Abstract
RECQ1 is the most abundant of the five human RecQ helicases, but little is known about its biological significance. Recent studies indicate that RECQ1 is associated with origins of replication, suggesting a possible role in DNA replication. However, the functional role of RECQ1 at damaged or stalled replication forks is still unknown. Here, for the first time, we show that RECQ1 promotes strand exchange on synthetic stalled replication fork-mimicking structures and comparatively analyze RECQ1 with the other human RecQ helicases. RECQ1 actively unwinds the leading strand of the fork, similar to WRN, while RECQ4 and RECQ5β can only unwind the lagging strand of the replication fork. Human replication protein A modulates the strand exchange activity of RECQ1 and shifts the equilibrium more to the unwinding mode, an effect also observed for WRN. Stable depletion of RECQ1 affects cell proliferation and renders human cells sensitive to various DNA damaging agents that directly or indirectly block DNA replication fork progression. Consequently, loss of RECQ1 activates DNA damage response signaling, leads to hyper-phosphorylation of RPA32 and activation of CHK1, indicating replication stress. Furthermore, depletion of RECQ1 leads to chromosomal condensation defects and accumulation of under-condensed chromosomes. Collectively, our observations provide a new insight into the role of RECQ1 in replication fork stabilization and its role in the DNA damage response to maintain genomic stability.
Introduction
RecQ helicases are highly conserved from bacteria to humans. Germ line mutations in three of the five human RecQ helicases, BLM, WRN and RECQ4 are associated with genetic syndromes that are characterized by premature aging and carcinogenesis.Citation1 RECQ1 and RECQ5β are not yet associated with any disease but are abundantly expressed throughout the cell cycle. RecQ helicases are considered caretakers of the human genome and are proposed to have multiple roles in DNA metabolism, including DNA replication, repair and recombination in addition to their emerging roles in telomeric maintenance.Citation2
WRN plays a significant role in DNA replication and S-phase progression and is also implicated in telomeric G-rich strand replication.Citation3,Citation4 BLM forms an evolutionarily conserved complex with topoisomerase III α-RMI1 and plays an important role in dissolution of double Holliday junctions (DHJs) and suppression of sister chromatid exchanges (SCEs).Citation5 RECQ5β was proposed to have both non-redundant and overlapping roles with that of BLM in the regulation of homologous recombination and suppression of SCEs by disruption of Rad51 presynaptic filaments.Citation6,Citation7 While RECQ1 was the first identified human RecQ helicaseCitation8 and the most abundant one,Citation9 little is known about its biological significance. Both RECQ1 and RECQ4 were proposed to be associated with DNA replication origins in a cell cycle-dependent manner.Citation10 More specifically, RECQ4 associates with the origins in the late G1 phase and is involved in the initiation of DNA replication and loading of replication factors such as RPA, PCNA and polymerase α.Citation11,Citation12 RECQ1 associates with the origins specifically in S-phase and is speculated to play an important role in replication fork elongation,Citation10 but its functional role in the stabilization and progression of stalled replication forks has not been described.
Replication fork progression is very important for cell viability, and any stalled or collapsed replication forks should therefore be handled with high priority. The progression of replication forks can be impaired by DNA damage, particularly if the blocking lesion is located on the leading-strand template.Citation13 While the synthesis of the leading strand gets halted at the lesion, the lagging-strand synthesis continues beyond the lesion site and results in an extensive widening of the gap on the leading strand.Citation14 Replication fork stalling can result in genomic instability and can trigger unscheduled DNA recombination events or SCEs that might lead to gross chromosomal re-arrangements and, ultimately, tumorigenesis. To avoid these detrimental consequences, cells often switch to different DNA damage bypass modes that can permit replication across the lesion.Citation13,Citation15 One such mechanism is template switching to the undamaged sister chromatid, thereby allowing a replicative polymerase to synthesize the complementary or the lesion strand in an error-free manner. It is believed that such template switching is achieved by strand exchange at the stalled forks, involving a movement of the fork backward so as to re-anneal the original template strands and displace the newly synthesized strands, which then anneal to generate a Holliday junction (HJ) structure with a short arm.Citation16,Citation17 Indeed, such structures have been observed to accumulate upon replication stalling both in prokaryotic and in eukaryotic cells.Citation18,Citation19
RecQ helicases are implicated in the processing of aberrant DNA structures that arise during DNA replication, and previous studies indicate that BLM, WRN and RECQ5β can all promote strand exchange on synthetic stalled replication forks.Citation20,Citation21 Previous biochemical studies indicate that RECQ1 preferentially unwinds forked DNA duplexes and homologous recombination (HR) intermediates such as the HJs and D-loops.Citation22 Conversely, RECQ1, but not BLM, is able to resolve immobile HJs.Citation22 Recent biochemical analyses indicate that RECQ1 also possesses DNA branch migration activity.Citation23,Citation24 In addition to its helicase activity, RECQ1 efficiently catalyzes strand annealing of complementary single-stranded DNA (ssDNA) in a reaction that is modulated by ATP-binding.Citation25 On the other hand, the crystal structure of human RECQ1 has recently been solved,Citation26 and it was shown that RECQ1 assumes different oligomeric forms to perform its helicase and strand-annealing activities.Citation27
In this study, we characterize the potential involvement of RECQ1 in the processing of synthetic stalled replication forks and analyze it mechanistically with that of the other human RecQ helicases. Our study is the first demonstration of the role of RECQ1 at stalled replication forks. It also provides new insights into the mechanistic similarities and differences between the human RecQ helicases, elucidating their differential roles in replication fork maintenance. We observe that stable depletion of RECQ1 leads to induction of replication stress and activation of DNA damage response signaling. Further, loss of RECQ1 leads to chromosomal condensation defects and accumulation of under-condensed chromosomes.
Results
RECQ1 promotes strand exchange on synthetic DNA structures mimicking a stalled replication fork
To investigate the role of RECQ1 in replication fork progression, we first tested its ability to promote strand exchange on synthetic stalled replication fork-like structures. Oligonucleotide-based stalled replication fork-like structures were generated as described previously.Citation20 Homologous arms were designed with a 5 nt region of heterology adjacent to the three-way junction to prevent spontaneous branch migration. Kanagraj et al. previously characterized the strand exchange of RECQ5β using these synthetic oligonucleotide-based model substrates.Citation20 In this study, we characterize the previously unknown strand exchange activity of RECQ1 and compared it with that of the other human RecQ helicases.
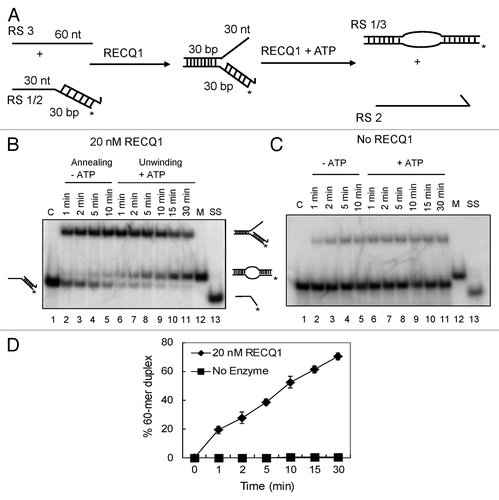
Strand exchange is a combination of both strand-annealing and unwinding activities. Initially, we prepared a partial forked duplex lacking the leading strand that represents a 3′-flap duplex. A 60 mer oligonucleotide, representing the leading strand template (RS3) was annealed to a 3′-tailed duplex [composed of a 30 mer oligonucleotide (RS2) and the complementary 32P-labeled 60mer oligonucleotide (RS1) that represents the lagging strand and the corresponding template strand, respectively] in the presence of RECQ1 (but in the absence of ATP) (). shows the representative images of kinetic experiments performed in the presence and absence of RECQ1 (20 nM), respectively. Strand annealing in the presence of RECQ1 resulted in the rapid formation of the 3′-flap duplex (within 2 min) (, lanes 2–5). Remarkably, upon the addition of ATP to this mixture, a robust strand exchange activity was observed over time (lanes 6–11), resulting in the formation of the 32P-labeled 60 mer duplex. This duplex contains a small bubble due to the short region of heterology at the junction. Almost 75–80% of the substrate was converted into 60 mer bubble duplex in about 30 min (), indicating efficient strand exchange in the presence of RECQ1. A small amount of the 60 mer duplex was also detected without the addition of ATP, which had presumably arisen from spontaneous branch migration. Similar kinetic experiments were performed in the absence of RECQ1 but did not show any traces of 60 mer bubble duplex formation, indicating that the activity observed is due to the presence of RECQ1 ().
