Abstract
Macroautophagy is a self-cannibalistic process that enables cells to adapt to various stresses and maintain energy homeostasis. Additionally, autophagy is an important route for turnover of misfolded proteins and damaged organelles, with important implications in cancer, neurodegenerative diseases and aging. Resveratrol and spermidine are able to induce autophagy by affecting deacetylases and acetylases, respectively, and have been found to extend the life-span of model organisms. With the aim to reveal the signaling networks involved in this drug-induced autophagic response, we quantified resveratrol and spermidine-induced changes in the phosphoproteome using SILAC and mass spectrometry. The data were subsequently analyzed using the NetworKIN algorithm to extract key features of the autophagy-responsive kinase-substrate network. We found that two distinct sequence motifs were highly responsive to resveratrol and spermidine and that key proteins modulating the acetylation, phosphorylation, methylation and ubiquitination status were affected by changes in phosphorylation during the autophagic response. Essential parts of the apoptotic signaling network were subjected to post-translational modifications during the drug-induced autophagy response, suggesting potential crosstalk and balancing between autophagy and apoptosis. Additionally, we predicted cellular signaling networks affected by resveratrol and spermidine using a computational framework. Altogether, these results point to a profound crosstalk between distinct networks of post-translational modifications and provide a resource for future analysis of autophagy and cell death.
Introduction
Macroautophagy is a cellular process of self-cannibalism, in which portions of the cytoplasm are sequestered in autophagosomes that then fuse with lysosomes to form autolysosomes, where the luminal content is digested. It is widely known that macroautophagy (which we refer to as “autophagy”) is heavily involved in cellular energy and metabolic homeostasis. Dysfunctional autophagy is involved in multiple human diseases, including cancer and neurodegeneration.Citation1 A normal autophagic capacity has been associated with longevity via caloric restriction, inhibition of TOR with rapamycin and the activation of sirtuins, which constitutes a family of non-histone and histone deacetylases.Citation2,Citation3 Sirtuin 1 (Sirt1) is known to be activated by the small-molecule drug resveratrol (a polyphenol) found in grapes and red wine. Resveratrol can prolong the lifespan of model organisms through the activation of Sirt1, resulting in the stimulation of autophagy,Citation4 which subsequently initiates several signaling events. In a recent study, it was shown that resveratrol has profound effects on the transcriptional network leading to tissue-specific induction of gene expression of generic transcription factors such as PDX1 and HNF1 in pancreatic β cells and FOXO1 in liver cells.Citation5 In pancreatic β cells, the downstream effect was changes in metabolic pathways via glucokinase and Glut2.Citation6 The beneficial effect on longevity is observed in various organisms, and recently it has been shown that there is a significant overlap of genes affected by resveratrol in both invertebrates and mammals, and that it, to a significant degree, mimics caloric restriction.Citation7 A model of the resveratrol-dependent network has recently been suggested by Park et al. involving signal mediators such as PKA, SIRT1, CamKII and AMPK.Citation8 Additionally, we have previously shown that the acetylase inhibitor spermidine (a cationic polyamine) is able to increase longevity and to activate the autophagic response as well, but independent of Sirt1, in multiple model organisms including yeast, nematodes, flies and mice.Citation9-Citation11 Spermidine affects several key processes in the cells including cell proliferation, gene regulation, apoptosis and autophagy by binding to negatively charged molecules, such as DNA, RNA and proteins, by affecting the acetylation status of proteins and by entering metabolic pathways.Citation12 Despite the fact that resveratrol and spermidine increase longevity via different pathways, we have shown recently that the two drugs converge in their mode of action by similarly affecting the human acetylprotome via deacetylase-activating and acetylase-inactivating effects, respectively.Citation13
Recently, a number of mass spectrometry-based proteomic studies of the autophagic system have been reported in references Citation14–Citation17. However, until now, no large-scale study has been conducted with the aim of deciphering the phosphorylation-dependent autophagic response induced by the longevity-related drugs resveratrol and spermidine. We hypothesize that co-regulatory events following cell stimulation by both drugs could reveal potential generic features of the autophagic response. To follow this idea, we performed a large-scale experiment where cells were cultured in medium containing amino acids of different isotopic composition (SILAC),Citation18 followed by phosphopeptide analysis using high-resolution mass spectrometry. The phoshosite-specific data were analyzed by network prediction algorithms and bioinformatic tools to obtain functional and contextual information about autophagy-responsive proteins. This analysis revealed that > 800 phosphorylation sites were co-regulated in response to resveratrol and spermidine treatment, and that dephosphorylation events were more frequent than phosphorylation. Additionally, we show that distinct kinase recognition sequence motifs were found to be subject to co-regulation, mostly RxxS motifs that were phosphorylated and SP motifs that were dephosphorylated. Such SP motifs were predicted to be dephosphorylated as a result of CDK2 inactivation and/or as a result of phosphatase activation. Finally, the analysis revealed that networks of proteins involved in multiple distinct posttranslational modifications were affected by phosphorylation-dependent signaling, pointing to extensive signaling crosstalk that involves distinct types of posttranslational modifications.
Results and Discussion
Identification and quantitation of regulated phosphorylation sites.
To study the activation of autophagy by resveratrol and spermidine by mass spectrometry-based phosphoproteomics, human colorectal cancer HCT116 cells were cultured in the presence of three distinct isotope-labeled amino acids. The SILAC-labeled cells were then either left untreated or treated with resveratrol or spermidine for 2 h for optimal autophagy induction. Quantitative high-resolution mass spectrometric analysis of phosphopeptides derived from these cells resulted in the identification of 4,310 proteins and 6,809 phosphorylation sites (peptide and protein false discovery rate < 0.01) (Figs. S1 and S2). Among the 5,067 sites assigned with high localization scores, 1,871 were regulated > 2-fold after at least one treatment (which is a conservative regulation criterion). For further analysis, a site was classified as regulated by one treatment if the ratio was changed by a ratio of at least 2.0-fold compared with the untreated control or if it was changed by a ratio of at least 1.75-fold when the other treatment was changed by a ratio of 2.0-fold, thus providing an example of correlated regulation.
