Abstract
It is now largely accepted that ribosomal proteins may be implicated in a variety of biological functions besides that of components of the translation machinery. Many evidences show that a subset of ribosomal proteins are involved in the regulation of the cell cycle and apoptosis through modulation of p53 activity. In addition, p53-independent mechanisms of cell cycle arrest in response to alterations of ribosomal proteins availability have been described. Here, we identify human rpL3 as a new regulator of cell cycle and apoptosis through positive regulation of p21 expression in a p53-independent system. We demonstrate that the rpL3-mediated p21 upregulation requires the specific interaction between rpL3 and Sp1. Furthermore, in our experimental system, p21 overexpression leads to a dual outcome, activating the G₁/S arrest of the cell cycle or the apoptotic pathway through mitochondria, depending on its intracellular levels. It is noteworthy that depletion of p21 abrogates both effects. Taken together, our findings unravel a novel extraribosomal function of rpL3 and reinforce the proapoptotic role of p21 in addition to its widely reported ability as an inhibitor of cell proliferation.
Introduction
p21 (also known as p21waf1/cip1), a main inhibitor of cyclin-dependent kinases (CDKs), is a negative regulator of cell cycle progression; various regulatory mechanisms contribute to finely coordinate multiple biological functions of p21 and determine its intracellular levels. They include transcriptional regulation, epigenetic silencing, mRNA translation and stability and ubiquitin-dependent as well as ubiquitin-independent degradation of the protein.Citation1,Citation2
In addition to growth-inhibiting activities, p21 can induce or repress p53-dependent or p53-independent apoptosis depending on the cellular system or the nature of the stress.Citation3-Citation5 In fact, p21 gene overexpression in human laryngeal squamous carcinoma cells lacking p53 protein expression enhances apoptosis in cisplatin-treated cells, whereas it attenuates apoptotic signals in methotrexate-treated cells.Citation6
A variety of transcription factors can modulate p21 in addition to p53.Citation7 The p21 promoter consists of proximal and distal regulatory elements. The majority of p21 gene regulatory proteins interact with the proximal region of p21 promoter that includes six GC-rich binding sites recognized by Sp1, Sp3 and Kruppel-like transcription factors.Citation8,Citation9 Numerous evidences support the essential role of Sp1 in the upregulation of p21 gene by several protein factors such as c-MycCitation10 or drugs, such as Mithramycin A, Sulforaphane and 5-FU.Citation11,Citation12
In eukaryotes, many data indicate that r-proteins exert a variety of extra-ribosomal functions. Alteration in the expression of r-proteins and protein factors involved in the ribosome biogenesis correlate with tumorigenesis, since many of these proteins are overexpressed in solid tumors and leukemia cells.Citation13,Citation14 On the other hand, some data suggest that several r-proteins might have a role as tumor suppressors, as described in zebrafish.Citation15 Recently, an increasing number of r-proteins have been proposed as additional important components of the tumor suppressor p53’s autoregulatory feedback loop, activating p53 and triggering cell cycle arrest and apoptosis.Citation16 In fact, either drug treatment or enforced expression of rpL11, rpL5, rpL23 causes p53-dependent cell-cycle arrest through HDM2 (human double minute 2) interaction.Citation17-Citation19 In addition to r-proteins, several nucleolar proteins such as NPM (Nucleophosmin, also known as B23, numatrin or NO38) and Nucleostemin are able to modulate p53 activity.Citation20,Citation21 Furthermore, emerging evidences indicate a functional correlation between NPM and some r-proteins, independent from ribosome biogenesis. It has been shown that rpL23 regulates NPM and Miz1-dependent p21 gene promoter transactivation by sequestering NPM into the nucleolus.Citation22 We have studied in recent years post-transcriptional regulatory strategies of mammalian r-proteins,Citation23 and we demonstrated that human r-protein L3 (rpL3) autoregulates, in a complex with the transacting factors hnRNP H1, NPM and KHSRP, its own expression through the association of alternative splicing and nonsense-mediated mRNA decay (AS-NMD).Citation24,Citation25 In this paper, we demonstrate that rpL3 is a positive regulator of p21 expression. We show that rpL3 is able to modulate the protein amounts of p21 through a novel pathway independent from p53 involving Sp1 and NPM. Furthermore, we observed that rpL3-mediated p21 upregulation results in cell cycle arrest or apoptosis depending on the intracellular levels of p21 protein.
Table 1. Primers used in PCR reactions
Results
rpL3 upregulates p21 expression at transcriptional levels independently from p53
In order to investigate whether rpL3 could control p21 expression in a cell environment devoid of p53, we analyzed p21 mRNA and protein levels by western blotting and semiquantitative RT-PCR upon rpL3 overexpression in Calu-6 cells lacking p53 protein expression.Citation23 The enforced expression of the HA-rpL3 protein resulted in a dose-dependent increase of p21 protein and mRNA ().