Figure 1. Strand-exchange by RECQ1 on synthetic forked duplexes lacking the leading strand. (A) Scheme of the assay. Strand exchange is a combination of both strand annealing and unwinding activities. Strand annealing of the homologous oligonucleotides by RECQ1 forming the stalled replication fork, which was eventually unwound after the addition of ATP. The lengths of individual arms are indicated in nucleotides (nt) or base pairs (bp). The homologous leading and lagging arms have a 5 nt heterology at the fork junction to prevent spontaneous strand exchange. The 3′ end of the lagging oligonucleotide is indicated by an arrow, and the position of the 5′-PCitation32 label is marked by an asterisk. (B) As indicated above, 1 nM PCitation32-labeled 30/60 mer duplex (RS1/2) was incubated with 1 nM 60 mer complementary oligonucleotide (RS3) in the presence of 20 nM RECQ1 to form the forked DNA structure. After 10 min, ATP was added to a final concentration of 5 mM. Ten μl aliquots were taken out at indicated time points and analyzed by non-denaturing PAGE. “C” represents the control substrate P32-labeled RS1/2; “M” represents the marker for strand exchange product, the 60mer bubble duplex; and “SS” represents the ssDNA unwound. (C) Similar strand exchange reactions were performed in the absence of RECQ1. (D) Quantification of the kinetics performed in the presence and absence of RECQ1. Relative concentration of the 60 mer duplex product was plotted vs. reaction time. The data points represent the average values from three independent experiments.
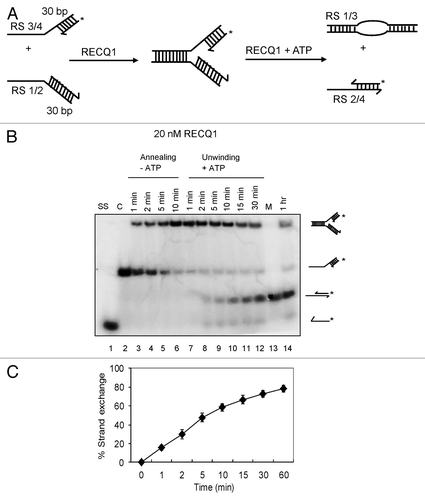
Next, we wanted to investigate whether RECQ1 can promote strand exchange if the forked structure contains both the leading and lagging strands. A synthetic stalled replication fork-like structure (RS1/2:RS3/4) was generated with a 10 nt gap, representing a lesion on the leading-strand. The length of the leading strand (RS4) was designed 10 nt shorter than that of the lagging strand (RS2). The DNA substrate was prepared as described above by RECQ1-mediated annealing of 32P-labeled partial duplex (RS3/RS4) with the unlabeled RS1/RS2 partial duplex (). Kinetics of strand annealing in the presence of RECQ1 indicated efficient formation of the stalled replication fork within 5 min (, lanes 3–6). Upon the addition of ATP, similar robust strand exchange activity was observed, as evidenced by the formation of the 20/30 mer duplex, formed by the annealing of the displaced 32P-labeled leading strand (RS4) to the lagging strand (RS2) (, lanes 7–14). About 75% strand exchange was observed in 30 min (), indicating that RECQ1 could efficiently bypass the leading strand gap. Collectively, our observations indicate that RECQ1 has the ability to promote the displacement of both arms of the fork and subsequent re-annealing of the displaced leading and lagging strands.
Figure 2. Strand-exchange by RECQ1 on synthetic stalled replication fork duplexes with a leading-strand gap. (A) Scheme of the assay. The oligonucleotides used are the same as in , except for an additional 20 mer representing the leading strand (RS4). (B) As indicated above, 1 nM P32-labeled 20/60 mer duplex (RS3/4) was incubated with 1 nM 30/60 mer duplex (RS1/2) in the presence of 20 nM RECQ1 to form the forked DNA structure. After 10 min, ATP was added to a final concentration of 5 mM. Ten μl aliquots were taken out at indicated time points and analyzed by non-denaturing PAGE. “C” represents the control substrate P32-labeled RS3/4; “M” represents the marker for strand exchange product and “SS” represents the ssDNA unwound. (C) Quantification of the kinetics performed in the presence of 20 nM RECQ1. Relative concentration of the subsequent leading/lagging strand duplex (RS2/4) was plotted vs. reaction time. The data points represent the average values from three independent experiments.
RPA modulates strand exchange of RECQ1 in a manner similar to WRN
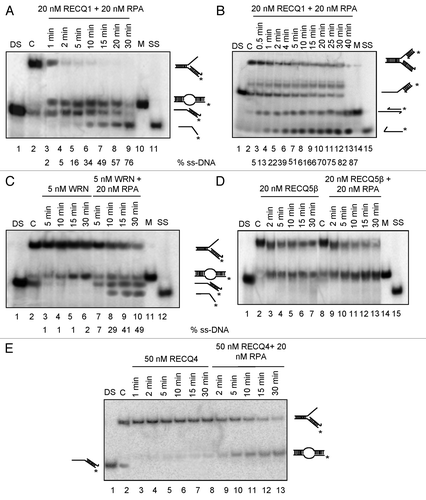
Human replication protein A (RPA) binds ssDNA, and RPA-coated ssDNA acts as a common intermediate for signaling-stalled replication forks and/or initiating DNA damage response and subsequent recruitment of repair proteins.Citation28,Citation29 Therefore, we investigated the effect of RPA on the strand-exchange activity of RECQ1 using the synthetic forked structures described above and further compare with that of the other human RecQ helicases. DNA substrates were prepared by spontaneous annealing of the constituent oligonucleotides, with the lagging oligonucleotide being present in a 4-fold molar excess over the parental strands. Previous studies indicated that RPA stimulated the strand exchange activity of RECQ5β on these substrates.Citation20 Similarly, the kinetics of strand exchange for RECQ1 is dramatically stimulated in the presence of RPA. About 80% activity was observed in about 2 min on both the model substrates irrespective of the leading-strand gap (), while a similar level of activity was observed in 30 min in the absence of RPA ( and ). Consistent with the previous observations,Citation20 RPA alone did not show any significant strand displacement activity using these oligonucleotide-based substrates (Fig. S1A). Although, the strand exchange kinetics of RECQ1 on a 3ˈ-flap duplex were dramatically stimulated, RPA rather shifted the activity more to the unwinding mode with accumulation of single-stranded DNA (compare and ), similar to that of WRN (, compare lanes 3–6 and 7–10) during the course of the reaction. Further, similar results were obtained in the presence of RPA using the stalled replication fork with a 10 nt gap on the leading strand (RS1/2:RS3/4; compare and ). Greater unwinding efficiency with relatively higher accumulation of ssDNA was observed using the stalled fork with a 10 nt gap on leading strand (compare , % ssDNA). Similar effect of RPA was previously reported on the strand exchange activity of both BLM and WRN.Citation20 On the other hand, no such shifting to the unwinding mode was observed with RECQ5β; only an increase in the accumulation of strand exchange product (60 mer bubble duplex, , lanes 3–7 and 9–13) was observed as previously described.Citation20 RECQ4, on the other hand, has weaker unwinding capacity,Citation30 and this likely limited its capacity to function in this assay. Nonetheless, we did observe very weak strand exchange activity for RECQ4 (, lanes 3–8) that was significantly stimulated in the presence of RPA (, lanes 9–13). Collectively, these observations indicate that although these RecQ helicases could efficiently catalyze the conversion of these stalled replication fork-like and 3′-flap DNA structures into the 60 mer duplex, in the presence of RPA, the action of both RECQ1 and WRN, unlike RECQ4 or RECQ5β was strongly biased toward unwinding the parental duplex, as evidenced by the accumulation of the single-stranded 32P-labeled 60 mer. Collectively, our results indicate that RECQ1 can perform strand exchange at a stalled replication fork structure, and that this activity is modulated by the presence of RPA. Moreover, RECQ1 behaves in a manner similar to WRN but distinct from that of RECQ4 and RECQ5β.
