Proteins are generally believed to adopt a unique fold, defined by their amino acid sequence, under specific environmental conditions.Citation1 These unique structures, in turn, endow proteins with one specific function. However, not all proteins obey the “1 amino acid sequence → 1 fold → 1 function” scheme. Moonlighting proteinsCitation2 that adopt one distinct three-dimensional structure but can accomplish two or more alternative functions, such as NusE/ S10,Citation3,Citation4 and proteins that obtain two slightly different folds in identical environments have been described. Examples of the latter are the Aquifex aeolicus ribosomal protein L20Citation5 and the chemokine lymphotactin (Ltn).Citation6 In either protein, the difference between the two conformational states is modest: in L20, a short nine-residue unstructured region alternatively folds into an α-helix, and in Ltn an N-terminal loop changes to a β-strand, while the C-terminal α-helix becomes unstructured. Differences in small parts of protein structures even in identical solution are, however, not uncommon. In fact, all molecules are subject to dynamic changes, but those have to be distinguished from genuine structural changes, a term which should be restricted to cases where the alternative states are different to a large extent; that is, they either have different stable standard secondary structures or different tertiary topologies. However, even the minor unstructured-structured transitions, as observed for L20 and Ltn, gave rise to the term metamorphic proteins.Citation7
Gross structural changes were observed for prion proteins.Citation8 They can adopt two structures; one state is soluble and functional, whereas another is insoluble and pathogenic. Yet, the two states do not coexist in identical solution, and the transition to the insoluble state is basically irreversible.
Thus, the concept of a protein that adopts two stable, largely distinct structures which are both appreciably populated in solution under identical environmental conditions seemed remote even after these initial observations. It seemed even more remote to consider a scenario in which a protein exists in dramatically different structures in the same environment, but also uses these distinct structures to fulfill entirely different functions.
Recently, we used solution nuclear magnetic resonance (NMR) spectroscopy to explore hypothetical structural changes of the bacterial two-domain transcription factor RfaH.Citation9 In crystals, RfaH forms a compact structure, in which the two domains closely interact.Citation10 The structure of the RfaH N-terminal domain (NTD) was identical to that of a closely related two-domain protein, NusG,Citation11 consistent with the identical roles of the NTDs in binding to RNA polymerase (RNAP). In contrast, the RfaH C-terminal domain (CTD) was a structural antipode to the NusG CTD in solution: while the RfaH CTD in the crystal was all-α-helical, the NusG CTD was all-β, despite similar amino acid sequences (~17% identity).
Altogether, the solution structure we determined for RfaH was identical to the structure captured in the crystal.Citation9,Citation10 In both RfaH and NusG, the two domains are separated by a flexible linker and fold independently when expressed separately. To our great surprise, the isolated RfaH CTD folded into an all-β structure, virtually identical to NusG CTD. Thus, the same CTD sequence folds into an all-α topology when it interacts with the NTD or into an all-β topology in the absence of NTD.
To obtain both CTD structures coexisting in the context of the intact protein, we weakened a salt bridge that stabilizes the domain interaction in wild-type RfaH by an amino acid substitution in the NTD, leaving the CTD unaltered. This protein variant existed in a 1:1 equilibrium between two states, one with an all-α CTD and the other with an all-β CTD, as observed by NMR.Citation9
RfaH CTD not only appears in two different folds, but also accomplishes two unrelated, alternative functions.Citation9 In the all-α state, the main function of the CTD is to mask the RNAP-binding site in the absence of a specific DNA recruitment signal. The all-β state is thought to recruit a ribosome to an mRNA lacking a ribosome-binding site, thereby dramatically stimulating translation. Thus, RfaH simultaneously changes both topology and function, constituting the new paradigm of a transformer protein (TFP). The concept of TFPs allows those proteins to play a role in two separate processes, as exemplified by RfaH. First, RfaH is a transcription regulator that undergoes a complete structural and functional transformation to become a translation activator. By this definition, Ltn is a borderline case—it has two alternative functions that are regulated by a transition between unstructured and structured states.