Although the overall phosphosite ratios correlated well between the two treatments (R = 0.63; Pearson correlation coefficient), distinct regions of differential regulation were observed (Fig. S3C). In particular, more than 600 dephosphorylation events were detected for both of the drug-induced autophagy responses in a correlated manner, whereas more than 550 specific dephosphorylation sites were found to be regulated exclusively following spermidine treatment (Fig. S3D). Moreover, approximately 200 phosphorylation events occurred in a correlated fashion in response to both spermidine and resveratrol. Again, more regulatory phosphorylation events (approximately 200) were identified exclusively upon spermidine treatment. Thus, although part of the phosphorylation-dependent responses to these two autophagy inducers were different, a core set of > 600 dephosphorylation and > 200 phosphorylation events were found to be shared and hence might constitute generic characteristics of the autophagy response. Strikingly, it turned out that the signaling power difference (ratio distribution difference) for the co-dephosphorylation events was significantly lower (p < 0.05; Wilcoxon test) than that of the co-phosphorylation events (Fig. S3E), suggesting that both agents must activate a common set of phosphatases or inactivate a common set of kinases.
Two distinct kinase motifs point at signal hubs in the autophagic response.
Several essential regulated proteins known to be involved in autophagy were found to be phosphorylated in response to resveratrol and spermidine (Table S1). This applies, for instance, to SIRT1, the target of resveratrol, p62/SQSTM1, an adaptor that facilitates autophagy of ubiquitinated proteins, and the elongation factor EF2α, where phosphorylation on T57 is essential for the induction of autophagy.Citation19 To visualize common and differential effects of resveratrol and spermidine on the phosphoproteome, hierarchical clustering of all regulated sites was performed, followed by cluster-specific kinase motif enrichment testing using the MotifX algorithm.Citation20 Only phosphorylation sites with high confidence localization assignment (localization probability > 0.9 Citation21) (; Table S1) were used for this analysis. Strikingly, two highly significant motifs were found among the co-regulated sites (p < 0.000001; binomial testing against all identified high-confidence sites). The RxxS motif was observed for 50% of the co-phosphorylated serine residues, and the SP motif was observed for 79% of the co-dephosphorylated serine residues (71% of the exclusively spermidine-responsive serine residues were located within an SP motif as well). These data suggest a common set of autophagy-relevant “master kinases,” “master-phosphatases” and/or “master kinase inactivators.” It should be noted that the same two motifs were enriched among regulated phosphorylation sites in yeast cells exposed to fatty acid starvation (Pultz D, et al. In press), suggesting that they reflect a generic, evolutionary conserved response to cellular stress.
Figure 1. Determination of kinase motifs within phosphorylation sites identified from cells stimulated with resveratrol and spermidine. Hierarchical clustering of all responsive sites (sites regulated more than 2-fold upon at least one of the treatments). Black color indicates “non responsive” upon the given treatment. Cluster-specific significant linear motifs related to very high-confidence phosphorylation sites (localization probability > 0.9) are shown in the margin (requirement: at least 20 occurrences, p < 1E-6; Binomial testing).
With the scope of identifying autophagy-relevant kinases, the NetworKIN algorithmCitation22 was subsequently applied, and hypergeometric kinase enrichment testing was performed using a Benjamini-Hochberg adjusted p-value < 0.05 as a criterion for significance (). The predicted kinases used for these statistical tests were pre-filtered using a NetworKIN score of at least 3 and predictions for each site having a NetworKIN score above 90% of the highest scoring (the NetworKIN score indicates how much the STRING scoreCitation23 amplifies the initial NetPhorest probability scoreCitation24). This strategy led to the removal of a majority of likely false-positive predictions.
Figure 2. (A) Distribution of kinase motifs associated with phosphorylation sites on selected regulated substrates. High-confidence phosphorylation sites (localization probability > 0.9) were analyzed using the NetworKIN algorithm. Fisher’s exact test and Benjamini-Hochberg adjustment were performed on the kinase-substrate predictions. The color code indicates the BH-adjusted P-value. Selected weblogos are shown in the margin. (B) Cluster-specific distribution of kinase-prediction frequencies corresponding to the enriched kinases shown in (A). (C) Cell cycle arrest assay on cells treated with resveratrol and spermidine at three time points as indicated.
We observed a highly significant enrichment in CDK2 substrates among the co-dephosphorylated sites, of which the vast majority were located within the SP/TP motif. This correlated with dephosphorylation of the CDK2 activator CDC25B on S374 in response to both resveratrol and spermidine. In support of CDK2 inactivation, we observed a significant G1-arrest (where CDK2 is a key-regulator) in HCT116 cells treated with resveratrol (). However, no predominant G1-arrest was observed for spermidine-treated cells, suggesting that the co-regulated SP sites are dephosphorylated by a different mechanism. One explanation might be that a group of phosphatases are activated by spermidine treatment without affecting CDK2, leading to protein dephosphorylation without cell cycle arrest, whereas CDK2 and other kinases might be inactivated by resveratrol treatment, leading to spontaneous/background dephosphorylation with cell cycle arrest. The hypothesis that phosphatases are activated by spermidine treatment is further supported by a significantly higher dephosphorylation power in spermidine-treated cells among co-dephosphorylated sites (Fig. S3E), as well as the identification of > 550 dephosphorylation events (207 of which are SP sites) that occurred exclusively in spermidine-treated cells. In the phosphoproteomic screen, we identified two S/T phosphatases that were selectively phosphorylated following spermidine treatment. These phosphatases, PPP1R14B (phosphorylated on T195 within the regulatory subunit) and PPM1F (phosphorylated on T448), suggest that they might participate in the dephosphorylation of the SP site.