Figure 3. Effect of RPA on the strand exchange activity of human RecQ helicases. (A) Effect of RPA on RECQ1-mediated strand exchange on forked DNA duplex lacking the leading strand (B) and forked DNA duplex with 10 nt leading-strand gap. Effect of RPA on (C) 5 nM WRN, (D) 20 nM RECQ5β and (E) 50 nM RECQ4-mediated strand exchange on forked duplexes lacking the leading strand. Kinetic experiments were performed with 5 nM WRN, 20 nM RECQ5β and 50 nM RECQ4 in the presence or absence of 20 nM RPA. “DS” represents the control duplex substrate PCitation32 labeled RS1/2; “C” represents the control stalled replication fork-like structure; “M” represents the marker for strand exchange product; “SS” represents the ssDNA unwound.
RECQ1 can unwind the leading arm of the fork
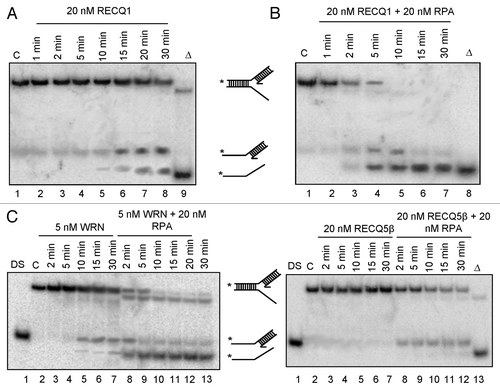
Next, we wanted to assess whether RECQ1 can actively unwind the leading arm of the fork. This we accomplished by examining its activity on a partial forked duplex DNA substrate lacking the lagging strand (resembling a 5′-flap duplex). We observe that RECQ1 could catalyze the concomitant unwinding of the parental arm on this structure, as evidenced by the accumulation of 5′-tailed duplex and ssDNA during the course of the reaction (). This unwinding was further stimulated in the presence of RPA (). RPA alone did not display significant strand exchange activity (Fig. S1B). WRN, but not RECQ5β, was also able to unwind these replication fork duplexes lacking the leading strand and was further stimulated in the presence of RPA (, compare lanes 3–7 and 8–13). Consistent to the previous observations,Citation20 RECQ5β could resolve the 5′-flap duplex only in the presence of RPA (, compare lanes 3–7 and 8–12). On the other hand, RECQ4 did not display any significant unwinding of the 5′-flap DNA structures either in the presence or absence of RPA (Fig. S1C). Our observations are in agreement with previous reports indicating that RECQ1 and WRN,Citation25,Citation31 but not Drosophila RECQ4 or RECQ5β,Citation32,Citation33 can unwind simpler 5′-flap-containing duplex substrates and further highlight mechanistic similarities and differences between RECQ1 and other human RecQ helicases in the processing of forked DNA structures. Collectively, our results indicate that the processing of these synthetic replication forks by RECQ1 differs from that of RECQ4 and RECQ5β, but is more similar to that of WRN.
Figure 4. Both RECQ1 and WRN, but not RECQ4 or RECQ5β, can unwind the leading strand of the replication fork duplexes. Replication fork duplex lacking the lagging strand was generated by annealing the oligo nucleotides RS1 and RS5/6. Unwinding kinetics of the replication fork duplexes lacking the lagging strand using 20 nM RECQ1 in the absence (A) or in the presence of 20 nM RPA (B). Unwinding kinetics of the replication fork duplexes lacking the lagging strand using 5 nM WRN (C), 20 nM RECQ5β (D) in the presence and absence of 20 nM RPA. “DS” represents the control duplex substrate PCitation32 labeled RS5/6; “C” represents the control stalled replication fork-like structure; Δ represents the heat denatured substrate.
Depletion of RECQ1 renders cells sensitive to replication blocking agents
To further investigate the in vivo importance of processing such synthetic stalled replication fork-like structures, we wanted to test the potential role of RECQ1 in preventing replication stress. To accomplish this, we set out to generate human cells that were stably depleted of RECQ1 using a lentivirus approach. Stable depletion of RECQ1 was performed and analysis of RECQ1-depleted cells was undertaken. Stable RECQ1 lentiviral-knockdown cells were generated by using two independent shRNA from Sigma Aldrich. The two shRNA targeting the coding region of RECQ1 are described in Materials and Methods. A dose-dependent depletion was observed with the two shRNA employed: shRECQ1–1 resulted in a partial depletion (of about ~40%), while transfection with shRECQ1–2 resulted in about 95% depletion on average by western blot analysis ().
Figure 5. Generation and characterization of stable knockdown cells of RECQ1. (A) Stable depletion of RECQ1 was performed in HeLa cells by using the two independent and dose-dependent lentiviral shRNA, shRECQ1–1 and shRECQ1–2. (B) Growth assays were performed to assess the cell proliferation and cell growth for RECQ1 stable knockdown cells over a period for 5 d. MTT cell proliferation experiments were performed to evaluate the cell proliferation in the increasing concentrations of hydroxyurea (C), camptothecin (D) and 8-methoxypsoralen (E).
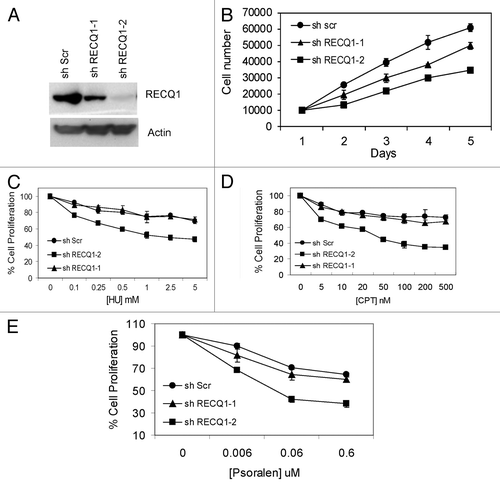
We first wanted to analyze the proliferative capacity of the stable RECQ1-knockdown cells. Stable depletion of RECQ1 was initially performed in HeLa cells, and growth assays were performed 48 h after selection of the knockdown cells in puromycin following lentiviral transduction. Upon reduction of the endogenous RECQ1 protein levels, the knockdown cells failed to proliferate at the same rate as their scrambled shRNA counterparts, as evidenced by the growth assay, and displayed a significant decrease in the cell number (). A correlated dose-dependent effect was observed, which is consistent with the depletion efficiency using the two independent lentiviral shRNA (). Our results are consistent with the previous observations that transient depletion of RECQ1 affected cell proliferation.Citation34-Citation37
Next, we wanted to analyze the sensitivity patterns of RECQ1 depletion. Using the two independent lentiviral shRNA constructs, depletion of RECQ1 was performed in both HeLa () and U2OS cells (Fig. S2A). Our results indicate that RECQ1 depletion in HeLa cells renders them sensitive to replication blocking agents (). MTT cell proliferation experiments indicate that these RECQ1 stable knockdown cells are sensitive to hydroxyurea (HU, ), camptothecin (CPT, ) and psoralen-induced interstrand cross-links (), all of which directly or indirectly block DNA replication fork progression. Similar results were also obtained with U2OS cells (Fig. S2). Consistent with the observations in HeLa cells, RECQ1-depleted U2OS cells displayed a defect in cell proliferation (Fig. S2B) and sensitivity toward replication-blocking agents (Fig. 2C–E). Psoralen produces covalent adducts with DNA bases on both strands of DNA and leads to the formation of DNA interstrand cross-links upon exposure with near-UV light. Interstrand cross-links prevent DNA strand separation and can thus act as absolute blocks to DNA replication and transcription.Citation38 While the partial knockdown of RECQ1 by using shRECQ1–1 did not show any significant differences in the sensitivity, the shRECQ1–2-transfected HeLa and U2OS cells are sensitive to these replication-blocking agents, further supporting a possible role of RECQ1 in DNA replication fork processing. For further experiments, shRECQ1–2 was employed for achieving complete depletion.
