A common hallmark of many fungal species is the capacity to undergo cellular morphogenesis programs, which, for fungal pathogens, play critical roles in sexual reproduction, nutrient acquisition and virulence.Citation1 Fungal morphogenesis comprises a diversity of processes,Citation1,Citation2 ranging from spore germination and branching in filamentous fungi such as the pathogen Aspergillus fumigatus, arthroconidia production in dermatophyte fungal pathogens, filamentous mold to yeast morphogenesis of dimorphic fungal pathogens, such as Histoplasma capsulatum, and the morphogenetic transition from yeast to filamentous growth in the model yeast Saccharomyces cerevisiae and pathogenic yeast Candida albicans. Morphogenesis can also influence fungal mating in the pathogenic fungus Cryptococcus neoformans,Citation3 or control nutrient acquisition under starvation conditions, as observed for S. cerevisiae.Citation4 Importantly, for C. albicans, morphological changes can facilitate tissue invasion, enhance biofilm formation and promote host immune evasion, making morphogenesis a crucial component of fungal virulence.Citation2
Given that morphogenesis is fundamental to fungal development and virulence traits, it is perhaps not surprising that it is subject to elaborate molecular regulation.Citation2 Even in the well-characterized S. cerevisiae model system, our understanding of the regulatory circuitry involved remains incomplete. Therefore, we undertook a global analysis of the genetic determinants that govern the key morphogenetic transition from yeast to filamentous growth in two distinct fungal species: the model yeast S. cerevisiae, and the leading fungal pathogen of humans, C. albicans, which are separated by ~200–800 million years of evolution.Citation5 We constructed a genome-wide collection of deletion mutants in the S. cerevisiae Σ1278b strain and screened this library, covering almost the entire S. cerevisiae genome, for genes involved in three distinct aspects of S. cerevisiae filamentation: haploid invasive growth, diploid pseudohyphal growth and biofilm formation.Citation6 We similarly screened two C. albicans homozygous deletion mutant libraries,Citation7,Citation8 representing ~13% of the C. albicans genome, for genes involved in two facets of C. albicans filamentation: filamentous growth in liquid medium containing serum and wrinkly colony morphology on solid Spider medium.Citation6 Together, this work provided the first global and comparative analysis of filamentation between two fungal species and revealed unique sets of genes underpinning filamentation under different conditions, highlighting striking examples of conservation and divergence in signaling between S. cerevisiae and C. albicans.
Environmental signals that govern morphogenesis are distinct between S. cerevisiae and C. albicans. For instance, S. cerevisiae pseudohyphal growth occurs in response to nitrogen-limiting conditions,Citation9 while C. albicans filamentation is induced by a diversity of environmental cues in addition to nutrient limitation, including alkaline pH, elevated CO2 and elevated temperature.Citation2 Surprisingly hundreds of genes influence specific filamentous growth programs. For instance, 474 of 680 (~70%) of genes involved in S. cerevisiae diploid pseudohyphal growth are unique for this process, including a specific set of polyamine biosynthetic genes, and overall 970 of 1415 (~50%) of all genes found to influence S. cerevisiae filamentation are unique to one aspect of filamentation. The same is observed in C. albicans, where ~52% and ~61% of genes are involved uniquely in liquid filamentation or solid filamentation, respectively. Similarly, a study that screened C. albicans transcription factor mutants for filamentation defects under a variety of conditions found that many genes had an impact in only a limited set of conditions.Citation7 This suggests that a majority of genes involved in morphogenesis have specialized functions for enabling filamentous growth in response to specific environmental cues rather than more global functions in enabling polarized growth.
