Heterochromatin is a special type of chromatin that is well-known for its negative impact on genome expression. This downregulation is accomplished by several complementary pathways, which are, in turn, regulated by region-specific epigenetic modifications of the histone tails. Despite intense research activity in this field, the underlying molecular mechanisms and their mutual interdependencies are not well understood.Citation1,Citation2
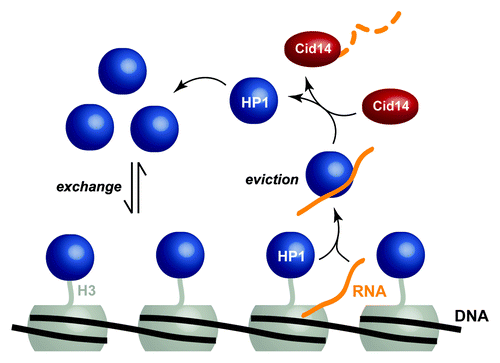
Whereas heterochromatin is classically perceived to be a static, inaccessible chromatin structure and therefore refractory to transcription, a number of recent findings have challenged this view, and heterochromatin has thus emerged to be more dynamic than previously anticipated.Citation2-Citation5 We have complemented this view with novel insights into gene silencing in the fission yeast Schizosaccharomyces pombe (S. pombe). By combining in vivo and in vitro experiments, our research groups elucidated a mechanism of heterochromatin repression in S. pombe that includes transient and dynamic interactions between the HP1Swi6 protein, the methylated amino acid residue lysine 9 of the histone H3 tail (H3K9) and any RNA that may be transcribed from adjacent heterochromatic DNA.Citation6 In combination with our previous finding that the RNA processing factor Cid14 is required for tight repression of certain heterochromatic regions in the S. pombe genome,Citation7 a comprehensive mechanism of suppression arises (). Importantly, this new RNA degradation pathway involves fast dynamics of the heterochromatin protein HP1Swi6, as revealed by measurements of its interaction kinetics with RNA and methylated H3K9.
Figure 1. Model for HP1Swi6-mediated degradation of heterochromatic RNA. HP1Swi6 proteins (dark blue) associate with H3K9 methylated nucleosomes (gray) only transiently and readily exchange with the cellular bulk HP1Swi6 population. In case transcription within heterochromatin occurs, HP1Swi6 binds the newly synthesized RNA (orange) and dissociates from H3K9 methylated nucleosomes as a result of competitive binding between RNA and the histone tail. Subsequently, the RNA is passed on to Cid14 (red), which, in turn, initiates RNA degradation.
The highly conserved HP1 proteins consist of an N-terminal chromodomain (CD) and a C-terminal chromoshadow domain (CSD) separated by a flexible hinge region. Numerous in vitro studies of the interaction between the CD of HP1 and the histone tail were focused on the binding affinity, which is in the low μM range for isolated CDs binding to methylated H3K9.Citation8,Citation9 In contrast, not much emphasis has been put on the binding kinetics of the full-length protein. Using solution NMR spectroscopy and surface plasmon resonance (SPR), we observed that the kinetic off-rate constant for dissociation of the HP1Swi6-H3K9me3 complex is in the range of 10–1,000 sec−1, corresponding to a lifetime of 1–100 ms. These measurements are in line with determinations of the lifetime of the HP1Swi6 ensemble on heterochromatin in vivo by FRAP experiments, which showed a value of about 400 ms in S. pombe.Citation10 Thus, it turns out that the HP1Swi6 ensemble acts as a dynamic “swarm” approaching chromatin at locations that are specifically marked by K9 methylated histone H3 tails ().
In addition to methylated H3K9, we found that HP1Swi6 also binds directly to RNA. By defining interaction surfaces for the molecular interactions with RNA or H3K9me3 peptides using solution NMR spectroscopy, we found an unexpected overlap: both RNA and H3K9me3 peptide binding involve the N-terminus of HP1Swi6, including the CD, suggesting that RNA and H3K9me3 binding might be competitive. Subsequent NMR and SPR experiments confirmed that the HP1Swi6-RNA and HP1Swi6-H3K9me3 interactions are indeed competitive processes. Thus, RNA binding to HP1Swi6 will lead to repulsion of HP1Swi6 from heterochromatin on the ensemble level. Finally, RNA binding further involves the hinge region, which encodes positively charged amino acids that abolish, when mutated to alanine, the HP1Swi6-RNA interaction and, thus, the competition with H3K9me3 binding. When tested in vivo, the HP1Swi6 RNA-binding mutant was not able to repress the expression of certain heterochromatic genes, although the structural integrity of heterochromatin remained unaffected.
In conclusion, the highly specific but dynamic H3K9me3 binding properties of HP1Swi6 ensure continuous recognition and repulsion of heterochromatic RNA. In contrast to RNA of euchromatic origin, heterochromatic RNA complexed with HP1Swi6 is destined for degradation, warranting tight repression of heterochromatin and preventing saturation of HP1Swi6 with RNA. Whereas this pathway appears to play a dominant role at the mating type locus and at telomeres, it may function redundantly with the RNAi pathway to repress centromeric heterochromatin on a co- or posttranscriptional level.Citation7
Our findings raise several intriguing questions: do other HP1 proteins share similar biophysical properties? Is RNA-mediated eviction of chromatin-binding proteins a more general mechanism to shape the epigenome? Are there other molecular functions that could be explained by the dynamic nature of heterochromatin? We are excited about addressing these and related issues in our future research.
References
- Beisel C, et al. Nat Rev Genet 2011; 12:123 - 35; http://dx.doi.org/10.1038/nrg2932; PMID: 21221116
- Cheutin T, et al. Science 2003; 299:721 - 5; http://dx.doi.org/10.1126/science.1078572; PMID: 12560555
- Bühler M, et al. Cell 2006; 125:873 - 86; http://dx.doi.org/10.1016/j.cell.2006.04.025; PMID: 16751098
- Volpe TA, et al. Science 2002; 297:1833 - 7; http://dx.doi.org/10.1126/science.1074973; PMID: 12193640
- Grewal SI, et al. Nat Rev Genet 2007; 8:35 - 46; http://dx.doi.org/10.1038/nrg2008; PMID: 17173056
- Keller C, et al. Mol Cell 2012; 47:215 - 27; http://dx.doi.org/10.1016/j.molcel.2012.05.009; PMID: 22683269
- Bühler M, et al. Cell 2007; 129:707 - 21; http://dx.doi.org/10.1016/j.cell.2007.03.038; PMID: 17512405
- Schalch T, et al. Mol Cell 2009; 34:36 - 46; http://dx.doi.org/10.1016/j.molcel.2009.02.024; PMID: 19362535
- Kaustov L, et al. J Biol Chem 2011; 286:521 - 9; http://dx.doi.org/10.1074/jbc.M110.191411; PMID: 21047797
- Cheutin T, et al. Mol Cell Biol 2004; 24:3157 - 67; http://dx.doi.org/10.1128/MCB.24.8.3157-3167.2004; PMID: 15060140