Keywords: :
In mammalian cells, the response to double-strand DNA breaks (DSBs) is crucial to maintain cell viability and prevent oncogenic transformation. A complex mechanism has evolved to achieve a timely response to these lesions characterized by post-translational modification events. The modifications, including phosphorylation, ubiquitination and SUMOylation, contribute to the recruitment of factors, either through specific binding modules in DNA-damage response proteins, or by the unmasking of cryptic signals on chromatin to which the proteins bind. This mechanism amplifies the damage signal and offers an opportunity to the cell to regulate the strength, longevity and spread of the response. In a recent report we elucidated the means by which the mammalian proteasome plays a role in dampening DSB signaling and regulates the repair of DSBs.Citation1
In considering the degree and complexity of Ub conjugation involved in the DSB response,Citation2 we supposed that deubiqutinating ezymes (DUBs) not currently implicated would be likely to be important. To test this, we performed an siRNA screen to identify enzymes required to reduce Ub conjugates following release from exposure to hydroxyurea (HU), i.e., on recovery from S-phase DSBs. This identified POH1/PSMD14/rpn11, the obligate DUB of the 19S proteasome activating complex. The 19S activates the 20S core and is required to degrade Ub-modified proteins. In addition to an influence on global Ub conjugates, we found the enzymatic activity of POH1 was also required to restrict Ub accumulation at sites of DNA damage following HU and irradiation (IR).
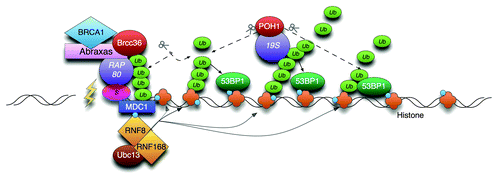
The Ub ligases RNF8 and RNF168 recruit to sites of DNA damage and are required for the subsequent accumulation of repair mediators 53BP1 and BRCA1 (in the BRCA1-A complex). 53BP1 binds to dimethylated histones but requires RNF8-mediated removal of competing histone binding proteins and RNF8-RNF168-mediated local generation of K63-linked poly-Ub to do so.Citation3 The BRCA1-A complex contains the K63-Ub binding protein RAP80, which directs localization of the complex to sites of damage. This complex also contains K63-Ub-specific DUB, BRCC36, able to hydrolyse K63-chains and restrict the amount of 53BP1 at damaged chromatin.Citation4
We found that while the ability of 53BP1 to form IR-induced foci (IRIF) was inhibited in cells with low expression of RNF8 or RNF168, they were permitted when POH1 was also depleted, and even 53BP1 expressed at very low levels could form IRIF when POH1 expression was reduced. Thus POH1 is a powerful antagonist to 53BP1, preventing 53BP1 from recruiting to DSB sites.
The regulation was not at the level of protein expression of RNF8, RNF168 or 53BP1. Instead POH1 acted in both mechanisms associated with 53BP1 accumulation. It promoted the occupation of chromatin by JMJD2A, a protein that competes with 53BP1 for the dimethyl histone mark, and restricted the degree of K63-Ub at sites of damage.
Intriguingly the 19S and BRCA1-A complexes have been likened due to the number of protein modules in common,Citation6 and both POH1 and BRCC36 are Jab1/Mov34/Mpr1 Pad1 N-terminal+ (MPN+) (JAMM) proteases with K63-Ub linkage specificity.Citation5 Importantly co-depletion of POH1 and BRCC36 did not elicit 53BP1 foci larger than POH1 depletion alone, suggesting these DUBs act in the same mechanistic pathway to restrict 53BP1 assemblies.
The influence of POH1 on DNA repair through non-homologous end joining correlated with its regulation of 53BP1 accumulation. Reduced DNA repair in RNF8-, RNF168- or 53BP1-depleted cells, in which no 53BP1 IRIF form, could be countered by co-depletion of POH1, which restored both repair and 53BP1 IRIF. Intriguingly, in cells depleted for POH1 and exhibiting enlarged 53BP1 foci end joining was reduced. This correlated with poor recruitment of the NHEJ factor Artemis. We speculate the block on DNA repair might be brought about by inappropriate proximity of 53BP1 to the DNA ends.