Figure 1. Role of rpL3 in the regulation of p21 expression. (A) Protein samples from Calu-6 cells untransfected or transiently transfected with increasing amounts (0, 0.5, 1, 2 μg) of a plasmid expressing rpL3 fused to HA tag (pHA-rpL3) were analyzed by WB assay, with antibodies directed against the HA tag and p21, 24 h after transfection. Loading in the gel lanes was controlled by detection of α-tubulin protein. (B) Representative RT-PCR analysis of total RNA from the same samples. β-actin was used as a control of RNA loading. Quantification of p21 protein and mRNA levels by PhosphorImager (Bio-Rad) is shown. (C) Protein extracts from Calu-6 cells transiently transfected with the full-length p21 promoter luciferase reporter plasmid (pWWP) alone and cotransfected with pWWP and increasing amounts of pHA-rpL3 or pL7a-His (0, 0.5, 1, 2 and 4 μg) were analyzed by WB assay with antibodies directed against the HA and His tags, 24 h after transfection. Loading in the gel lanes was controlled by detection of α-tubulin protein. Analysis of the relative luciferase activity, normalized against Renilla Luciferase (pRL) activity, in the same samples. (D) Protein extracts from Calu-6 cells transiently transfected with CMV-Luc plasmid alone, or cotransfected with pCMV-Luc and 2 μg of pHA-rpL3 or pL7a-His, were analyzed by WB assay with antibodies directed against the HA and His tags. Loading in the gel lanes was controlled by detection of α-tubulin protein. The relative luciferase activity, normalized against Renilla Luciferase (pRL) activity, in the same samples was detected. Results illustrated in – are representative of three independently performed experiments.
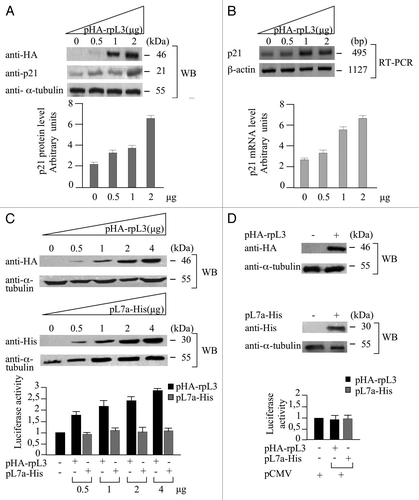
To investigate the possibility that p21 upregulation is due to the ability of rpL3 to control the activity of p21 promoter, we performed a reporter luciferase assay in conditions of rpL3 overexpression.
For the specificity of the assay, a plasmid expressing rpL7a-His (pL7a-His), an arbitrary ribosomal protein, was used as control.Citation26 As shown in , the enforced expression of rpL3, in contrast to rpL7a, is associated to the upregulation of p21 gene promoter transactivation in a dose-dependent manner.
In addition, we performed analogous experiments by using pCMV, a plasmid containing the CMV promoter fused to luciferase DNA, as a further control (). The results showed that rpL3, as rpL7a, failed to enhance reporter gene expression driven by CMV promoter.
rpL3 binds the p21 gene promoter
We investigated whether rpL3 was able to interact with p21 gene promoter. To this aim, Calu-6 cells were collected and subjected to ChIP by using anti-rpL3 and anti-IgG as specificity control.
The presence of rpL3 was investigated by western blotting in the DNA-immunoprecipitated complexes (). Quantitative PCR assay on the samples was performed by using primers specific for proximal region of p21 promoter (-194–+88),Citation27 which is the region interacting with the majority of p21 regulatory proteins, and primers for control loci.
Figure 2. Analysis of the interaction between rpL3 and p21 gene promoter. Protein samples from DNA-rpL3 or DNA-IgG immunocomplexes were analyzed through WB assay with antibody against rpL3. Note the absence of signal in DNA-IgG immunocomplex. The same DNA-immunoprecipitates were subjected to quantitative PCR (qPCR) with primers specific for the proximal region of p21 gene promoter or control loci.
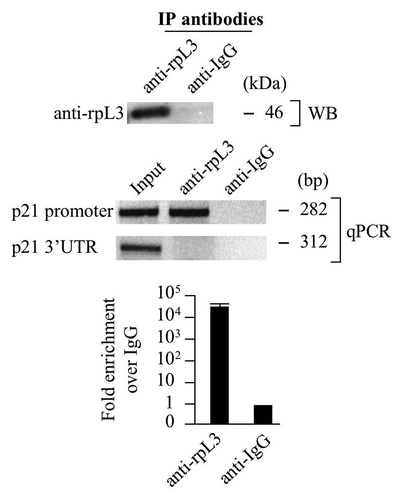
The presence of a specific signal for p21 promoter in the DNA-rpL3 immunocomplex, absent in the control challenged with anti-IgG antibodies, indicated that rpL3 is able to bind p21 promoter ().
Next, in order to evaluate whether rpL3 was able to interact directly with p21 proximal promoter region, an EMSA experiment was performed using purified recombinant GST-tagged rpL3 and GST as control. No mobility shift was observed when recombinant GST-rpL3 was challenged with the probe, suggesting that the binding of rpL3 to the p21 gene promoter is indirect, and it may require additional protein factors (data not shown).