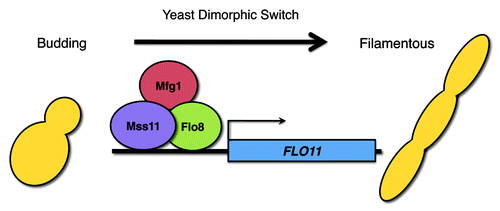
Despite the distinct genetic architecture underlying each of the different morphogenetic growth programs, we also identified a core set of S. cerevisiae genes involved in all of the filamentous growth programs tested.Citation6 This includes the previously uncharacterized transcriptional regulator Mfg1, which we found to be a key regulator of morphogenesis in both S. cerevisiae and C. albicans under all environmental conditions tested.Citation6 Mfg1 forms a complex with two known transcriptional regulators, Flo8 and Mss11, to control the expression of hundreds of genes, including some key morphogenetic determinants, such as the S. cerevisiae flocculin encoded by FLO11 (). Our analysis raises the question as to whether core genes are more predictive of being involved in morphogenesis across these two species. However, of the 43 orthologous genes we identified that were involved in morphogenesis in both S. cerevisiae and C. albicans, the majority are not core morphogenetic regulators. In fact, 26 of 43 (~60%) of S. cerevisiae genes with a conserved role in morphogenesis between species are only involved in a specific facet of S. cerevisiae filamentation: haploid invasive growth, pseudohyphal growth or biofilm formation. This suggests that morphogenetic regulators that are conserved across evolutionary time include specialized regulators that control one aspect of morphogenesis in response to a specific cue, as well as core regulators that play a more universal role in morphogenesis.
Figure 1. Transcriptional regulators Flo8, Mss11 and the newly identified Mfg1 form a complex and control expression of key morphogenetic determinants in both C. albicans and S. cerevisiae, thereby regulating the transitions from yeast to filamentous growth. This model depicts these three transcription factors regulating FLO11 in S. cerevisiae, but C. albicans lacks a FLO11 ortholog, suggesting that the targets of this complex have been rewired between the two species.
Given the diversity of fungal morphogenetic growth programs, and the range of conditions that can influence fungal morphogenesis,Citation1 it will be of interest to determine to what degree morphogenetic regulation is conserved among other fungal species. Certain pathways, such as the cAMP-protein kinase A (PKA) pathway, play a conserved role in morphogenesis in species as diverse as S. cerevisiae, C. albicans, C. neoformans and A. fumigatus;Citation2 however, there has been no large-scale comparative analysis between other fungal species to date. As additional functional genomic resources become available for important fungal pathogens, such as C. neoformans,Citation10 our work will provide a powerful platform to assess how morphogenetic regulatory circuitry has been conserved or rewired among divergent fungal species, offering broad insights into biology, disease and evolution.
References
- Moore D. Fungal Morphogenesis. Cambridge University Press; 2003.
- Shapiro RS, et al. Microbiol Mol Biol Rev 2011; 75:213 - 67; http://dx.doi.org/10.1128/MMBR.00045-10; PMID: 21646428
- Lin X, et al. Annu Rev Microbiol 2006; 60:69 - 105; http://dx.doi.org/10.1146/annurev.micro.60.080805.142102; PMID: 16704346
- Cullen PJ, et al. Proc Natl Acad Sci USA 2000; 97:13619 - 24; http://dx.doi.org/10.1073/pnas.240345197; PMID: 11095711
- Heckman DS, et al. Science 2001; 293:1129 - 33; http://dx.doi.org/10.1126/science.1061457; PMID: 11498589
- Ryan O, et al. Science 2012; 337:1353 - 6; http://dx.doi.org/10.1126/science.1224339; PMID: 22984072
- Homann OR, et al. PLoS Genet 2009; 5:e1000783; http://dx.doi.org/10.1371/journal.pgen.1000783; PMID: 20041210
- Noble SM, et al. Nat Genet 2010; 42:590 - 8; http://dx.doi.org/10.1038/ng.605; PMID: 20543849
- Gimeno CJ, et al. Cell 1992; 68:1077 - 90; http://dx.doi.org/10.1016/0092-8674(92)90079-R; PMID: 1547504
- Liu OW, et al. Cell 2008; 135:174 - 88; http://dx.doi.org/10.1016/j.cell.2008.07.046; PMID: 18854164