RfaH not only constitutes a first example of a TFP but will also serve as a very useful model for protein folding. The complete CTD refolding gives us a unique opportunity to study the kinetics and dynamics of fold conversion by a wide variety of methods, including NMR and optical spectroscopy. Perhaps the most pressing question, both in a biological and in a protein folding context, relates to a quantitative reversal of the domain-opened to the domain-closed structure upon dissociation from RNAP. The domain-opened protein is only marginally soluble, presenting a challenge for NMR and other, more intricate methods will have to be used to study its refolding.
Thus, we did not only have the privilege to observe an exciting new biological principle in action and find a true transformer protein, but we also could supply the protein-folding community with an additional model and shed light on the regulation principles of bacterial gene expression.
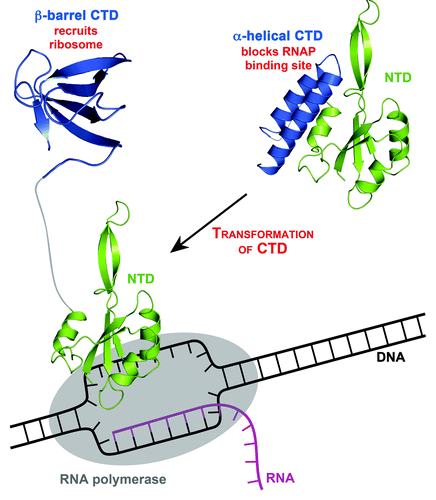
Figure 1. The transformation of RfaH. RfaH in ribbon representation; C-terminal domain (CTD), blue; N-terminal domain (NTD), green. In the closed form of RfaH (right), CTD and NTD are tightly interacting, and RfaH works as a transcription factor. In this state, the α-helical CTD masks the area of the NTD, which has the ability to bind RNAP (RNAP, gray oval), thus preventing this interaction. Binding to a specific DNA site, the operon polarity suppressor site, leads to domain separation (left), enabling the NTD to bind to RNAP in the transcription complex. The subsequent (or simultaneous) refolding of the CTD into a β-barrel, which can bind to ribosomal protein S10 to recruit a ribosome, transforms RfaH into a translation factor. PDB codes: 2OUG, full-length RfaH; 2LCL, RfaH CTD
References
- Anfinsen CB. Science 1973; 181:223 - 30; http://dx.doi.org/10.1126/science.181.4096.223; PMID: 4124164
- Copley SD. Bioessays 2012; 34:578 - 88; http://dx.doi.org/10.1002/bies.201100191; PMID: 22696112
- Burmann BM, et al. Science 2010; 328:501 - 4; http://dx.doi.org/10.1126/science.1184953; PMID: 20413501
- Squires CL, et al. Annu Rev Microbiol 2000; 54:775 - 98; http://dx.doi.org/10.1146/annurev.micro.54.1.775; PMID: 11018144
- Timsit Y, et al. EMBO Rep 2006; 7:1013 - 8; http://dx.doi.org/10.1038/sj.embor.7400803; PMID: 16977336
- Tuinstra RL, et al. Proc Natl Acad Sci USA 2008; 105:5057 - 62; http://dx.doi.org/10.1073/pnas.0709518105; PMID: 18364395
- Murzin AG. Science 2008; 320:1725 - 6; http://dx.doi.org/10.1126/science.1158868; PMID: 18583598
- Surewicz W, et al. 2011:135-167.
- Burmann BM, et al. Cell 2012; 150:291 - 303; http://dx.doi.org/10.1016/j.cell.2012.05.042; PMID: 22817892
- Belogurov GA, et al. Mol Cell 2007; 26:117 - 29; http://dx.doi.org/10.1016/j.molcel.2007.02.021; PMID: 17434131
- Mooney RA, et al. J Mol Biol 2009; 391:341 - 58; http://dx.doi.org/10.1016/j.jmb.2009.05.078; PMID: 19500594