NetworKIN predicted four kinases (PAK4, PAK7, DMPK and CLK1) to be significantly associated with co-phosphorylation on serine residues located in the RxxS motif (). The sites predicted to be phosphorylated by PAK4 and PAK7 are more or less the same, all having an arginine (R) in position 2 from the central serine or threonine and, in > 40% of the cases, an additional R in the -3 position. Sites predicted to be phosphorylated by DMPK (enrichment score > 3) were all serine residues and in almost all cases with arginine located in the -3 position. Thus, DMPK constitutes a master kinase candidate that phosphorylates the generic stress motif RxxS. Interestingly, the DMPK isoform A has been reported to induce autophagy and to stimulate the mitochondrial pathway of apoptosis.Citation25 Thus, the NetworKIN-derived prediction is supported by previously reported experimental evidence, and we propose that this kinase may participate in the spermidine- and resveratrol-induced autophagic response. CAM kinase-like kinase CLK1 were predicted to phoshorylate serine residues located close to an R in the -3 position in approximately 60% of the cases (). Although CLK1 has not yet been linked to autophagy, it is known to preferentially phosphorylate SR-rich proteins that often are involved in RNA processing. We thus propose that CLK1 might be involved in the regulation of RNA processing during the autophagy response.
Finally, a significant proportion of substrates was predicted to be phosphorylated by CK2 specifically during spermidine treatment. This is supported by experiments that show that CK2 is regulated by polyamines, including spermidine, that bind physically to a defined polyamine binding domain.Citation26 Indeed, it has been suggested that CK2 is able to biochemically sense the relative polyamine levels in the cell and initiate appropriate responses.Citation27
Deciphering pathways affected by the resveratrol-
and spermidine-responsive phosphoproteome. To identify biological networks associated with the resveratrol- and spermidine-modulated phosphoproteome, a Gene Ontology (GO)Citation28 term enrichment test was performed for groups of proteins with similar changes in phosphorylation using p-values < 0.05 in Benjamini-Hochberg adjusted Fishers Exact test (Fig. S4).
This analysis revealed several transport proteins to be co-dephosphorylated with similar signal power (Fig. S5). These proteins included p62/SQSTM1 (dephosphorylated on S366 and known to be involved in autophagosome formation),Citation29 VPS26 (dephosphorylated on S302 and is associated with lysosomal transport)Citation30 and EPS8 (dephosphorylated > 4-fold on S625 and T629 and is recruited to lysosomes during autophagy in cancer cells).Citation31 Strikingly, three nuclear pore complexes (NUP214, NUP160 and NUP98) were dephosphorylated, suggesting that the autophagic response may affect the nuclear transport of macromolecules. Even though some of the proteins were (de)phosphorylated in a differential fashion in response to resveratrol and spermidine, the cellular function of differentially regulated proteins was still related. In particular, proteins that were (de) phosphorylated by one or the other treatment were significantly associated with microtubule-based processes, intracellular transport and the cell cycle (p < 0.001; Fishers Exact test). This is consistent with prior knowledge that transport processes along microtubules are pivotal for autophagy.Citation32,Citation33
A similar analysis of (de)phosphorylated proteins using the KEGG-pathway enrichment test (Fisher test, BH adjustment and enrichment score calculation by -log(P), Fig. S6A) yielded results that were consistent with the GO-term analysis (Fig. S4). Pathways associated with focal adhesion, tight junctions, actin skeleton regulation and insulin signaling were found to be co-regulated by resveratrol and spermidine (p < 0.05; Fishers Exact test). Many of the proteins associated with the enriched pathways were indeed (de)phosphorylated in response to both treatments (Fig. S7).
Next, we used the STRING algorithmCitation23 to model an association network based on all co-regulated proteins (co-phosphorylated and co-dephosphorylated) plus one extended node layer, irrespective of its identification in the proteomic screen (high confidence score, > 0.7, Fig. S6C). KEGG pathway enrichment testing of the modeled network (Fig. S6B) led to the identification of additional enriched pathways, including the apoptosis pathway. Thus, even though the modified proteins themselves are not enriched in pro- and anti-apoptotic proteins, they are associated with a significant number of apoptosis-relevant proteins within the network, suggesting some level of crosstalk between autophagy and apoptosis.
Identification of essential regulated phosphoproteins of the autophagosomal network.
Behrends et al. recently performed a mass spectrometry-based interaction study on the autophagosomal interaction network (AIN) in cells exposed to the classical autophagy inducer rapamycin, which targets mTOR and mimics nutrient starvation. We studied the overlap between the resveratrol- and spermidine-responsive phosphoproteome with the AIN subdivided into “autophagy interactome” (AI), “central autophagy interactome” (CAI) and “autophagy core network” (ACN). Moreover, we evaluated the relationship between the resveratrol- and spermidine-modulated phosphoproteome and a series of putative regulators of autophagy (PRA) identified by an siRNA screen.Citation34
More than 30% of all proteins with phosphorylation sites regulated in response to spermidine or resveratrol were part of the AI, CAI or ACN (), while rather few (< 1.3%) mapped to the PRA. In particular, > 150 of all co-regulated proteins mapped to the AI and > 30 proteins to the CAI (). Three co-regulated phosphoproteins belonged to the ACN. These proteins comprise SQSTM1 and TRAF2 (both dephosphorylated) and PIK3C2A [phosphorylated > 4-fold on tyrosine residues Y1436 and Y1444 (localization probability > 0.9)]. We hypothesize that the co-regulated sites associated with CAI and CAN constitute a high-confidence autophagosomal core phosphorylation network (ACPN). Hence, we applied the NetworKIN algorithmCitation22 to these sites (with a cut-off of > 0.9 applied to the confidence localization probability) to predict kinases acting on the ACPN (). We found for example that the yeast kinase homolog TJP was modulated on four sites (S201, S205, S275 and Y1149) in response to both resveratrol and spermidine, suggesting that this kinase might play a key role in signal propagation during the generic autophagy response. Though little is known about this kinase, TJP has been reported to be involved in tight junction processes and to undergo stress-modulated cytoplasmic-nuclear shuttling.Citation35 NetworKIN predicted the Y1149 residue of TJP to be phosphorylated by a receptor tyrosine kinase and TJP serine residues to be substrates of AKT and CLK kinases. The general translation initiation factor EIF5B was co-phosphorylated in response to resveratrol and spermidine on S113, likely by a CK2 kinase. EIF5B promotes binding of the formylmethionine-tRNA to ribosomes, suggesting that the CK2 kinase family might be involved in translational regulation during the autophagy response. SEC22B, which is known to be involved in vesicle trafficking,Citation36 was dephosphorylated on T140 in response to resveratrol and spermidine.