Loss of RECQ1 alters cell cycle distribution and activates DNA damage response signaling
To further investigate the cellular proliferation defect after RECQ1 depletion, cell cycle analysis of asynchronous cultures was performed (). Consistent with the growth assays, our FACS analysis indicated that depletion of RECQ1 in HeLa cells increased accumulation of cells in subG1 phase, indicating cell death with a decrease in G1 phase and an additional mild decrease in S-phase cells (). A slight increase in the number of G2/M cells was also observed. Similarly, transient depletion of RECQ1 in T98G cells (glioblastoma cell lines) was previously shown to alter cell cycle distribution, with an accumulation of G0/G1 cells and a decrease in S-phase population, with an effect on the rate of DNA synthesis with lower BrdU incorporation.Citation10 Consistently, BrdU staining in the RECQ1-depleted cells is relatively lower than in the scrambled cells (Fig. S3A).
Figure 6. Characterization of stable knockdown cells of RECQ1. (A) FACS analysis or cell cycle distribution of shScrambled and shRECQ1–2-depleted HeLa cells. (B) Stable depletion of RECQ1 accumulates spontaneous replication-associated double-strand breaks (DSBs). Immunostaining of RECQ1-knockdown cells for accumulation of 53BP1 at BrdU sites. (C) Stable depletion of RECQ1 leads to hyper phosphorylation of RPA32. (D) Western analysis showing activation of DNA damage response proteins like p-ATM (1981) and activation of CHK1 (Ser 345) by stable depletion of RECQ1.
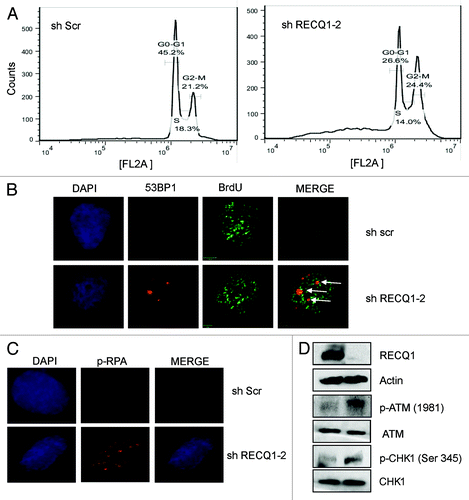
Depletion of RECQ1 leads to spontaneous accumulation of DNA double-strand breaks (DSBs), as evidenced by the accumulation of 53BP1 foci (Fig. S3B). About 5-fold more 53BP1 foci were observed upon stable depletion of RECQ1 compared with the control RNA-treated cells (Fig. S3C). More than 50% of the 53BP1 foci were positive and co-localized with the BrdU sites, indicating accumulation of replication-associated DSBs (). Further, we wanted to directly analyze whether a replication stress response was activated in the absence of RECQ1. It is well-known that RPA phosphorylation and CHK1 activation occurs in response to DNA replication fork stalling.Citation39 Interestingly, our immunofluorescence and western analysis indicated that stable depletion of RECQ1 leads to hyper-phosphorylation of RPA32 and activation of CHK1, respectively, possibly indicating accumulation of stalled replication forks in the absence of RECQ1. Phosphorylation of CHK1 (at ser 345) was shown to be increased by RPA binding to the ssDNA.Citation40 CHK1 suppresses origin firing during S-phase in response to replication blocksCitation41 and is required for continuous replication fork elongation.Citation42 Consistently, a defect in the frequency of origin firing was reported after transient depletion of RECQ1 along with a decrease in replication fork progression rate.Citation10 Phosphorylation of RPA and activation of CHK1 are also considered important consequences for the cellular signaling in response to DNA damage. More interestingly, RECQ1 depletion also led to activation of phospho-ATM (ser 1981), a primary DNA damage-response protein, and is consistent with the previous observations.Citation43 Collectively, our observations indicate possible accumulation of stalled or collapsed replication forks in RECQ1-depleted cells, which would eventually trigger DNA damage signaling.
Loss of RECQ1 leads to accumulation of under-condensed chromosomes
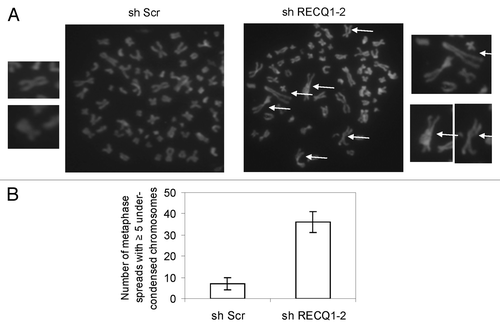
DNA replication is essential for proper condensation of chromosomes during mitosis, and aberrant DNA replication timing can result in defective chromosomal condensation.Citation44 We wanted to analyze if depletion of RECQ1 induces any defects in chromosomal condensation. Interestingly, we observed that stable depletion of RECQ1 accumulates under-condensed chromosomes (). Depletion of RECQ1 was performed in HeLa cells using shRECQ1–2 and resulted in nearly 5-fold greater accumulation of such under-condensed chromosomes (). Such abnormally condensed chromosomes with larger loops and reduced lateral condensation are typically observed in DNA replication mutants like ORC mutants and PCNA-depleted cells.Citation44 Reduced chromatin loading of PCNA was recently reported upon transient depletion of RECQ1.Citation10
Figure 7. Stable depletion of RECQ1 leads to accumulation of under-condensed chromosomes. (A) Representative images of the metaphase spreads analyzed from RECQ1-depleted cells. The arrows indicate under-condensed chromosomes. (B) Quantification of metaphase spreads from shScrambled and shRECQ1–2-depleted HeLa cells. Results are an average of two independent experiments with a minimum of 50 metaphase spreads analyzed.
