Abstract
Comment on: Lorenz A, et al. Science 2012; 336:1585-8.
In order to segregate homologous chromosomes (homologs) properly during meiosis, they have to first find each other and then become physically linked. This is achieved by inducing hundreds of DNA double-strand breaks (DSBs) during meiotic prophase, which are subsequently repaired by homologous recombination catalyzed by the central recombinase Rad51 and/or its meiosis-specific variant Dmc1. One outcome of this process is crossovers (COs) between homologs, which depend on DNA endonucleases for their formation. These COs provide the physical connections to ensure proper chromosome segregation and contribute to genetic diversity by re-shuffling parental genes. Theoretically, only one CO per homolog pair is sufficient for guiding correct chromosome segregation, and one to several per homolog pair are actually observed. This leaves between 60–95% of DSBs (depending on the organism) to be repaired as non-crossovers (NCOs), using the homolog as a template, or to be redirected for repair using the sister chromatid.
Molecular models for homologous recombination resulting specifically in a NCO outcome were first proposed over 30 years ago, and it was envisaged that activities capable of unwinding DNA strands (i.e., DNA helicases) would be crucially important for this process.Citation1 Indeed, in mitotically growing cells, several DNA helicases have been implicated in directing this type of recombination. However, until recently, it was unclear which of these drove NCO formation in meiosis. The picture that is emerging is that some, but not all, fulfill this function. Moreover, the helicase that has been selected during evolution to perform meiotic NCO formation appears to vary between different organisms.
In the budding yeast, Saccharomyces cerevisiae, the RecQ-type DNA helicase Sgs1 has long been suspected to drive meiotic NCO recombination, but evidence has been ambiguous. Intriguingly, recent work by the Lichten and Hunter labs demonstrated that Sgs1 is not only in charge of producing NCOs, but is also a master regulator of all meiotic recombination required for the removal of multi-chromatid invasions and promotion of CO formation by the Zip1–4, Mer3 and Msh4–5 (ZMM)-dependent pathway.Citation2,Citation3
In Caenorhabditis elegans, the DEAH-family helicase RTEL-1 has been implicated in controlling the CO outcome of meiotic recombination. However, it probably does this by redirecting interhomolog invasions toward intersister repair rather than driving them into NCO products.Citation4,Citation5
Recently, the FANCM orthologs in Schizosaccharomyces pombe (Fml1) and in Arabidopsis thaliana were identified as the helicases curbing CO formation during meiosis in these organisms. In Arabidopsis, as in many other organisms studied, there are two pathways of meiotic CO recombination: the class I COs, which depend on the ZMM proteins and are subject to interference (where a CO prevents the formation of other COs in its vicinity), and the non-interfering class II COs, which largely depend on the DNA endonuclease Mus81-Eme1/Mms4. If one disables the ZMM pathway, the number of COs is reduced, and those that do form become randomly distributed. This will leave at least some homolog pairs without the required CO to guide their segregation. In Arabidopsis it was shown that abolishing FANCM function in a msh4 mutant background restored CO formation (at least partially), and that the formation of these COs now depends on MUS81.Citation6,Citation7 This indicates that FANCM directs recombination intermediates, which have the potential of becoming MUS81-dependent COs, toward interhomolog NCOs or repair from the sister chromatid.
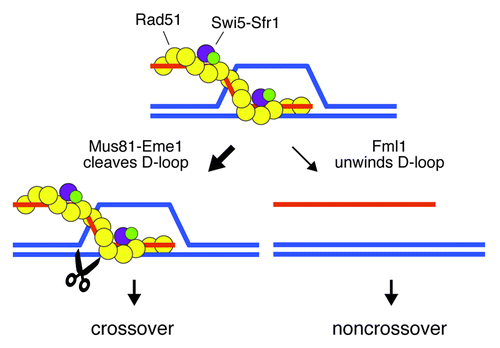
In fission yeast, we could determine that Fml1, supported by its accessory factors Mhf1 and Mhf2, indeed transforms COs between homologs into NCOs between homologs, and appears to do so by competing with Mus81-Eme1 for processing the same type of recombination intermediate.Citation8 This intermediate is likely to be the displacement (D)-loop formed by the invasion of a DNA duplex by a homologous single-stranded DNA tail coated with Rad51/Dmc1. Our in vitro analysis of purified Mus81-Eme1 and Fml1 shows that both enzymes favor working on this substrate.Citation8,Citation9 Mus81-Eme1 can cleave it and, in doing so, generates a CO, whereas Fml1 can unwind it, which promotes a NCO-specific pathway called synthesis-dependent strand annealing (SDSA) (). Because S. pombe lacks the ZMM pathway, it relies entirely on Mus81-Eme1 to ensure that enough COs are made. Indeed, there is a strong bias in favor of Mus81-Eme1 processing recombination intermediates, which, at least in part, depends on the Swi5-Sfr1 complex.Citation8 Swi5-Sfr1 is a Rad51/Dmc1 mediator that is perfectly shaped to nestle into the groove of the Rad51-DNA filament. There it stimulates the recombination reaction by stabilizing the filament and enhancing Rad51’s ATPase activity.Citation10 Getting rid of Sfr1 decreases the stability of the filament and also enables Fml1 to act on recombination intermediates, which are normally processed by Mus81-Eme1. How exactly Swi5-Sfr1 blocks Fml1’s ability to process recombination intermediates, while allowing Mus81-Eme1 to operate, is currently unclear. One possibility is that only Mus81-Eme1 is capable of accessing the D-loop when its DNA is encased within a Swi5-Sfr1-stabilized Rad51 filament.
Figure 1. Model showing alternative mechanisms of D-loop processing during meiosis in fission yeast. The presence of Swi5-Sfr1 strongly biases processing of the D-loop toward cleavage by Mus81-Eme1. DNA strands are represented by the orange and blue lines.
In humans, FANCM is best known for its involvement in a mitotic DNA repair network, which, when defective, can give rise to the genetic disorder Fanconi anemia, characterized by heightened cancer susceptibility, anemia and developmental problems. It will be interesting to see whether it has also been selected from the many candidate helicases to limit CO formation in the mammalian germline.
References
- Resnick MA. J Theor Biol 1976; 59:97 - 106; http://dx.doi.org/10.1016/S0022-5193(76)80025-2; PMID: 940351
- Zakharyevich K, et al. Cell 2012; 149:334 - 47; http://dx.doi.org/10.1016/j.cell.2012.03.023; PMID: 22500800
- De Muyt A, et al. Mol Cell 2012; 46:43 - 53; http://dx.doi.org/10.1016/j.molcel.2012.02.020; PMID: 22500736
- Youds JL, et al. Science 2010; 327:1254 - 8; http://dx.doi.org/10.1126/science.1183112; PMID: 20203049
- Rosu S, et al. Science 2011; 334:1286 - 9; http://dx.doi.org/10.1126/science.1212424; PMID: 22144627
- Crismani W, et al. Science 2012; 336:1588 - 90; http://dx.doi.org/10.1126/science.1220381; PMID: 22723424
- Knoll A, et al. Plant Cell 2012; 24:1448 - 64; http://dx.doi.org/10.1105/tpc.112.096644; PMID: 22547783
- Lorenz A, et al. Science 2012; 336:1585 - 8; http://dx.doi.org/10.1126/science.1220111; PMID: 22723423
- Osman F, et al. Mol Cell 2003; 12:761 - 74; http://dx.doi.org/10.1016/S1097-2765(03)00343-5; PMID: 14527420
- Kuwabara N, et al. Structure 2012; 20:440 - 9; http://dx.doi.org/10.1016/j.str.2012.01.005; PMID: 22405003