Figure 3. (A) Mapping of the regulated phosphoproteome to the autophagosomal interaction network (AIN) as reported by Behrends et al.Citation14 The AIN was subdivided into the ‘autophagy interactome’ (AI), the ‘central autophagy interactome’ (CAI) and the ‘autophagy core network’ (ACN). Possible false-positives retrieved fromCitation34 were included for validation. (B) Heatmap of cluster-specific autophagy responsive phosphoproteins mapped to CAI and ACN.
Figure 4. Phosphorylation-dependent signaling network of co-regulated phosphoprotein mapped to the high confidence autophagosomal interaction network from . The NetworKIN algorithm was applied to high confidence sites of this high-confidence autophagy responsive subset of phosphoproteins (localization probability > 0.9).
The autophagy-responsive kinome.
Resveratrol and spermidine affect the acetylproteome through direct effects on acetyltransferases and deacetylases.Citation3 Apparently, these effects have a major impact on the phosphoproteome, as shown here. Therefore, it seems reasonable to argue that both drugs propagate kinase-dependent signals. Indeed, we identified 109 (de)phosphorylated sites within kinases or direct kinase regulators. Among these, 14 were co-phosphorylated, 22 co-dephosphorylated () and 73 differentially regulated (Fig. S8).
Table 1. Co-regulated kinases and kinase regulators
Several of these co-regulated kinases and kinase-regulators are known to be involved in the autophagy response. This applies to TJP (see above), MAPK13 (p38), the p38 pathway activator TAOK1, PIK3C2A and yet another enzyme involved in the phosphoinositol pathway, the PI4K kinase and PI4K2A. In addition, several kinases that have not yet been implicated in autophagy were found to be (de)phosphorylated during the spermidine and resveratrol response. This applies to key proteins from the MAPK-signaling pathway, such as RAF1, EPHA/B4 receptors and EGF-signaling regulator EPS8. Moreover, the microtubule kinase MAST4 (which was dephosphorylated on S1697) might be relevant for autophagy (). Many other kinases were regulated in a drug-specific manner, in line with the existence of drug-specific branches of the kinase-substrate network (; Table S1).
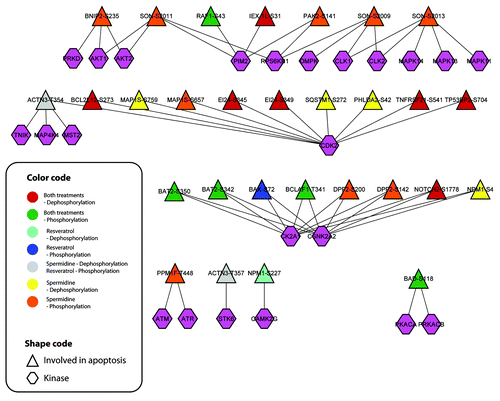
Figure 5. Autophagy responsive phosphorylation networks interfere with networks of posttranslational/posttranscriptional modifiers suggesting extensive crosstalk. The phosphorylation network (localization probability > 0.9) of posttranslational/posttranscriptional modifiers including associated kinases was modeled by the NetworKIN algorithm. The color code indicates the cluster in which the sites are associated and the node shape indicate the class of the posttranslational/posttranscriptional modifier. Responsive kinases and co-regulated phosphoproteins are highlighted at the top and bottom.
Phosphorylation-mediated signaling propagates into other protein modifier networks, indicating signal conversion and crosstalk.
Resveratrol and spermidine led to the regulation of proteins involved in the post-transcriptional modification of proteins with with acetyl and methyl groups and ubiquitin. Intriguingly, SIRT1, the primary target of resveratrol and a major regulator of autophagy and longevity, was found to be co-phosphorylated at T719, probably by a CK2 kinase family member (). Additionally, two histone deacetylases, HDAC1 and HDAC2, were co-dephoshorylated; HDAC1 on S393 and HDAC2 on S488. Two histone acetylases, MYST2 and GTF3C4, were co-dephosphorylated on T88 (a CDK2 motif) and co-phosphorylated on T514 and Y508, respectively. The fact that such key (de)acetylases are subjected to coordinated modulation by phosphorylation suggests an intricate link between the kinome and the networks of (de)acetylating enzymes (). Of note, only few phosphosites on acetyl transferases were affected differentially in response to resveratrol and spermidine, underscoring the convergence of both agents on the regulation of the acetylproteome.
Ubiquitination reactions are known to have a profound impact on autophagy.Citation37 We found ten co-(de)phosphorylated sites on proteins involved in ubiquitin transfer. This applies to two central ubiquitin-ligases, HECTD1 (on S358) and CBL (on S483), several USP-proteins (ubiquitin hydrolases) and the ubiquitin ligase UBE2T (on S184, a site predicted to be phosphorylated by ATM or ATR). In contrast to the acetyl transferases, which were, in general, co-regulated (see above), several ubiquitin modifiers were regulated differentially in response to resveratrol and spermidine.
Although (de)methylases have not previously been associated with autophagy, several were found to be regulated by changes in phosphorylation in response to autophagy inducers. Specifically, we identified co-regulation of two RNA-methylases, FBL (phosphorylated on S124 and S126) and NOP2 (dephosphorylated on T817, S67 and S819, the two latter located in a CDK2 motif), and of the histone-demethylase EZH2 (dephosphorylated on T492 and S371). The functional impact of (de)methylases on the transmission of pro-autophagic signals requires further investigation.
Phosphatases and phosphatase regulators were (de)phosphorylated during the spermidine and resveratrol response in a highly drug-dependent way, correlating with the existence of major differences in the changes of the phosphoproteome induced by resveratrol vs. spermidine. Indeed, spermidine triggered significantly more dephosphorylation reactions than resveratrol (; Fig. S3), and more phosphatases/regulators were (de)phosphorylated in cells exposed to spermidine. Nonetheless, some phosphatases, like the apoptosis-regulating phosphatase 2A inhibitor SET (S7), CDC25B (S374) and PPP1CA (T331), were found to be co-regulated by dephosphorylation (with all three sites located within a CDK2-motif) ().