NPM modulates rpL3-dependent p21 gene promoter transactivation
The absence of a direct binding between rpL3 and p21 promoter prompted us to search for rpL3-interacting regulatory proteins involved in the rpL3-mediated regulation of p21 transcription. We have previously reported a direct protein-protein interaction between rpL3 and NPM and demonstrated that this interaction is functional to the regulation of the rpL3 gene alternative splicing.Citation24
Here, we explored the possibility that the rpL3-NPM complex was involved in the transactivation of the p21 gene transcription.
To this purpose, we performed analogous ChIp experiments by using anti-rpL3, anti-NPM and anti-IgG as control. As shown in , the presence of NPM and rpL3 proteins (WB), and a signal for p21 gene promoter (qPCR) in the reciprocally DNA-immunoprecipitated complexes, absent in the control obtained with anti-IgG antibodies, were consistent with the presence of rpL3 and NPM on the p21 gene promoter.
Figure 3. Role of NPM in the rpL3-mediated p21 gene transcriptional transactivation. (A) Analysis of the interaction between NPM and rpL3 on p21 gene promoter. Protein samples from DNA-rpL3, DNA-NPM or DNA-IgG immunocomplexes were analyzed through WB assay with antibodies against the indicated proteins. Note the absence of signal in IgG immunocomplex. The same DNA-immunoprecipitates were subjected to quantitative PCR (qPCR) with primers specific for the proximal region of p21 gene promoter or control loci. (B) Effect of NPM overexpression on rpL3-mediated p21 gene promoter activity. Protein extracts from Calu-6 cells transiently transfected with pWWP alone or cotrasfected with pWWP, and the plasmids expressing the recombinant Flag-NPM (pFlag-NPM), with or without pHA-rpL3, were analyzed by WB assay with antibodies directed against the HA and Flag tags. Loading in the gel lanes was controlled by detection of α-tubulin protein. The relative luciferase activity, normalized against Renilla Luciferase (pRL) activity, in the same samples was detected. (C) Effects of RNAi-mediated depletion of NPM on rpL3-mediated p21 gene promoter activity. Protein extracts from Calu-6 cells transiently transfected with pWWP alone or cotrasfected with pWWP and siRNA directed against endogenous NPM mRNA (siRNA-NPM) in presence or absence of pHA-rpL3 were analyzed by WB assay with antibodies directed against the HA tag and NPM. Loading in the gel lanes was controlled by detection of α-tubulin protein. The relative luciferase activity, normalized against Renilla Luciferase (pRL) activity, in the same samples was detected.
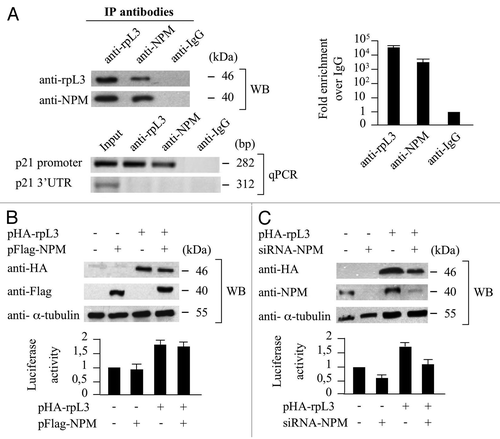
To investigate whether the interaction between rpL3 and NPM on p21 promoter affects its activity, a reporter luciferase assay was performed by altering intracellular levels of NPM in conditions of HA-rpL3 overexpression. The results demonstrated that either in rpL3 physiological conditions or rpL3 overexpression, the enforced expression of NPM did not affect the activity of p21 gene promoter (). Whereas, in physiological conditions of rpL3 expression, the NPM silencing was associated with the inhibition of p21 transcriptional activity as indicated by a significant decrease (about 40%) in p21 gene promoter activity (). Of interest, in rpL3 overexpression conditions, NPM silencing resulted in a decrease of rpL3-dependent transactivation of p21 gene promoter. These data suggest a positive role of NPM in rpL3-mediated activity on p21 gene promoter.
rpL3-dependent p21 gene promoter transactivation requires Sp1
In the attempt to identify other proteins involved in the rpL3-mediated activation of p21 transcription, we focused our study on the possible role of Sp1, a protein factor that regulates p21 transcription.Citation8 We first determined whether Sp1 was present in the rpL3-p21 gene promoter immunocomplex. To this purpose, a ChIp experiment in condition of mythramycin A (MTM) treatment was performed. MTM is a specific Sp1 inhibitor that is able to displace Sp1 from its binding sites in numerous genes.Citation11 The shows the presence of Sp1 and rpL3 proteins (WB), and a signal for p21 gene promoter (qPCR), in the reciprocally DNA-immunoprecipitated complexes, not detected in the control obtained with anti-IgG antibodies, in absence of MTM, indicating that a specific association between Sp1 and rpL3 occurs on the p21 gene promoter.