Discussion
Extensive studies within the last decade indicate significant roles for human RecQ helicases in various DNA repair processes induced by DNA replication defects. Targeting DNA replication fork progression has been one of the recent developments in cancer therapy.Citation45 Interestingly, both RECQ1 and WRN are identified as potential therapeutic targets for various carcinomas.Citation46 Hence, it is important to study the differential or mechanistic involvement of the human RecQ helicases in replication fork progression. Here, we show that the human RECQ1 possesses the ability to promote strand exchange on synthetic forked DNA structures that mimic a stalled replication fork. This is an early demonstration of a potential role of RECQ1 in the processing of stalled replication forks. Previous studies indicate that BLM and WRN can unwind both the leading and lagging strands, indicating their significant roles in the reversal of stalled replication forks.Citation20,Citation47,Citation48 We observe that RECQ1 (but not RECQ4 or RECQ5β) can unwind both the leading and lagging arms of the stalled replication forks, similar to that of WRN, indicating that RECQ1 might have a similar role to that of WRN in helping cells deal with stalled replication forks. We also observed that RPA modulates the strand exchange activity of RECQ1 in a manner similar to that of WRN. Although the kinetics of strand exchange activity of RECQ1 are dramatically stimulated in the presence of RPA, the activity is more shifted to the unwinding mode (unlike RECQ4 or RECQ5β, but similar to that of WRN) irrespective of the leading-strand gap, which would trigger an efficient bypass and eventual removal of the lesion on the leading strand. These observations provide insight into understanding the comparative and differential involvement of the human RecQ helicases in DNA replication fork progression. Such studies would help us understand the underlying molecular basis of the clinical features of these syndromes in relation to their mechanistic roles in DNA replication and the maintenance of genomic stability. For example, it is interesting to note that, in vivo, lower rates for replication fork progression were observed in the absence of RECQ1Citation10 or WRN,Citation49 but not in the absence of RECQ4Citation10 or RECQ5β.Citation50
We observed that stable depletion of RECQ1 affects cell proliferation and renders the cells sensitive to replication-blocking agents like HU and CPT. Psoralen forms covalent monoadducts and interstrand cross-links in DNA upon exposure to near-UV (UVA) light. Interstrand cross-links (ICL) inhibit DNA replication by terminating chain elongation at the cross-link sites and thereby induce generation of replication-mediated double-strand breaks.Citation51 It was proposed that DNA replication is inhibited near the cross-link sites for two reasons: one is due to the inability of the two strands to unwind and another a physical blockage to polymerase progression.Citation51 In addition, the stalled DNA polymerase generates a unique DNA structure, where a structure-specific endonuclease can incise one of the template strands and results in a DNA double-strand break at one side of the growing fork and a gap on the other side. A major role of DNA replication in cross-link repair is formation of a “Y-shape” DNA structure at a stalled site.Citation51 These partially unwound structures exposing the single-strand/duplex junction near a cross-link can result in formation of stalled replication forks. Both HeLa and U2OS cells stably depleted of RECQ1 are sensitive to psoralen-induced interstrand cross-links. We speculate that RECQ1 might have a potential functional role at stalled replication forks created by ICL, but further studies are required to gain more insight into this interesting avenue. Further, our hypothesis of possible roles of RECQ1 in the repair of damaged replication forks and thereby accumulation of stalled/collapsed replication forks in the absence of RECQ1 would eventually trigger recombination repair and an increase in HR events (). Consistently, downregulation of RECQ1 was shown to trigger spontaneous Rad51 foci formation and an increase in SCEs,Citation35,Citation36 and this is also consistent with the finding that RECQ1 mouse embryonic fibroblasts (MEFs) are associated with chromosomal instability and an increase in the frequency of SCEs.Citation52 HR-mediated repair of broken replication forks can particularly result in the blockage of leading-strand extensions in the presence of bulky lesions.
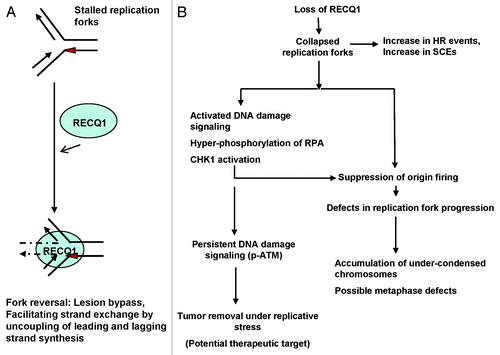
Figure 8. Model for the action of RECQ1 at stalled replication forks and its potential role in the maintenance of genomic stability. (A) The ability of RECQ1 to perform strand exchange at stalled replication fork structures may promote fork reversal, leading to fork stabilization. Red triangle represents a lesion encountered on the leading strand. (B) In vivo implication with a proposed biological role of RECQ1 in the maintenance of genomic stability. RECQ1 deficiency has biological consequences and induces defects in replication fork progression and activates DNA damage signaling.
Further, stable depletion of RECQ1 accumulates spontaneous DSBs and activates the DNA damage signaling cascade. Accumulation of phospho-ATM (ser 1981) was observed, indicating activation of DNA damage response signaling. More than 50% of the 53BP1 foci co-localized with the BrdU sites indicating accumulation of replication-associated DSBs. Hyper-phosphorylation of RPA32 and activation of CHK1 were typically observed indicating a possible defect in the replication fork progression. It would be interesting to investigate the role of RECQ1 in S-phase checkpoint and to further analyze the progression of RECQ1-depleted cells through S-phase in response to replicative stress.
Consistent with the previous observations,Citation10 depletion of RECQ1 alters cell cycle distribution, with lower numbers of S-phase cells being observed. Loss of RECQ1 would result in accumulation of stalled or collapsed replication forks that would eventually trigger DNA damage response signalingCitation29,Citation53 (). Chronic or persistent activation of DNA damage signaling (in specific phospho-ATM) has an important role in elimination of tumor cells that undergo excessive DNA replication stress.Citation54 Consistently, it was reported that RECQ1 silencing has anticancer effects, and that RECQ1 is considered a new suitable target for cancer therapyCitation34,Citation43,Citation55 ().
It is well-established that the density of functional replication origins and completion of S-phase are requirements for proper chromosomal condensation. Delay in the replication of an entire chromosome produces delays in chromosome condensation and results in under-condensed chromosomes at mitosisCitation56 (). A certain level of condensation occurs during S-phase and is also dependent on the distribution of origin firing.Citation44 Interestingly, RECQ1 was shown to accumulate at DNA replication origin sites, and transient depletion of RECQ1 was shown to reduce the probability of origin firing and further reduce the origin-sequence DNA.Citation10 If the majority of replication origins are inactive, more DNA is being pulled though fewer DNA polymerase complexes, which results in much larger chromosomal loops.Citation44 Such under-condensed chromosomes with larger loops and reduced lateral condensation is a phenotype of DNA replication mutants like MCM or ORC mutants, which undergo cell death by mitotic arrest.Citation44 Interestingly, we observe a similar phenotype of accumulation of under-condensed chromosomes upon stable depletion of RECQ1, indicating possible metaphase defects. Defects in DNA replication can trigger mitotic arrest, and improper or incomplete S-phase may influence the degree of chromosomal folding and mitotic checkpoints.Citation44 On the other hand, it was shown that activation of CHK1 blocks premature mitotic entry.Citation57 We speculate that RECQ1-deficient cells could be slower to traverse the mitotic phase of the cell cycle post-replication. Consistently, depletion of RECQ1 was recently proposed to kill the cancer cells (but not normal cells) by mitotic catastrophe.Citation43 Future studies are required to analyze spindle assembly and other possible metaphase defects in the absence of RECQ1. Collectively, our cellular studies implicate a unique and important role of human RECQ1 in replication fork progression and, consequently, a role in DNA damage response signaling and genome homeostasis.
