lee MS, Green R, Marsillac SM, Coquelle N, Williams RS, Yeung T, et al. Comprehensive analysis of missense variations in the BRCT domain of BRCA1 by structural and functional assays. Cancer Res 2010; 70:4880 - 4890; PMID: 20516115; http://dx.doi.org/10.1158/0008-5472.CAN-09-4563