Figure 4. Analysis of the interaction between rpL3 and Sp1 on p21 gene promoter. Protein samples from DNA-rpL3, DNA-Sp1 or DNA-IgG immunocomplexes from Calu-6 cells untreated or treated with 200 nM of mythramycin A (MTM) for 16 h were analyzed through WB assay with antibodies against rpL3 or Sp1. Note the absence of signal in DNA-IgG immunocomplex. The same DNA immunoprecipitates were subjected to quantitative PCR (qPCR) with primers specific for the proximal region of p21 gene promoter or control loci.
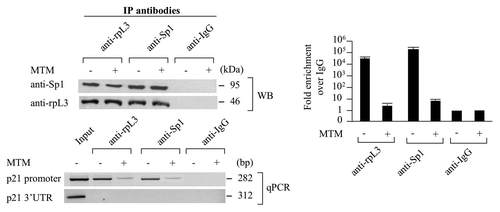
Of interest, the presence of MTM prevents the binding of both rpL3 and Sp1 on the p21 gene promoter, indicating that rpL3 is able to bind p21 gene promoter only in the presence of Sp1.
In order to study the function of Sp1 in rpL3-dependent transactivation of p21 gene promoter, a reporter luciferase assay was performed in conditions of rpL3 and Sp1 overexpression, with or without MTM. shows that in absence of MTM, rpL3 or Sp1 overexpression increased the activity of p21 gene promoter, as previously observed (see sub-section “rpL3 upregulates p21 expression at transcriptional level independently from p53” in this paper and ref. Citation28). In addition, rpL3 and Sp1 had a synergistic effect on the transactivation of p21 gene promoter. Interestingly, the ability of both rpL3 and Sp1 to activate the transcription of luciferase was strongly reduced in the presence of MTM. These data suggest that the ability of rpL3 to activate p21 transcription requires the presence of Sp1 on the promoter.
Figure 5. Role of Sp1 in the rpL3-mediated p21 gene transcriptional transactivation. (A) Protein extracts from Calu-6 cells untreated or treated with 200 nM of mythramycin A (MTM) and transiently transfected with pWWP alone, or in the presence of pHA-rpL3 or the plasmid expressing the recombinant Sp1-His (pSp1-His) or pHA-rpL3 and pSp1-His were analyzed by WB assay with antibodies directed against the HA and His tags, 24 h after transfection. Loading in the gel lanes was controlled by detection of α-tubulin protein. The relative luciferase activity, normalized against Renilla Luciferase (pRL) activity, in the same samples was detected. (B) Analysis of p21 promoter regions involved in rpL3-mediated p21 gene transcriptional activation. On the left, schematic representation of the deletion constructs of the p21 gene promoter used in the luciferase transfection assay. Calu-6 cells were transiently transfected with the pWP124, pWP101 or pWPdel-SmaI in presence or absence of pHA-rpL3. Proteins were analyzed by WB assay with antibody directed against the HA tag 24 h after transfection. Loading in the gel lanes was controlled by detection of α-tubulin protein. Analysis of the relative luciferase activity, normalized against Renilla Luciferase (pRL) activity, in the same samples is shown.
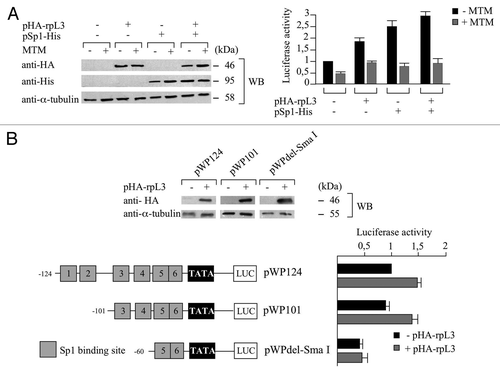
Sp1-binding boxes 3 and 4 are involved in rpL3-dependent transactivation of p21 gene promoter
The proximal region of the human p21 promoter carries six GC-rich Sp1-binding sites (numbered 1–6 in plasmid pWP124, ). To determine which of Sp1-binding elements mediate rpL3 responsiveness, Calu-6 cells were transiently transfected with pW124 or mutant plasmids in which two or four Sp1-binding sites were simultaneously deleted (pWP101 and pWPdel-SmaI)Citation29 in presence or absence of pHA-rpL3 (). As shown in , upon the overexpression of rpL3, deletion of Sp1-binding sites 1 and 2 (pWP101) led to an increase in the luciferase activity comparable to that obtained when all the six Sp1 binding motifs were present (pWP124), whereas additional deletion of Sp1-binding sites 3 and 4 (pWPdel-SmaI) abrogated completely rpL3’s effect on p21 gene promoter transactivation. These data suggest that the region spanning from -60–-101 upstream from the transcription start site, including Sp1-binding sites 3 and 4, corresponds to rpL3-responsive element, and it is involved in rpL3-mediated activation of p21 gene promoter.