Materials and Methods
DNA substrates
All oligonucleotides used for the preparation of DNA substrates were PAGE-purified and purchased from Integrated DNA Technologies (IDT). The sequences of the oligonucleotides (RS1–6) were shown in Table S1, and the schemes of the DNA substrates are shown where required. Oligonucleotides were labeled at the 5′-end using the γPCitation32-ATP with T4 polynucleotide kinase (New England Biolabs), and the substrates were generated as described , by annealing the respective oligonucleotides. Strand exchange and helicase assays were carried (in a 10 μl reaction volume) out at 37°C in the buffer containing 20 mM Tris-HCl, pH 7.5, 8 mM DTT, 5 mM MgCl2, 10 mM KCl, 10% glycerol and 80 μg/ml BSA. For the strand exchange assays, 1 nM RSK1/RS2 partial duplex was incubated with 20 nM RECQ1 and either 1 nM RS3 oligonucleotide or 1 nM RS3/RS4 partial duplex to generate forked DNA structures, as depicted in and , respectively. After 10 min, ATP was added to a final concentration of 5 mM, and 10 μl aliquots were removed at indicated time points (both before and after the addition of ATP) and analyzed. Control experiments were performed under similar conditions in the absence of RECQ1. Similar kinetic strand exchange assays were performed with the 3′-flap duplex using RECQ4 (50 nM), RECQ5β (20 nM) and WRN (5 nM). The substrate for these experiments was pre-formed by the spontaneous annealing of the constituent oligonucleotides RS1, RS2 and RS3, with the RS2 oligonucleotide being present in a 4-fold molar excess over the others. Helicase reactions were performed as described previously.Citation22 Where indicated, RPA (20 nM) was added to DNA before the addition of the RecQ proteins and ATP. The reactions were terminated by adding equal volumes of stop buffer (35 mM EDTA, 0.9% SDS, 25% Glycerol, 0.04% Bromophenol blue, 0.04% Xylene cyanol) and separated by non-denaturing PAGE.
Recombinant proteins
Recombinant His-tagged RECQ1 protein was overexpressed using a baculovirus/Sf9 insect system as previously described.Citation25 Recombinant RECQ5β was overexpressed in E. coli BL21(DE3) CodonPlus RIPL strain (Stratagene, Agilent Technologies) as a fusion protein with a self-cleaving intein-chitin-binding domain (CBD) and purified as previously described.Citation58 Recombinant RECQ4 with a C-terminal 9-histidine tag in the pGEX6p1 vector (GE Healthcare) was expressed and purified from E. coli Rosetta2 (DE3) (Novagen, EMD Millipore) as described previously.Citation30 Dual tagged WRN protein with N-terminal 6 × His and C-terminal InteinCBD tags was expressed and purified from High FiveTM insect cells as previously described.Citation59
Cell culture and generation of stable knockdown cells of RECQ1
Both HeLa and U2OS cells (from ATCC) were cultured in complete DMEM medium supplemented with 10% FBS, 1% Pen-Strep. We have previously used lentiviral shRNA to stably knockdown RECQ5β.Citation60 Here, we generate stable knockdown cells of RECQ1 by using similar approach of lentiviral transduction. pLKO.1 vector harboring either of the two lentiviral shRNA targeting the coding region of human RECQ1 were purchased from Sigma Aldrich and used for lentiviral transduction. The sequences used were: shRECQ1–1: CCGGGCCAATGAAATTCAGGTAGTACTCGAGTACTACCTGA
ATTTCATTGGCTTTTTG (TRCN0000051998) and shRECQ1–2: CCGGGCACATGCTATTACTATGCAACTCGAGTTGCATAGTAATAGCATGTGCTTTTTG (TRCN0000289591). shRNA construct expressing scrambled sequence (Plasmid #1864, deposited by Sabatini lab) was purchased from Addgene. For lentiviral transduction, 2 × 105 cells were seeded in 10-cm culture plates and transfected the following day with the lentivirus. Cells were split 48 h after transfection and selected in the presence of 2 µg/ml puromycin.
Growth assays
Scrambled and RECQ1-depleted HeLa cells were counted and plated 96h after transduction and 48h post-selection into 18 dishes, 10,000 cells/dish. Three dishes were harvested every 24 h and counted using a Coulter counter. The first set of three harvested on day 1 were used to normalize as plating controls. The data points represent the mean of three independent experiments with standard deviations.
Flow cytometry
To analyze the cell cycle distribution, the cells were harvested (96h after transduction and 48 h post-selection in puromycin) by trypsinization using the pre-used media to account for floating (mitotic) cells. The cells were then washed twice with PBS, fixed by 70% ice-cold ethanol and stored at -20°C. The cells were then washed twice with ice-cold PBS and resuspended with 10μg/ml propidium iodide (PI) solution with 1 mg/ml RNaseA. FACS was performed by Accuri C6 Flow cytometer (BD Biosciences) and analyzed using FlowJO software.
Analysis of metaphase chromosomes
Metaphase spreads were prepared from RECQ1-depleted HeLa cells. Cells were harvested and then incubated in 75mM KCl for 20min at 37°C, followed by fixation in ice-cold (3:1) methanol and glacial acetic acid. Metaphase spreads were then made by dropping the cells onto a glass slide and stained with DAPI. Images were captured using CytovisionTM software (Applied Imaging Corp.).
Cell proliferation assays
RECQ1 depletion was performed in both HeLa and U2OS cells as described above. RECQ1-depleted and scrambled cells were plated at 5,000 cells per well in 96-well plate along with the scrambled cells. Twenty-four hours after plating, the cells were incubated with variable concentrations of hydroxyurea (HU, 0–5 mM for 18 h), camptothecin (CPT, 0–500 nM for 18 h) or 8-methoxypsoralen (0–0.6 μM for 30 min) at 37°C. For psoralen treatment, cells were further subjected to exposure to 1.8 J/cm2 UVA in a Rayonet chamber, as previously described.Citation61 The cell proliferation was analyzed 48 h following the addition of DNA damaging agents using the Cell Proliferation Kit I (MTT) (Roche Applied Sciences) following manufacturer’s protocol. Briefly, 10 μl of MTT (lysis) reagent was added to the wells and incubated for 4–5 h at 37°C and then 100 μl of detergent (solubilizing buffer) was added followed by overnight incubation in dark at 37°C. The absorbance was read at dual wavelengths (570/650). The plots represent the normalized data and a mean of two independent experiments performed in triplicates with standard deviations.
Immunofluorescence and list of antibodies used
For immunofluorescence experiments, both shScrambled and shRECQ1–2-depleted cells (25,000 cells each) were seeded on four chambered slides (MatTek corporation) and grown overnight. Cells were then incubated with 50 μM BrdU for a 30 min pulse, where indicated, and immediately fixed with 3.7% formaldehyde in PBS for 10 min at room temperature. Primary antibodies used in the immunofluorescence experiments were rabbit anti-53BP1 antibody from Novus Biologicals (1:200), mouse monoclonal anti-BrdU antibody from BD Biosciences (1:50) and rabbit anti-phospho-RPA32 (S4/S8) antibody from Bethyl Labaratories (1:200). Other antibodies used are rabbit anti-RECQ1 antibody (H-110) from Santa Cruz, rabbit ant-phospho-Chk1 (Ser 345) antibody from Abcam, rabbit anti-ATM antibody from Epitomics, rabbit anti-Chk1 antibody from Santa Cruz.
| Abbreviations: | ||
| WRN | = | Werner syndrome helicase |
| SCEs | = | sister chromatid exchanges |
| RMI1 | = | RecQ-mediated genome instability 1 |
| HR | = | homologous recombination |
| HJ | = | Holliday junction |
| RPA | = | human replication protein A |
| HU | = | hydroxyurea |
| CPT | = | camptothecin |
| ICLs | = | interstrand cross-links |
| DSBs | = | double-strand breaks |
| MEFs | = | mouse embryonic fibroblasts |
Additional material
Download Zip (635.3 KB)Acknowledgments
We thank Drs. Chandrika Cangovi and Takashi Tadokoro for critical reading of the manuscript. We thank Dr. Mahesh Ramamoorthy for his technical expertise in the preparation of lentivirus. We thank Dr. Sommers Joshua for the expression and purification of recombinant RECQ1. This work was supported by funds from the Intramural Research Program of the National Institute on Aging, NIH, AG000726–20.
