Abstract
Comment on: Takeda K, et al. Sci Signal 2012; 5:2
Chemotaxing eukaryotic cells can detect very shallow gradients in which the concentration difference between the front and the back of the cells is less than ∼2%.Citation1 Moreover, these cells are able to determine the direction of the gradient in ambient concentrations that span several orders of magnitude.Citation1 Although multiple mechanisms for this accurate gradient sensing have been proposed, it is believed that adaptation, during which the output returns to a fixed base-amount following a change in the input, is crucial in regulating eukaryotic chemotaxis.
To determine the adaptive properties of a signaling chemotactic pathway, we measured the response of the genetically tractable model system Dictyostelium discoideum to abrupt changes in uniform chemoattractant concentrations.Citation2 These concentration modifications were applied using a microfluidic device that was able to change concentration in <1 sec. We focused on the response of Ras, a protein that is immediately downstream from the G protein-coupled chemoattractant receptors and activates a range of downstream effectors. Ras proteins are activated by RasGEFs (guanine nucleotide exchange factors), which exchange Ras-bound GDP for GTP and are inactivated by a slow, intrinsic GTPase activity that can be stimulated >103 fold by RasGAPs (GTPase-activating proteins). The dynamics of the changes in the levels of Ras-GTP in response to chemoattractant stimulation was measured using the fluorescent reporter RBD-GFP.
In the absence of a stimulus, RBD-GFP is distributed uniformly within the cytosol. Following a sudden increase in the chemoattractant concentration, RBD-GFP translocates rapidly to the cell membrane by binding to Ras-GTP, followed by a slower return to the cytosol. Quantifying the dynamics of the reporter revealed that RBD-GFP returned to its pre-stimulus level after ∼35 s, indicating an adaptive response. This adaptive response was observed for concentration increases ranging from 0.1 nM to 1 µM, demonstrating that Ras-GTP adaptation was near perfect over a wide range of stimuli. In addition, we found a similar adaptive response for sudden decreases in concentration, and discovered that the time to reach the maximal response decreased as the size of the stimulus increased.
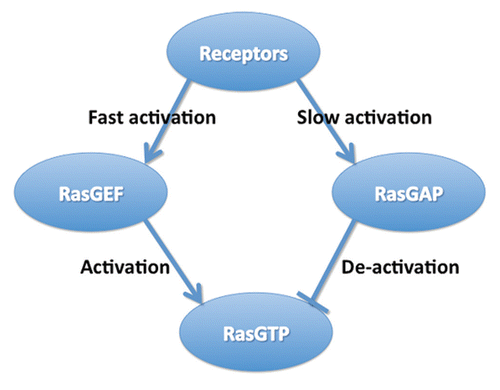
We then simulated the chemotactic pathway using a mathematical model for adaptation that contained only Ras-GTP, RasGEF and RasGAP. Previous mathematical analysis has shown that only two topologies containing three elements are able to achieve perfect adaption.Citation3,Citation4 One of these topologies, the integral control topology, contains a negative feedback loop and is the adaptive mechanism employed in bacterial chemotaxis and some other biological systems.Citation5–Citation7 The second possible topology does not contain feedback loops and has not previously been identified in any biological system analyzed to date. In this incoherent feedforward topology, shown in , both the RasGEF and the RasGAP are activated by the chemoattractant signal acting through the receptors, with a faster activation of RasGEF leading to a transient increase of RasGTP. When applied to our chemotactic pathway, we found that the integral control mechanism is not able to reproduce the experimental data. Specifically, the time scales of reaching the maximum response and the subsequent return to basal levels increase significantly in the integral control mechanism. The incoherent feedforward topology, on the other hand, is able to accurately describe the experimental results, suggesting that adaptation in the chemotactic pathway is achieved via a feedforward pathway and not through negative feedback loops.
The topology of our network is consistent with the local excitation, global inhibition (LEGI) model for gradient sensing.Citation8 Central in this model is the proportional activation of an intracellular membrane-bound activator and a diffuse inhibitor throughout the cell. Our model suggests that the activator RasGEF is the local, membrane-bound component, whereas the inhibitor RasGAP is the diffuse cytosolic component. A RasGAP, DdNF1, having these properties has been previously identified in reference Citation9. As expected, loss of DdNF1 leads to highly extended adaptation of Ras-GTP, as measured using RBD-GFP, leading to aberrant chemotaxis. To further explore the role of this eukaryotic pathway in gradient sensing, it will be necessary to quantify the Ras response in cells exposed to rapidly established gradients.Citation10 Furthermore, future work should also focus on the long-time response following the adaptive phase. During this phase, cells form membrane extensions that are closely correlated with membrane areas of increased concentration of activated Ras (“patches”).Citation11 We expect that the combination of quantitative experiments with modeling, as employed in the adaptation study,Citation2 will shed light on the mechanisms that underlie eukaryotic chemotaxis ().
Figures and Tables
Figure 1 A cartoon representation of the incoherent feedforward network topology capable of accurately reproducing the experimental results. The chemoattractant signal is transmitted to the chemotactic pathway via the binding of ligands to the receptors. These receptors activate both the Ras activator (RasGEF) and Ras de-activator (RasGAP) in a linear fashion, ensuring perfect adaptation. A measurable increase in activated Ras can be accomplished by making the RasGEF activation faster than the RasGAP activation.
Acknowledgments
This work was supported by the US National Institutes of Health (PO1 GM078586)
Comment on: Takeda K, et al. Sci Signal 2012; 5:2; PMID: 22215733; http://dx.doi.org/10.1126/scisignal.2002413
References
- Song L, et al. Eur J Cell Biol 2006; 85:981 - 989; PMID: 16529846; http://dx.doi.org/10.1016/j.ejcb.2006.01.012
- Takeda K, et al. Sci Signal 2012; 5:2; PMID: 22215733; http://dx.doi.org/10.1126/scisignal.2002413
- Ma W, et al. Cell 2009; 138:760 - 773; PMID: 19703401; http://dx.doi.org/10.1016/j.cell.2009.06.013
- Behar M, et al. Biophys J 2007; 93:806 - 821; PMID: 17513354; http://dx.doi.org/10.1529/biophysj.107.107516
- Muzzey D, et al. Cell 2009; 138:160 - 171; PMID: 19596242; http://dx.doi.org/10.1016/j.cell.2009.04.047
- Barkai N, et al. Nature 1997; 387:913 - 917; PMID: 9202124; http://dx.doi.org/10.1038/43199
- El-Samad H, et al. J Theor Biol 2002; 214:17 - 29; PMID: 11786029; http://dx.doi.org/10.1006/jtbi.2001.2422
- Parent CA, et al. Science 1999; 284:765 - 770; PMID: 10221901; http://dx.doi.org/10.1126/science.284.5415.765
- Zhang S, et al. Curr Biol 2008; 18:1587 - 1593; PMID: 18948008; http://dx.doi.org/10.1016/j.cub.2008.08.069
- Sasaki AT, et al. J Cell Biol 2004; 167:505 - 518; PMID: 15534002; http://dx.doi.org/10.1083/jcb.200406177
- Hecht I, et al. PLoS Comput Biol 2011; 7:1002044; PMID: 21738453; http://dx.doi.org/10.1371/journal.pcbi.1002044