rpL3 overexpression induces cell cycle arrest or apoptosis in Calu-6 cells
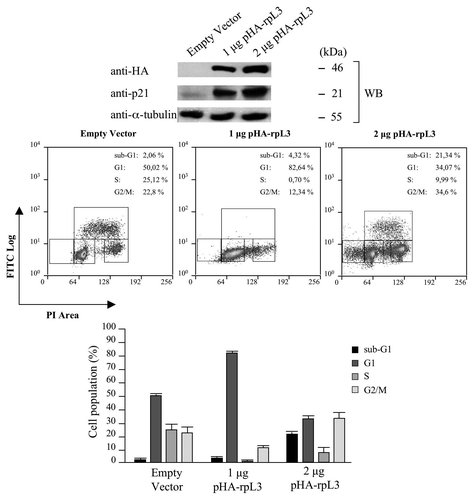
In order to investigate the effect of rpL3-mediated upregulation of p21 transcription on cell proliferation, we performed 5-Bromodeoxyuridine (BrdU) incorporation to monitor DNA synthesis during cell division and propidium iodide (PI) staining to analyze DNA content of Calu-6 cells transiently transfected with 1 μg or 2 μg of pHA-rpL3. As expected, rpL3 overexpression induced an increase of p21 expression in a dose-dependent manner. shows that BrdU incorporation in cells transfected with 1 μg of pHA-rpL3 was significantly decreased vs. that observed in control (about 0.7% vs. 25%, respectively). This indicated that the enforced expression of rpL3 produced a strong reduction in the percentage of cells in S-phase of the cell cycle. Analysis of cell cycle using the counterstaining with PI indicated that the observed decrease of cells in S-phase was associated with an increase of cell population in the G1-phase of the cell cycle (about 80% vs. control 50%). Surprisingly, in the presence of higher rpL3 levels (2 µg of pHA-rpL3), we observed only a mild effect on the cell cycle, but rather the presence of a significant sub-G1 population, indicative of apoptosis (about 21% vs. control 2%).
Figure 6. Effects of rpL3-mediated upregulation of p21 gene expression on the cell cycle. (A) Protein samples from Calu-6 cells transfected with 2 μg of pcDNAHAV3 (empty vector), 1 μg or 2 μg of pHA-rpL3 were analyzed by WB assay with antibodies directed against the HA tag and p21. Loading in the gel lanes was controlled by detection of α-tubulin protein. (B) The same samples were stained with FITC conjugated anti-5-bromodeoxyuridine antibody and counterstained with propidium iodide and analyzed for DNA synthesis and for DNA content by flow cytometry 48 h after transfection. On the bottom, the percentage of cells in different phases of cell cycle is shown.
Intracellular levels of rpL3 affect the choice between G1/S arrest or mitochondrial apoptosis in Calu-6 cells
To gain further insight into the mechanism by which rpL3 causes cell cycle arrest and apoptosis, we proceeded to perform a time-course analysis of the nuclei DNA content of Calu-6 cells transiently transfected with 1 μg or 2 µg of pHA-rpL3 by flow cytometry ().
Figure 7. Effects of rpL3-mediated upregulation of p21 gene expression on the cell viability. Calu-6 cells transfected with 2 μg of pcDNAHAV3 (empty vector) or 1 μg and 2 μg of pHA-rpL3 were analyzed for DNA content by propidium iodide staining 24 h, 48 h and 72 h after transfection. On the bottom, a quantification of percentage of cells in sub-G1 phase is shown.
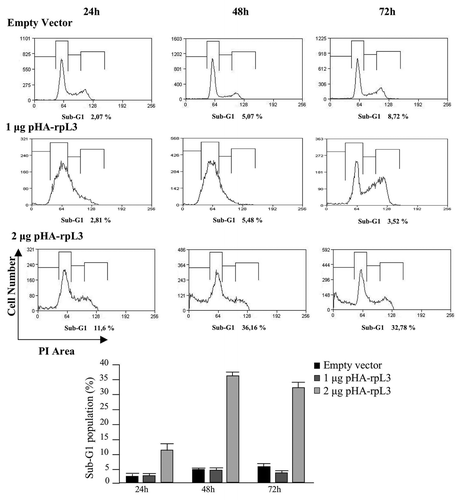
In cells transfected with 2 μg of pHA-rpL3, an increase in sub-G1 cell population vs. control was observed at all time points, although the strongest effect was obtained after 48 h. At this time point, the percentage of cells transfected with 2 µg of pHA-rpL3 in the sub-G1 phase of the cell cycle increased approximately 7-fold as compared with control (about 36% vs. 5%, respectively). This effect was not observed with 1 μg of pHA-rpL3. Transfection of cells with this amount of pHA-rpL3 did not induce apoptosis even 72 h after transfection. Furthermore, we observed a G1/S arrest at 24 and 48 h (as already shown), and more interestingly, at 72 h, the dividing ability of Calu-6 cells transfected with 1 µg pHA-rpL3 was restored, as evidenced by the presence of a G2/M peak ().