Disclsoure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem Sci 2008; 33:609 - 20; http://dx.doi.org/10.1016/j.tibs.2008.09.003; PMID: 18926708
- Singh DK, Ghosh AK, Croteau DL, Bohr VA. RecQ helicases in DNA double strand break repair and telomere maintenance. Mutat Res 2012; 736:15 - 24; http://dx.doi.org/10.1016/j.mrfmmm.2011.06.002; PMID: 21689668
- Cheng WH, Muftic D, Muftuoglu M, Dawut L, Morris C, Helleday T, et al. WRN is required for ATM activation and the S-phase checkpoint in response to interstrand cross-link-induced DNA double-strand breaks. Mol Biol Cell 2008; 19:3923 - 33; http://dx.doi.org/10.1091/mbc.E07-07-0698; PMID: 18596239
- Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science 2004; 306:1951 - 3; http://dx.doi.org/10.1126/science.1103619; PMID: 15591207
- Raynard S, Bussen W, Sung P. A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIalpha, and BLAP75. J Biol Chem 2006; 281:13861 - 4; http://dx.doi.org/10.1074/jbc.C600051200; PMID: 16595695
- Hu Y, Lu X, Barnes E, Yan M, Lou H, Luo G. Recql5 and Blm RecQ DNA helicases have nonredundant roles in suppressing crossovers. Mol Cell Biol 2005; 25:3431 - 42; http://dx.doi.org/10.1128/MCB.25.9.3431-3442.2005; PMID: 15831450
- Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev 2007; 21:3073 - 84; http://dx.doi.org/10.1101/gad.1609107; PMID: 18003859
- Puranam KL, Blackshear PJ. Cloning and characterization of RECQL, a potential human homologue of the Escherichia coli DNA helicase RecQ. J Biol Chem 1994; 269:29838 - 45; PMID: 7961977
- Kawabe T, Tsuyama N, Kitao S, Nishikawa K, Shimamoto A, Shiratori M, et al. Differential regulation of human RecQ family helicases in cell transformation and cell cycle. Oncogene 2000; 19:4764 - 72; http://dx.doi.org/10.1038/sj.onc.1203841; PMID: 11032027
- Thangavel S, Mendoza-Maldonado R, Tissino E, Sidorova JM, Yin J, Wang W, et al. Human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation. Mol Cell Biol 2010; 30:1382 - 96; http://dx.doi.org/10.1128/MCB.01290-09; PMID: 20065033
- Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, Dunphy WG, et al. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell 2005; 121:887 - 98; http://dx.doi.org/10.1016/j.cell.2005.05.015; PMID: 15960976
- Xu X, Rochette PJ, Feyissa EA, Su TV, Liu Y. MCM10 mediates RECQ4 association with MCM2-7 helicase complex during DNA replication. EMBO J 2009; 28:3005 - 14; http://dx.doi.org/10.1038/emboj.2009.235; PMID: 19696745
- Cordeiro-Stone M, Zaritskaya LS, Price LK, Kaufmann WK. Replication fork bypass of a pyrimidine dimer blocking leading strand DNA synthesis. J Biol Chem 1997; 272:13945 - 54; http://dx.doi.org/10.1074/jbc.272.21.13945; PMID: 9153257
- Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell 2006; 21:15 - 27; http://dx.doi.org/10.1016/j.molcel.2005.11.015; PMID: 16387650
- Barbour L, Xiao W. Regulation of alternative replication bypass pathways at stalled replication forks and its effects on genome stability: a yeast model. Mutat Res 2003; 532:137 - 55; http://dx.doi.org/10.1016/j.mrfmmm.2003.08.014; PMID: 14643434
- Bugreev DV, Rossi MJ, Mazin AV. Cooperation of RAD51 and RAD54 in regression of a model replication fork. Nucleic Acids Res 2011; 39:2153 - 64; http://dx.doi.org/10.1093/nar/gkq1139; PMID: 21097884
- Cox MM. Recombinational DNA repair of damaged replication forks in Escherichia coli: questions. Annu Rev Genet 2001; 35:53 - 82; http://dx.doi.org/10.1146/annurev.genet.35.102401.090016; PMID: 11700277
- Sogo JM, Lopes M, Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 2002; 297:599 - 602; http://dx.doi.org/10.1126/science.1074023; PMID: 12142537
- Zou H, Rothstein R. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell 1997; 90:87 - 96; http://dx.doi.org/10.1016/S0092-8674(00)80316-5; PMID: 9230305
- Kanagaraj R, Saydam N, Garcia PL, Zheng L, Janscak P. Human RECQ5beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res 2006; 34:5217 - 31; http://dx.doi.org/10.1093/nar/gkl677; PMID: 17003056
- Machwe A, Xiao L, Lloyd RG, Bolt E, Orren DK. Replication fork regression in vitro by the Werner syndrome protein (WRN): holliday junction formation, the effect of leading arm structure and a potential role for WRN exonuclease activity. Nucleic Acids Res 2007; 35:5729 - 47; http://dx.doi.org/10.1093/nar/gkm561; PMID: 17717003
- Popuri V, Bachrati CZ, Muzzolini L, Mosedale G, Costantini S, Giacomini E, et al. The Human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J Biol Chem 2008; 283:17766 - 76; http://dx.doi.org/10.1074/jbc.M709749200; PMID: 18448429
- Bugreev DV, Brosh RM Jr., Mazin AV. RECQ1 possesses DNA branch migration activity. J Biol Chem 2008; 283:20231 - 42; http://dx.doi.org/10.1074/jbc.M801582200; PMID: 18495662
- Mazina OM, Rossi MJ, Deakyne JS, Huang F, Mazin AV. Polarity and bypass of DNA heterology during branch migration of Holliday junctions by human RAD54, BLM, and RECQ1 proteins. J Biol Chem 2012; 287:11820 - 32; http://dx.doi.org/10.1074/jbc.M112.341347; PMID: 22356911
- Sharma S, Sommers JA, Choudhary S, Faulkner JK, Cui S, Andreoli L, et al. Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J Biol Chem 2005; 280:28072 - 84; http://dx.doi.org/10.1074/jbc.M500264200; PMID: 15899892
- Pike AC, Shrestha B, Popuri V, Burgess-Brown N, Muzzolini L, Costantini S, et al. Structure of the human RECQ1 helicase reveals a putative strand-separation pin. Proc Natl Acad Sci USA 2009; 106:1039 - 44; http://dx.doi.org/10.1073/pnas.0806908106; PMID: 19151156
- Muzzolini L, Beuron F, Patwardhan A, Popuri V, Cui S, Niccolini B, et al. Different quaternary structures of human RECQ1 are associated with its dual enzymatic activity. PLoS Biol 2007; 5:e20; http://dx.doi.org/10.1371/journal.pbio.0050020; PMID: 17227144
- Wang H, Guan J, Wang H, Perrault AR, Wang Y, Iliakis G. Replication protein A2 phosphorylation after DNA damage by the coordinated action of ataxia telangiectasia-mutated and DNA-dependent protein kinase. Cancer Res 2001; 61:8554 - 63; PMID: 11731442
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003; 300:1542 - 8; http://dx.doi.org/10.1126/science.1083430; PMID: 12791985
- Rossi ML, Ghosh AK, Kulikowicz T, Croteau DL, Bohr VA. Conserved helicase domain of human RecQ4 is required for strand annealing-independent DNA unwinding. DNA Repair (Amst) 2010; 9:796 - 804; http://dx.doi.org/10.1016/j.dnarep.2010.04.003; PMID: 20451470
- Brosh RM Jr., Waheed J, Sommers JA. Biochemical characterization of the DNA substrate specificity of Werner syndrome helicase. J Biol Chem 2002; 277:23236 - 45; http://dx.doi.org/10.1074/jbc.M111446200; PMID: 11956187