Crosstalk between autophagy and apoptosis circuits via phosphorylation signaling networks.
Driven by the consideration of crosstalk between apoptosis and autophagy signaling networks, we investigated resveratrol- and spermidine-driven (de)phosphorylaton reactions in apoptosis-relevant proteins using the NetworKIN algorithm. Among cell death-associated proteins, 27 sites were co-regulated (), although there were also drug-specific effects (Fig. S9).
Table 2. Co-regulated apoptosis-related proteins
Among the co-regulated proteins, several were important apoptosis regulators, including BAD, BCLAF1 and RAF1 (all co-phosphorylated). Three distinct kinase families were predicted to cause phosphorylation of the first three proteins: PKA for BAD-S118, CK2 kinase for BCLAF1-T341 and PIM2 for RAF-S43. The involvement of PKA is supported by the recent finding that resveratrol functions as a nonselective phosphodiesterase inhibitor, and resveratrol therefore increases the cellular pool of cAMP,Citation8 which is known to activate PKA by functioning as a second messenger. Spermidine and resveratrol stimulated the coordinated dephosphorylation of DIDO1, DAP, TP53BP2, PDCD2, TNFRSF21 and NOTCH2. Indeed, it has been shown that resveratrol can trigger NOTCH2-dependent apoptosis.Citation38 Among differentially regulated proteins, we identified BAX, which is able to lead to mitochondria membrane permeabilization,Citation39 to be phosphorylated on S72, presumably by CK2, in response to resveratrol but not spermidine. Moreover, the stress-responsive apoptosis signaling activator PAK2 was phosphorylated on S141 in response to spermidine but not resveratrol. We used this information to model a signaling network at the interface between apoptosis and autophagy (). The computational modeling predicts that CK2 is likely important for fine-tuning the autophagy-apoptosis balance due to its impact on multiple co-phosphorylated sites (including BAD-S118) and differentially phosphorylated sites (on BAX and DPF2).
Figure 7. Autophagy responsive phosphorylation networks interfere with the apoptotic network, suggesting a phosphorylation dependent balancing of apoptotic and autophagic cell signal events. The phosphorylation network (localization probability > 0.9) of posttranslational/posttranscriptional modifiers including associated kinases was modeled by means of the NetworKIN algorithm. The color code indicates the cluster to which the sites are associated.
Materials and Methods
Cell culture.
HCT 116 cells were grown for 2 weeks in three different SILAC media (Invitrogen) containing either (1) light isotopes of L-arginine and L-lysine (Arg0/Lys0), (2) L-arginine-13C6 HCl (Euroisotop) and L-lysine 2HCl 4,4,5,5-D4 (Arg6/Lys4; Euroisotop) or (3) L-arginine-13C615N4 HCl and L-lysine 13C615N4 HCl (Arg10/Lys8; Invitrogen) and complemented as reported in Blagoev et al.Citation40 Cells were treated for 2 h with 100 µM spermidine (Arg10/Lys8) or 100 µM resveratrol (Arg6/Lys4) and then lysed and subdivided in nuclear, cytosolic and mitochondrial fractions as previously described in reference Citation41. The precipitation of proteins in the cytosolic fraction was performed using ice-cold (-20°C) acetone (4 times the volume of the sample extract). The samples were then vortexed and placed for 2 h at -20°C. The resulting solution was centrifuged for 10 min at 16,000 g, and the supernatant was removed. The pellet was first washed twice with ice-cold 4:1 acetone/water and subsequently dried using a concentrator (SpeedVac; Thermo Fisher Scientific) for 10–15 min.
Peptide preparation.
Samples prepared for our previous report on the effect of resveratrol and spermidineCitation13 were used for this phosphorylation experiment as well. Briefly, the starting material was composed by three fractions of purified nuclei, mitochondria and cytoplasm from approximately 109 cells. The denaturant 6 M/2 M Urea/Thiourea in 50 mM ammonium bicarbonate were added to all fractions and, specifically, the nuclease benzonase (Merck) was added to the nuclei fraction. All steps were performed at room temperature to avoid carbamoylation of amines. Reduction of cysteines was performed using DTT (5 mM) for 30 min followed by alkylation with iodoacetamide (11 mM) for 20 min in the dark. The proteins were digested 1:100 protease:protein, first with LysC (Wako) for 3.5 h, diluted 4 times with 50 mM ABC and then with trypsin (Promega) over night. The mixture was spun at 10,000 rpm for 10 min, and the supernatant was filtered through a 0.45 µm filter (Millex HV). From each fraction approximately 100 µg digested protein were collected for isoelectric focusing (IEF), used for protein normalization.
Phosphopeptide enrichment and isoelectric focusing.
Phosphopeptides were enriched by TiO2 affinity chromatography, as described in Larsen et al. with minor modifications. Briefly, peptide sample was incubated with TiO2 beads, equilibrated in 50 mg/ml DHB, 80% ACN/1% TFA. After washing beads in 60% ACN/1% TFA, phosphopeptides were eluted on a C8 disc StageTip in 40% ACN/15% NH4OH.
The collected protein digest samples (for protein normalization) were desalted using SepPak C18 purification cartridges and 5% of each enriched eluate was collected for mass spectrometric (MS) analysis prior to IEF fractionation. The remaining part of the eluates were subsequently separated into 12 fractions by IEF using the Agilent 3100 OFFGEL Fractionator (Agilent, G3100) using the protocol described previously in reference Citation43. Gelstrips and amfolyte buffer were purchased from GE Healthcare. The peptides were focused for 20 kVh at a maximum current of 50 µA and maximum power of 200 mW. Ten µl of 10% TFA were added to each fraction and STAGE-tipped as described in reference Citation44, prior to MS analysis.
Mass spectrometric analysis.