These results suggest that rpL3 may induce G1/S arrest or apoptosis depending on its concentration inside the cells.
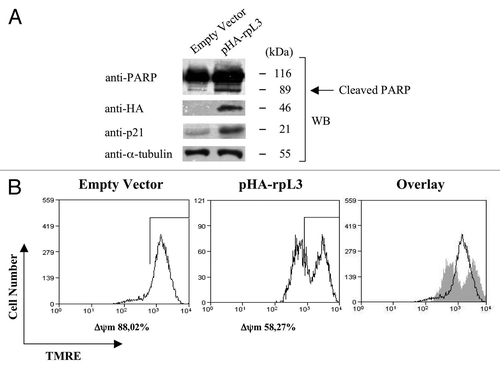
To further study the apoptotic effect of rpL3, we analyzed PARP protein cleavage by western blotting analysis and reduction of mitochondrial inner membrane potential (ΔΨm) by tetramethyl-rhodamine ethyl ester (TMRE) staining, hallmarks of mitochondrial apoptosis,Citation30,Citation31 in Calu-6 cells transfected with 2 μg of pHA-rpL3. As expected, in these cells, western blot for PARP revealed an increased amount of cleaved PARP (). Furthermore, shows that the percentage of Calu-6 cells with a physiological ΔΨm decreased from 88% to 58% (control cells vs. 2 μg of pHA-rpL3 transfected cells). Overall, these results indicate that higher levels of rpL3 protein obtained with 2 μg of pHA-rpL3 activate apoptosis through mitochondrial pathway in Calu-6 cells.
Figure 8. Characterization of rpL3-mediated apoptosis in Calu-6 cells. (A) Calu-6 cells transfected with 2 μg of pcDNAHAV3 (empty vector) or pHA-rpL3 were analyzed by WB assay with antibodies directed against PARP, the HA tag and p21, 48 h after transfection. Loading in the gel lanes was controlled by detection of α-tubulin protein. (B) The same samples were analyzed for mitochondrial membrane potential by tetramethyl-rhodamine ethyl ester (TMRE) staining, and fluorescence was measured by flow cytometry 48 h after transfection.
Cell cycle arrest and apoptosis upon rpL3 overexpression are p21-dependent
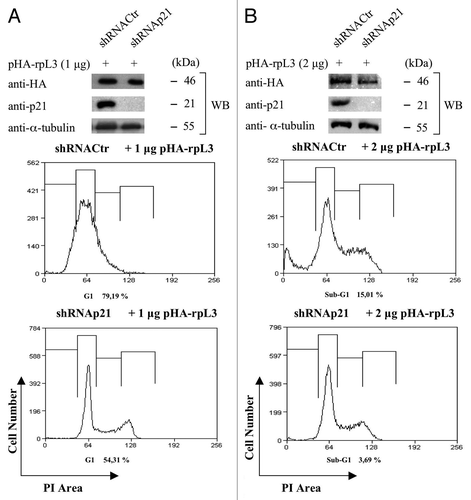
To verify that rpL3-induced cell cycle arrest or apoptosis in Calu-6 cells occurs through a specific pathway involving p21, we examined the effects of rpL3 overexpression on cell proliferation and cell death after p21 silencing.
We performed analysis of the nuclei DNA content of Calu-6 cells and p21-depleted Calu-6 (p21ΔCalu-6) cells transiently transfected with 1 μg or 2 μg of pHA-rpL3 by flow cytometry. (lower panels) shows that rpL3 was unable to induce cell cycle arrest in condition of p21 depletion. In addition, as shown in (lower panels), the higher dosage of rpL3 failed to mediate the induction of apoptosis in p21-depleted cells, thus demonstrating that the dual effect of rpL3 on cell cycle arrest or apoptosis is dependent from p21. All these results indicate that rpL3 is able to induce cell cycle arrest or activate the intrinsic apoptotic pathway in Calu-6 cells by regulating the intracellular amounts of p21.
Figure 9. Role of p21 on rpL3-mediated cell cycle arrest and apoptosis. (A) Protein extracts from Calu-6 cells and p21ΔCalu-6 cells transiently transfected with 1 μg of pHA-rpL3 were analyzed by WB assay with antibodies directed against the HA tag and p21. Loading in the gel lanes was controlled by detection of α-tubulin protein. The same samples were analyzed for DNA content by propidium iodide staining, and fluorescence was measured by flow cytometry 24 h after transfection. (B) Protein extracts from Calu-6 cells and p21ΔCalu-6 transiently transfected with 2 μg of pHA-rpL3 were analyzed by WB assay with antibodies directed against the HA tag and p21. Loading in the gel lanes was controlled by detection of α-tubulin protein. The same samples were analyzed for DNA content by propidium iodide staining, and fluorescence was measured by flow cytometry 24 h after transfection.