- Capp C, Wu J, Hsieh TS. Drosophila RecQ4 has a 3′-5′ DNA helicase activity that is essential for viability. J Biol Chem 2009; 284:30845 - 52; http://dx.doi.org/10.1074/jbc.M109.008052; PMID: 19759018
- Ozsoy AZ, Ragonese HM, Matson SW. Analysis of helicase activity and substrate specificity of Drosophila RECQ5. Nucleic Acids Res 2003; 31:1554 - 64; http://dx.doi.org/10.1093/nar/gkg243; PMID: 12595564
- Futami K, Ogasawara S, Goto H, Yano H, Furuichi Y. RecQL1 DNA repair helicase: A potential tumor marker and therapeutic target against hepatocellular carcinoma. Int J Mol Med 2010; 25:537 - 45; PMID: 20198302
- Mendoza-Maldonado R, Faoro V, Bajpai S, Berti M, Odreman F, Vindigni M, et al. The human RECQ1 helicase is highly expressed in glioblastoma and plays an important role in tumor cell proliferation. Mol Cancer 2011; 10:83; http://dx.doi.org/10.1186/1476-4598-10-83; PMID: 21752281
- Sharma S, Brosh RM Jr.. Human RECQ1 is a DNA damage responsive protein required for genotoxic stress resistance and suppression of sister chromatid exchanges. PLoS One 2007; 2:e1297; http://dx.doi.org/10.1371/journal.pone.0001297; PMID: 18074021
- Wu Y, Brosh RM Jr.. Distinct roles of RECQ1 in the maintenance of genomic stability. DNA Repair (Amst) 2010; 9:315 - 24; http://dx.doi.org/10.1016/j.dnarep.2009.12.010; PMID: 20061189
- Noll DM, Mason TM, Miller PS. Formation and repair of interstrand cross-links in DNA. Chem Rev 2006; 106:277 - 301; http://dx.doi.org/10.1021/cr040478b; PMID: 16464006
- Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev 2001; 15:2177 - 96; http://dx.doi.org/10.1101/gad.914401; PMID: 11544175
- Choi JH, Lindsey-Boltz LA, Kemp M, Mason AC, Wold MS, Sancar A. Reconstitution of RPA-covered single-stranded DNA-activated ATR-Chk1 signaling. Proc Natl Acad Sci USA 2010; 107:13660 - 5; http://dx.doi.org/10.1073/pnas.1007856107; PMID: 20616048
- Petermann E, Woodcock M, Helleday T. Chk1 promotes replication fork progression by controlling replication initiation. Proc Natl Acad Sci USA 2010; 107:16090 - 5; http://dx.doi.org/10.1073/pnas.1005031107; PMID: 20805465
- Elvers I, Hagenkort A, Johansson F, Djureinovic T, Lagerqvist A, Schultz N, et al. CHK1 activity is required for continuous replication fork elongation but not stabilization of post-replicative gaps after UV irradiation. [Epub ahead of print] Nucleic Acids Res 2012; 40:8440 - 8; http://dx.doi.org/10.1093/nar/gks646; PMID: 22753029
- Futami K, Kumagai E, Makino H, Goto H, Takagi M, Shimamoto A, et al. Induction of mitotic cell death in cancer cells by small interference RNA suppressing the expression of RecQL1 helicase. Cancer Sci 2008; 99:71 - 80; PMID: 17953710
- Pflumm MF, Botchan MR. Orc mutants arrest in metaphase with abnormally condensed chromosomes. Development 2001; 128:1697 - 707; PMID: 11290306
- Petermann E, Helleday T. DNA replication-associated lesions: importance in early tumorigenesis and cancer therapy. Biochem Soc Trans 2007; 35:1352 - 4; http://dx.doi.org/10.1042/BST0351352; PMID: 17956349
- Arai A, Chano T, Futami K, Furuichi Y, Ikebuchi K, Inui T, et al. RECQL1 and WRN proteins are potential therapeutic targets in head and neck squamous cell carcinoma. Cancer Res 2011; 71:4598 - 607; http://dx.doi.org/10.1158/0008-5472.CAN-11-0320; PMID: 21571861
- Machwe A, Xiao L, Groden J, Orren DK. The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry 2006; 45:13939 - 46; http://dx.doi.org/10.1021/bi0615487; PMID: 17115688
- Ralf C, Hickson ID, Wu L. The Bloom’s syndrome helicase can promote the regression of a model replication fork. J Biol Chem 2006; 281:22839 - 46; http://dx.doi.org/10.1074/jbc.M604268200; PMID: 16766518
- Sidorova JM, Li N, Folch A, Monnat RJ Jr.. The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle 2008; 7:796 - 807; http://dx.doi.org/10.4161/cc.7.6.5566; PMID: 18250621
- Popuri V, Jing H, Ramamoorthy V, Tadokoro T, Croteau DL, Bohr VA. RECQL5 plays both co-operative and complementary roles with WRN syndrome helicase. Nucleic Acids Res 2012; In press
- Bessho T. Induction of DNA replication-mediated double strand breaks by psoralen DNA interstrand cross-links. J Biol Chem 2003; 278:5250 - 4; http://dx.doi.org/10.1074/jbc.M212323200; PMID: 12473662
- Sharma S, Stumpo DJ, Balajee AS, Bock CB, Lansdorp PM, Brosh RM Jr., et al. RECQL, a member of the RecQ family of DNA helicases, suppresses chromosomal instability. Mol Cell Biol 2007; 27:1784 - 94; http://dx.doi.org/10.1128/MCB.01620-06; PMID: 17158923
- Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 2004; 73:39 - 85; http://dx.doi.org/10.1146/annurev.biochem.73.011303.073723; PMID: 15189136
- Jinushi M. Chronic activation of DNA damage signals causes tumor immune evasion in the chemoresistant niche. Oncoimmunology 2012; 1:400 - 2; http://dx.doi.org/10.4161/onci.19123; PMID: 22737630
- Futami K, Kumagai E, Makino H, Sato A, Takagi M, Shimamoto A, et al. Anticancer activity of RecQL1 helicase siRNA in mouse xenograft models. Cancer Sci 2008; 99:1227 - 36; http://dx.doi.org/10.1111/j.1349-7006.2008.00794.x; PMID: 18422747
- Breger KS, Smith L, Thayer MJ. Engineering translocations with delayed replication: evidence for cis control of chromosome replication timing. Hum Mol Genet 2005; 14:2813 - 27; http://dx.doi.org/10.1093/hmg/ddi314; PMID: 16115817
- Matsuyama M, Goto H, Kasahara K, Kawakami Y, Nakanishi M, Kiyono T, et al. Nuclear Chk1 prevents premature mitotic entry. J Cell Sci 2011; 124:2113 - 9; http://dx.doi.org/10.1242/jcs.086488; PMID: 21628425
- Garcia PL, Liu Y, Jiricny J, West SC, Janscak P. Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J 2004; 23:2882 - 91; http://dx.doi.org/10.1038/sj.emboj.7600301; PMID: 15241474
- Tadokoro T, Kulikowicz T, Dawut L, Croteau DL, Bohr VA. DNA binding residues in the RQC domain of Werner protein are critical for its catalytic activities. Aging (Albany NY) 2012; 4:417 - 29; PMID: 22713343
- Ramamoorthy M, Tadokoro T, Rybanska I, Ghosh AK, Wersto R, May A, et al. RECQL5 cooperates with Topoisomerase II alpha in DNA decatenation and cell cycle progression. Nucleic Acids Res 2012; 40:1621 - 35; http://dx.doi.org/10.1093/nar/gkr844; PMID: 22013166
- Muniandy PA, Thapa D, Thazhathveetil AK, Liu ST, Seidman MM. Repair of laser-localized DNA interstrand cross-links in G1 phase mammalian cells. J Biol Chem 2009; 284:27908 - 17; http://dx.doi.org/10.1074/jbc.M109.029025; PMID: 19684342