The TiO2 column eluates were dried using a Speed Vac and C18 Stage tip purified.Citation45 MS analysis was performed on an LTQ-OrbitrapCitation46 (Thermo Fisher Scientific) connected to an Agilent 1200 nanoflow HPLC system (Agilent) using a nanoelectrospray ion source (Proxeon Biosystems). The peptides were separated by reversed-phase chromatography using a in-house-made fused silica emitter (75 µm ID) packed with Reprosil-Pur C18-AQ 3 µm reversed phase material (Dr. Maisch GmbH). Peptides were loaded in 98% solvent A (0.5% acetic acid) followed by 100 min linear gradient to 50% solvent B (80% acetonitrile, 0.5% acetic acid). Survey full scan MS spectra (range 350–1,800, resolution 60,000@m/z 400) were acquired on an LTQ-Orbitrap XL (Thermo-Fisher Scientific), followed by fragmentation of intense multiply charged ions by multi-stage activation (MSA) with excitation at nominal mass losses of 97.97, 48.99 and 32.66 Da from the precursor m/z.Citation47 Ions selected for MS/MS were placed on a dynamic exclusion list for 45 sec. Real-time internal lock mass recalibration was used during the MS and MS/MS performance.Citation48 All samples used for protein normalization were analyzed on an LTQ FT ULTRA mass spectrometer (Thermo Finnigan), where the five most intense ions from each precursor scan were selected for fragmentation in the LTQ-no real-time lockmass recalibration. Reverse phase chromatography settings were the same as described for the analysis using an Orbitrap mass spectrometer.
Peptide and protein identification and quantitation.
All raw files were processed with MaxQuantv.1.0.13.13Citation49 into centroided data and submitted to database searching with Mascot v.2.2 (Matrix-Science). Pre-processing by MaxQuant was performed to determine charge states, mis-cleavages and SILAC states and to filter the MS/MS spectra, keeping the six most intense peaks within a 100 Da bin. Carbamidomethyl of cysteine was chosen, as fixed modification and acetylation of the protein N terminus, oxidation of methionine, deamidation of NQ, Gln- > pyro-Glu (N-term Q) and phospho (STY) were chosen to be variable modifications. The processed MS/MS spectra were searched against a concatenated target-decoy database of forward and reverse sequences from the IPI database (IPIv.3.69). For the search, Trypsin/P + DP was choosen for the in silico protein digestion, allowing three mis-cleavages. The mass tolerance for the MS spectra acquired in the Orbitrap was set to 7 ppm, whereas the MS/MS tolerance was set to 0.6 Da for the CID MS/MS spectra from the LTQ. Subsequent to the peptide search, protein and peptide identification was performed given an estimated maximal false discovery rate (FDR) of 1% on both the protein and peptide level. For the FDR calculation, posterior error probabilities were calculated based on peptides of at least six amino acids and their corresponding MASCOT-score (minimum 7). For the protein group identification, at least one unique peptide was required. For protein quantification, only unmodified peptides and peptides modified by acetyl (protein N-term), oxidation (M), deamidation (NQ) and Gln- > pyro-Glu (N-term Q) were considered.. If a counterpart to a given phosphorylated peptide is identified, this counter peptide is also not used for protein quantitation. According to the protein group assignment performed by MaxQuant, both razor and unique peptides were used for protein quantitation. A minimum of two ratio counts was required for the protein quantitation. For quantitation of phosphorylated sites, the least modified peptides were used. The ratios for the sites were normalized by the corresponding protein ratios to account for eventual changes in protein abundance. In case a protein ratio was not determined, normalization was done based on a logarithm-transformation algorithm, as described in reference Citation49.
Assessment of cell cycle distribution.
HCT116 cells were incubated in triplicate wells for 4, 8 and 16 h with 100 µm of resveratrol (Sigma) or 100 µm of spermidine (Sigma). For cell cycle analyses, cells were harvested, washed and stained with Hoechst 33342 (10 µg/ml; Invitrogen), followed by an incubation period of 30 min at room temperature. Subsequently, cell cycle distribution was determined by cytofluorometric analysis on a FACScan cytofluorometer (BD Biosciences). Statistical analysis was performed by using the CellQuestTM software (BD Biosciences) upon gating on the events characterized by normal forward scatter and sidescatter parameters ().
Conclusion and Perspectives
Using SILAC and mass spectrometry to decipher the changes in signaling networks in cells exposed to resveratrol and spermidine, we succeeded in uncovering the responsive phosphoproteome to a depth of 1,871 regulated phosphorylation sites, where > 800 were co-regulated. In particular, we found that dephosphorylation events outweighted phosphorylation events and occurred at higher fold changes, suggesting a global trend toward dephosphorylation upon treatment with resveratrol and spermidine. Using bioinformatic approaches, distinct kinase motifs were found to be co-regulated. This applies to RxxS motifs that were phosphorylated and SP motifs that were dephosphorylated. Such SP motifs were predicted to be dephosphorylated as a result of CDK2 inactivation and/or as a result of the activation of an CDK2-antagonistic phosphatase.
To further study the resveratrol- and spermidine-induced autophagy response, we applied systems biology methods, thus applying unbiased statistics to an ensemble of regulatory events with the objective of achieving a global snapshot of dynamic changes in cellular networks. Specifically, we used the versatile probabilistic modeling algorithms STRING and NetworKIN for network reconstruction. These modeling algorithms and bioinformatic tools allowed us to analyze the (de)phosphorylation events affecting five sub-proteomes, namely proteins involved in protein acetylation, methylation, phosphorylation, dephosphorylation and ubiquitination. These analyses suggest that networks of proteins involved in multiple distinct posttranslational modifications intersect in a highly connective fashion, suggesting that acetylation-dependent changes [which are the prime targets of resveratrol and spermidine] are closely linked to (de)phosphorylation-dependent signaling event, pointing to extensive crosstalk between signaling events based on distinct types of posttranslational modifications. We are confident that this study will serve as a resource for the scientific community interested in stress signaling and autophagy.
Additional material
Download Zip (8.2 MB)Acknowledgements
This project was supported by European Community’s Seventh Framework Program (FP7/2007-2013) under grant agreement n° HEALTH-F4-2007-200767 for the collaborative project APO-SYS (J.S.A. and G.K.). M.V.B. was supported by the Syddansk Universitets Forskningsfond and the Danish Ministry of Science, Technology and Innovation by the EliteResearch (“EliteForsk”) award 2010. G.K. was supported by the Ligue Nationale contre le Cancer (Equipes labelisée), Agence Nationale pour la Recherche (ANR), European Commission (ArtForce, ApopTrain, ChemoRes), Fondation pour la Recherche Médicale (FRM), Institut National du Cancer (INCa), Cancéropôle Ile-de-France and the AXA Chair for Longevity Research.