Discussion
In eukaryotes, ribosome biogenesis is a complex process that requires a number of coordinated events prior to nuclear export of the mature subunits to the cytoplasm.Citation32-Citation34 Recent studies have shed new light on the mechanisms underlying the regulation of these events and revealed new connections between ribosome biogenesis and cell cycle.Citation35 The signaling pathways able to connect these two processes have long been relatively unknown; however, more recently, the role of r-proteins-Mdm2-p53 stress response pathway in the coregulation of these events has been widely demonstrated.Citation36,Citation37 Alterations in processes such as rRNAs synthesis, rRNAs modification as well as r-proteins imbalance produce alterations in ribosome biogenesis and result in nucleolar stress. In response to this event, several r-proteins translocate to the nucleoplasm and bind to Mdm2, thus promoting p53 stabilization and subsequent p53-mediated cell cycle arrest or apoptosis.Citation38 Several p53-independent pathways that require nucleolar proteins, such as ARF, NPM or free r-proteins, have also been well-studied.Citation39,Citation40
Data reported in this paper allow us to define a new extraribosomal function for human rpL3 in addition to its role in the regulation of the alternative splicing of its own gene.Citation23 Specifically, we demonstrate that rpL3 is able to induce cell cycle arrest or apoptosis in Calu-6 cells by positively modulating the activity of p21 promoter in a p53-independent pathway. To our knowledge, the present study demonstrates for the first time a direct role of an r-protein, specifically rpL3, in the regulation of p21 transcription.
In an attempt to understand the mechanism of the rpL3-mediated upregulation of p21 expression, we focused our studies on the identification of protein partners of rpL3 protein involved in this activity. It is known that NPM is able to indirectly bind to p21 proximal promoter and activate its transcription.Citation22 Furthermore, our previous results demonstrated that NPM is able to interact directly with rpL3, and that this interaction is functional to the regulation of the rpL3 protein levels inside the cells.Citation24 Data reported in this paper clearly demonstrate that NPM represents a positive regulator of rpL3-mediated transactivation of p21 gene, although it is not a crucial player in this event ().
It is well-known that p21 proximal promoter contains six Sp1 binding sites, and that Sp1 is able to transcriptionally regulate the activity of different genes including those involved in the regulation of cell cycle.Citation8 Specifically, Sp factors have been proposed to be essential for transactivation of the p21 promoter by members of the p53 family proteins.Citation41 Here, we demonstrate that rpL3 associates in vivo with Sp1 on the p21 proximal promoter only in absence of MTM. Although treatment with MTM did not prevent the formation of a complex between rpL3 and Sp1 into the cells, it hindered its recruitment on p21 promoter (). Data from rpL3 and Sp1 overexpression in Calu-6 cells in absence of MTM indicated that expression of Sp1, in combination with rpL3, strongly increased rpL3-mediated p21 transactivation; these findings, together with the observation that rpL3, even in excess, was unable to transactivate p21 promoter in presence of MTM, strongly indicated that the binding of Sp1 to the proximal p21 promoter region is essential for the rpL3-mediated transactivation of p21 promoter. Thus, we propose that Sp1 is a key component of rpL3-mediated p21 transactivation; in fact, Sp1 binding to GC boxes 3 and 4 is a prerequisite for rpL3 recruitment on p21 proximal promoter ().
In the light of these results, we propose a working model in which overexpression of rpL3, which may mimic situations of alteration in ribosome biogenesis, leads to a formation of a multiprotein complex containing at least rpL3, Sp1 and NPM on the proximal region of p21 promoter with consequent upregulation of p21 expression. This upregulation leads to a dual effect; arresting the cell cycle progression at G1/S phase or promoting apoptosis in dependence of rpL3 and p21 protein levels into the cells (). To better understand the dynamics of these events, we performed a time-dependent analysis of the DNA content in Calu-6 cells transfected with different amounts of pHA-rpL3. Interestingly, we observed that 1 μg of pHA-rpL3 induced a reversible G1/S arrest of the cell cycle. In fact, Calu-6 cells restored their capability to progress through the G1/S transition at 72 h after transfection. Instead, transfection with 2 μg of pHA-rpL3 triggered mitochondrial apoptosis even at 24 h after transfection ( and ). Moreover, we show that p21 is essential for rpL3-induced cell cycle arrest or mitochondrial apoptosis, since stable depletion of p21 in Calu-6 cells completely abolished both events (). Taken together, these data demonstrate that rpL3 plays an important role in the activation, through modulation of p21 levels, of two independent pathways; i.e., reversible arrest of cell cycle or mitochondrial apoptosis.