These observations were all the more interesting when we examined the influence of POH1 depletion on BRCA1 and RAP80. We expected enlarged accumulations of these proteins, since the signal for their accumulation is also K63-Ub. However no increase was seen. The lack of BRCA1 spreading despite the increase in local K63-Ub, and, further, the inability to restore BRCA1 IRIF in RNF8-depleted cells by reduction of POH1, suggests that Ub-binding by RAP80 is insufficient for BRCA1 recruitment. Consistent with this conclusion RAP80 interaction with SUMO has recently been reported to be required for its recruitment.Citation7 Thus, the cell is able to separate the regulation of the mediators, 53BP1 and BRCA1, despite a shared signal for their accumulation.
K63-Ub binding proteins in the DSB response, such as 53BP1, inhibit DNA resection in homologous recombination (HR) repair. Thus we anticipated that HR might be defective in POH1-depleted cells in a manner dependent on 53BP1. However, although we demonstrated a requirement for POH1 in HR repair, this was not through, or not wholly through, 53BP1. Instead we found that overexpression of the small-nucleic acid-like protein, 19S component and BRCA2- co-factor, DSS1, could restore HR in cells with low POH1. DSS1 promotes BRCA2-medaited RAD51 loading,Citation8 and we suggest that the 19S may have a role enriching DSS1 at sites of damage to improve HR-repair.
Together with other recent reports of the proteasome at sites of DNA damage in mammalian cells (20S, 19S and other activators),Citation9,Citation10 these data elucidate a previously unappreciated role for the proteasome in DNA repair. Thus, to our appreciation of this complex as a protein macerator, we must now add the more subtle role of Ub-conjugate regulation. ()
Figure 1. Model of POH1-mediated restriction of 53BP1 through K63- poly-Ub cleavage. RNF8/168 modify histones with K63-linked Ub. The combined activity of the removal of chromatin binding proteins (JMJD2A/B, not shown) and K63-poly-Ub generation promotes the accumulation of 53BP1 to the mark. POH1 activity counters this by promotes JMJD2A residence in chromatin (not shown) and by hydrolysing K63-poly-Ub.Citation1 The BRCA1-A complex is tethered by SUMO interactionCitation7 and does not spread.
| Abbreviations: | ||
| DSBs | = | double strand breaks |
| Ub | = | ubiquitin |
| SUMO | = | small ubiquitin like modifier |
| DUBs | = | deubiqutinating enzymes |
| HU | = | hydroxyurea |
| RNF8 | = | Ring Finger 8 |
| RNF168 | = | Ring Finger 168 |
| 53BP1 | = | p53 Binding protein 1 |
| BRCA1 | = | Breast Cancer protein 1 |
| RAP80 | = | receptor associated protein |
| BRCC36 | = | BRCA1/BRCA2-containing complex subunit 3 |
| K63-Ub | = | Lysine 63-linked Ubiquitin |
| RAD51 | = | RAD51/RecA homolog |
| JMJD2A | = | Jumonji domain containing 2A |
| DSS1 | = | deleted in split-hand/split-foot 1 |
| HR | = | homologous recombination |
| NHEJ | = | Non homologous end joining |
References
- Butler LR, et al. EMBO J 2012; 31:3918 - 34; http://dx.doi.org/10.1038/emboj.2012.232; PMID: 22909820
- Al-Hakim A, et al. DNA Repair (Amst) 2010; 9:1229 - 40; http://dx.doi.org/10.1016/j.dnarep.2010.09.011; PMID: 21056014
- Mallette FA, et al. EMBO J 2012; 31:1865 - 78; http://dx.doi.org/10.1038/emboj.2012.47; PMID: 22373579
- Shao G, et al. Proc Natl Acad Sci U S A 2009; 106:3166 - 71; http://dx.doi.org/10.1073/pnas.0807485106; PMID: 19202061
- Cooper EM, et al. EMBO J 2009; 28:621 - 31; http://dx.doi.org/10.1038/emboj.2009.27; PMID: 19214193
- Wang B, et al. Genes Dev 2009; 23:729 - 39; http://dx.doi.org/10.1101/gad.1770309; PMID: 19261749
- Hu X, et al. J Biol Chem 2012; 287:25510 - 9; PMID: 22689573
- Liu J, et al. Nat Struct Mol Biol 2010; 17:1260 - 2; http://dx.doi.org/10.1038/nsmb.1904; PMID: 20729859
- Levy-Barda A, et al. Cell Cycle 2011; 10:4300 - 10; http://dx.doi.org/10.4161/cc.10.24.18642; PMID: 22134242
- Galanty Y, et al. Genes Dev 2012; 26:1179 - 95; http://dx.doi.org/10.1101/gad.188284.112; PMID: 22661229