Note
Supplemental materials can be found at:
www.landesbioscience.com/journals/cc/article/20233
References
- Kroemer G, Jäättelä M. Lysosomes and autophagy in cell death control. Nat Rev Cancer 2005; 5:886 - 97; http://dx.doi.org/10.1038/nrc1738; PMID: 16239905
- Dali-Youcef N, Lagouge M, Froelich S, Koehl C, Schoonjans K, Auwerx J. Sirtuins: the ‘magnificent seven’, function, metabolism and longevity. Ann Med 2007; 39:335 - 45; http://dx.doi.org/10.1080/07853890701408194; PMID: 17701476
- Madeo F, Tavernarakis N, Kroemer G. Can autophagy promote longevity?. Nat Cell Biol 2010; 12:842 - 6; http://dx.doi.org/10.1038/ncb0910-842; PMID: 20811357
- Morselli E, Maiuri MC, Markaki M, Megalou E, Pasparaki A, Palikaras K, et al. Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Dis 2010; 1:e10; http://dx.doi.org/10.1038/cddis.2009.8; PMID: 21364612
- Vetterli L, Maechler P. Resveratrol-activated SIRT1 in liver and pancreatic β-cells: a Janus head looking to the same direction of metabolic homeostasis. Aging (Albany NY) 2011; 3:444 - 9; PMID: 21483037
- Vetterli L, Brun T, Giovannoni L, Bosco D, Maechler P. Resveratrol potentiates glucose-stimulated insulin secretion in INS-1E beta-cells and human islets through a SIRT1-dependent mechanism. J Biol Chem 2011; 286:6049 - 60; http://dx.doi.org/10.1074/jbc.M110.176842; PMID: 21163946
- Antosh M, Fox D, Helfand SL, Cooper LN, Neretti N. New comparative genomics approach reveals a conserved health span signature across species. Aging (Albany NY) 2011; 3:576 - 83; PMID: 21775776
- Park SJ, Ahmad F, Philp A, Baar K, Williams T, Luo H, et al. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell 2012; 148:421 - 33; http://dx.doi.org/10.1016/j.cell.2012.01.017; PMID: 22304913
- Morselli E, Galluzzi L, Kepp O, Criollo A, Maiuri MC, Tavernarakis N, et al. Autophagy mediates pharmacological lifespan extension by spermidine and resveratrol. Aging (Albany NY) 2009; 1:961 - 70; PMID: 20157579
- Eisenberg T, Knauer H, Schauer A, Büttner S, Ruckenstuhl C, Carmona-Gutierrez D, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol 2009; 11:1305 - 14; http://dx.doi.org/10.1038/ncb1975; PMID: 19801973
- Madeo F, Eisenberg T, Büttner S, Ruckenstuhl C, Kroemer G. Spermidine: a novel autophagy inducer and longevity elixir. Autophagy 2010; 6:160 - 2; http://dx.doi.org/10.4161/auto.6.1.10600; PMID: 20110777
- Minois N, Carmona-Gutierrez D, Madeo F. Polyamines in aging and disease. Aging (Albany NY) 2011; 3:716 - 32; PMID: 21869457
- Morselli E, Mariño G, Bennetzen MV, Eisenberg T, Megalou E, Schroeder S, et al. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J Cell Biol 2011; 192:615 - 29; http://dx.doi.org/10.1083/jcb.201008167; PMID: 21339330
- Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature 2010; 466:68 - 76; http://dx.doi.org/10.1038/nature09204; PMID: 20562859
- Dengjel J, Kristensen AR, Andersen JS. Ordered bulk degradation via autophagy. Autophagy 2008; 4:1057 - 9; PMID: 18776736
- Kristensen AR, Schandorff S, Høyer-Hansen M, Nielsen MO, Jäättelä M, Dengjel J, et al. Ordered organelle degradation during starvation-induced autophagy. Mol Cell Proteomics 2008; 7:2419 - 28; http://dx.doi.org/10.1074/mcp.M800184-MCP200; PMID: 18687634
- Zimmermann AC, Zarei M, Eiselein S, Dengjel J. Quantitative proteomics for the analysis of spatio-temporal protein dynamics during autophagy. Autophagy 2010; 6:1009 - 16; http://dx.doi.org/10.4161/auto.6.8.12786; PMID: 20603599
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 2002; 1:376 - 86; http://dx.doi.org/10.1074/mcp.M200025-MCP200; PMID: 12118079
- Hait WN, Jin S, Yang JM. A matter of life or death (or both): understanding autophagy in cancer. Clin Cancer Res 2006; 12:1961 - 5; http://dx.doi.org/10.1158/1078-0432.CCR-06-0011; PMID: 16609004
- Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol 2005; 23:1391 - 8; http://dx.doi.org/10.1038/nbt1146; PMID: 16273072
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 2006; 127:635 - 48; http://dx.doi.org/10.1016/j.cell.2006.09.026; PMID: 17081983
- Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jørgensen C, Miron IM, et al. Systematic discovery of in vivo phosphorylation networks. Cell 2007; 129:1415 - 26; http://dx.doi.org/10.1016/j.cell.2007.05.052; PMID: 17570479
- von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, et al. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res 2005; 33:D433 - 7; http://dx.doi.org/10.1093/nar/gki005; PMID: 15608232
- Miller ML, Jensen LJ, Diella F, Jørgensen C, Tinti M, Li L, et al. Linear motif atlas for phosphorylation-dependent signaling. Sci Signal 2008; 1:ra2; http://dx.doi.org/10.1126/scisignal.1159433; PMID: 18765831
- Oude Ophuis RJ, Wijers M, Bennink MB, van de Loo FA, Fransen JA, Wieringa B, et al. A tail-anchored myotonic dystrophy protein kinase isoform induces perinuclear clustering of mitochondria, autophagy, and apoptosis. PLoS One 2009; 4:e8024; http://dx.doi.org/10.1371/journal.pone.0008024; PMID: 19946639
- Leroy D, Heriché JK, Filhol O, Chambaz EM, Cochet C. Binding of polyamines to an autonomous domain of the regulatory subunit of protein kinase CK2 induces a conformational change in the holoenzyme. A proposed role for the kinase stimulation. J Biol Chem 1997; 272:20820 - 7; http://dx.doi.org/10.1074/jbc.272.33.20820; PMID: 9252407
- Stark F, Pfannstiel J, Klaiber I, Raabe T. Protein kinase CK2 links polyamine metabolism to MAPK signalling in Drosophila. Cell Signal 2011; 23:876 - 82; http://dx.doi.org/10.1016/j.cellsig.2011.01.013; PMID: 21262350
- Ashburner M, Lewis S. On ontologies for biologists: the Gene Ontology--untangling the web. Novartis Found Symp 2002; 247:66 - 80, discussion 80-3, 84-90, 244-52; http://dx.doi.org/10.1002/0470857897.ch6; PMID: 12539950
- Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 2005; 171:603 - 14; http://dx.doi.org/10.1083/jcb.200507002; PMID: 16286508
- Shi H, Rojas R, Bonifacino JS, Hurley JH. The retromer subunit Vps26 has an arrestin fold and binds Vps35 through its C-terminal domain. Nat Struct Mol Biol 2006; 13:540 - 8; http://dx.doi.org/10.1038/nsmb1103; PMID: 16732284
- Welsch T, Younsi A, Disanza A, Rodriguez JA, Cuervo AM, Scita G, et al. Eps8 is recruited to lysosomes and subjected to chaperone-mediated autophagy in cancer cells. Exp Cell Res 2010; 316:1914 - 24; http://dx.doi.org/10.1016/j.yexcr.2010.02.020; PMID: 20184880
- Köchl R, Hu XW, Chan EY, Tooze SA. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic 2006; 7:129 - 45; http://dx.doi.org/10.1111/j.1600-0854.2005.00368.x; PMID: 16420522
- Xie R, Nguyen S, McKeehan WL, Liu L. Acetylated microtubules are required for fusion of autophagosomes with lysosomes. BMC Cell Biol 2010; 11:89; http://dx.doi.org/10.1186/1471-2121-11-89; PMID: 21092184
- Lipinski MM, Hoffman G, Ng A, Zhou W, Py BF, Hsu E, et al. A genome-wide siRNA screen reveals multiple mTORC1 independent signaling pathways regulating autophagy under normal nutritional conditions. Dev Cell 2010; 18:1041 - 52; http://dx.doi.org/10.1016/j.devcel.2010.05.005; PMID: 20627085
- Traweger A, Fuchs R, Krizbai IA, Weiger TM, Bauer HC, Bauer H. The tight junction protein ZO-2 localizes to the nucleus and interacts with the heterogeneous nuclear ribonucleoprotein scaffold attachment factor-B. J Biol Chem 2003; 278:2692 - 700; http://dx.doi.org/10.1074/jbc.M206821200; PMID: 12403786
- Zhang T, Wong SH, Tang BL, Xu Y, Hong W. Morphological and functional association of Sec22b/ERS-24 with the pre-Golgi intermediate compartment. Mol Biol Cell 1999; 10:435 - 53; PMID: 9950687
- Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell 2009; 34:259 - 69; http://dx.doi.org/10.1016/j.molcel.2009.04.026; PMID: 19450525
- Truong M, Cook MR, Pinchot SN, Kunnimalaiyaan M, Chen H. Resveratrol induces Notch2-mediated apoptosis and suppression of neuroendocrine markers in medullary thyroid cancer. Ann Surg Oncol 2011; 18:1506 - 11; http://dx.doi.org/10.1245/s10434-010-1488-z; PMID: 21184191
- Lalier L, Cartron PF, Juin P, Nedelkina S, Manon S, Bechinger B, et al. Bax activation and mitochondrial insertion during apoptosis. Apoptosis 2007; 12:887 - 96; http://dx.doi.org/10.1007/s10495-007-0749-1; PMID: 17453158
- Blagoev B, Mann M. Quantitative proteomics to study mitogen-activated protein kinases. Methods 2006; 40:243 - 50; http://dx.doi.org/10.1016/j.ymeth.2006.08.001; PMID: 17071406
- Gurbuxani S, Schmitt E, Cande C, Parcellier A, Hammann A, Daugas E, et al. Heat shock protein 70 binding inhibits the nuclear import of apoptosis-inducing factor. Oncogene 2003; 22:6669 - 78; http://dx.doi.org/10.1038/sj.onc.1206794; PMID: 14555980
- Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jørgensen TJ. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol Cell Proteomics 2005; 4:873 - 86; http://dx.doi.org/10.1074/mcp.T500007-MCP200; PMID: 15858219
- Hubner NC, Ren S, Mann M. Peptide separation with immobilized pI strips is an attractive alternative to in-gel protein digestion for proteome analysis. Proteomics 2008; 8:4862 - 72; http://dx.doi.org/10.1002/pmic.200800351; PMID: 19003865
- Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2007; 2:1896 - 906; http://dx.doi.org/10.1038/nprot.2007.261; PMID: 17703201
- Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 2003; 75:663 - 70; http://dx.doi.org/10.1021/ac026117i; PMID: 12585499
- Hu Q, Noll RJ, Li H, Makarov A, Hardman M, Graham Cooks R. The Orbitrap: a new mass spectrometer. J Mass Spectrom 2005; 40:430 - 43; http://dx.doi.org/10.1002/jms.856; PMID: 15838939
- Schroeder MJ, Shabanowitz J, Schwartz JC, Hunt DF, Coon JJ. A neutral loss activation method for improved phosphopeptide sequence analysis by quadrupole ion trap mass spectrometry. Anal Chem 2004; 76:3590 - 8; http://dx.doi.org/10.1021/ac0497104; PMID: 15228329
- Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, et al. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics 2005; 4:2010 - 21; http://dx.doi.org/10.1074/mcp.T500030-MCP200; PMID: 16249172
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 2008; 26:1367 - 72; http://dx.doi.org/10.1038/nbt.1511; PMID: 19029910