These data are in complete agreement with recent evidences demonstrating a much more complex scenario than had been expected regarding p21’s role in the regulation of cell cycle. In particular, p21 has proved to exert also a proapoptotic function under certain conditions in specific systems beyond the well-known role in growth inhibition.Citation42 Notably, in most of these cases, the cellular systems used express a non-functional p53 isoform. The mechanism by which p21 may promote apoptosis is poorly understood, and different working models have been proposed. However, the common assumption is that the cellular activities of p21 are tightly regulated by multiple factors that seem to be specific for each function in different systems. For example, the interaction of p21 with PCNA is implicated in sodium butyrate-induced apoptosis,Citation43 other studies indicate that the modulation of proapoptotic or antiapoptotic genes is responsible for p21-induced apoptosis.Citation44,Citation45
Data reported in this paper indicate that rpL3 in a complex with Sp1 and NPM is able to positively transactivate p21 promoter. The p21 overexpression induced by this complex could trigger two distinct pathways: cell cycle arrest or mitochondrial apoptosis in a context devoid of p53. Although the molecular details underlining these mechanisms remain to be clarified, we speculate that the intracellular levels of p21 achieved upon modulation of the rpL3 amounts are determinant of cell fate. In the lights of these observations, our working model predicts that the intracellular levels of rpL3 could be crucial in regulating the hierarchy of interactions of p21 with multiple factors, leading to the assembly of specific protein complexes, which could trigger two distinct effects: cell cycle arrest or apoptosis.
At the present, the challenge is to identify the p21 molecular partners involved in the cellular response to rpL3 overexpression. This will help to elucidate the role of ribosomal proteins in the control of cell cycle and cell death through p21-dependent molecular pathways.
Materials and Methods
Cell cultures, transfections and drug treatment
Human cell line Calu-6Citation23 was cultured in Dulbecco’s modified Eagle’s medium (DMEM) with glutamax (Invitrogen) supplemented with 10% fetal bovine serum (FBS), 0.1 mM non-essential amino acids (Euroclone), 2 mM L-glutamine and penicillin-streptomycin 50 U/ml.
p21ΔCalu-6 cell line, derived from Calu-6 cell line and stably silenced for p21, was grown in DMEM supplemented with 10% fetal bovine serum (FBS), 0.1 mM non-essential amino acids (Euroclone), 2 mM L-glutamine, penicillin-streptomycin 50 U/ml and 0.5 μg/ml puromycin (Sigma-Aldrich).
siRNA transfections were performed in Calu-6 cells (2.5 × 106 cells, 6 mm-well plate) at a concentration of 150 nM using Oligofectamine Reagent (Invitrogen) according to the manufacturer’s instructions.
Plasmids were transfected in Calu-6 cells and p21ΔCalu-6 cells (2.5 × 106 cells, 60 mm-well plate) by using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instruction.
After DNA transfections, RNA and proteins were extracted by using the Trizol procedure (Invitrogen) for RT-PCR analysis and western blot, respectively.
Transfection efficiency was assessed by cotransfecting a GFP-expressing vector and normalizing RNA levels against GFP mRNA levels (data not shown).
Calu-6 cells (2.5 × 106 cells, 60 mm-well plate) were treated with 200 nM Mithramycin A (Sigma-Aldrich) for 16 h before transfections or chromatin immunoprecipitation.
Plasmids and production of recombinant proteins
The cDNAs of NPM and Sp1 were obtained by RT-PCR from Calu-6 cells using specific primers () and cloned into a version of the eukaryotic expression vector pcDNA3.1/His C (Invitrogen) containing the FLAG epitope and into a version of the eukaryotic expression vector pcDNA3.1/HisMyc A (Invitrogen) containing the His epitope, respectively. The constructs were verified by DNA sequencing.
The plasmids encoding HA-rpL3, HA-NPM, rpL7a-His and GST-rpL3 were available.Citation24 The plasmids pWWP, pWP124, pWP101 and pWPdel-SmaI were kindly provided by Dr. Yoshihiro Sowa.Citation29
The recombinant proteins GST-rpL3 and GST were produced as previously reported.Citation24
RNA interference
The siRNAs targeting NPM were purchased from Santa Cruz Biotechnology (Santa Cruz Biotechnology).
To stably silence p21, Calu-6 cells were seeded the day before transfection in a 60-mm plate, and 24 h later they were transfected with 2 μg of a p21 short hairpin RNA (shRNA) expressing pSM2c vector contained in the Expression Arrest™ human shRNA library (Open Biosystems). Transfected cells were selected with 1 μg/ml of puromycin (Sigma-Aldrich) for 7 d, and then the p21 depletion was evaluated by western blotting.
The sense strand of the p21 shRNA sequence is: 5′- TGCTGTTGACAGTGAGCGACCAGCCTCTGGCATTAGAATTTAGTGAAGCCACAGATGTAAATTCTAATGCCAGAGGCTGGGTGCCTACTGCCTCGGA-3′.
A short hairpin non-silencing construct was used as control.
PCR
Reverse transcriptase-PCR analysis was performed as described previously.Citation24 The primers used to amplify p21 cDNA and β-actin, were reported in .
Real-time PCR was used to analyze DNA in ChIP experiments as described in this section (see sub-section “Chromatin immunoprecipitation”).
Western blotting
Western blotting assay was performed as described previously,Citation24 using antibodies anti-rpL3 (Primm), anti-FLAG (Sigma-Aldrich), anti-Sp1 (Millipore), anti-p21, anti-NPM, anti-HA, anti-His, anti-PARP and anti-α-tubulin (Santa Cruz Biotechnology).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assay was performed as previously reported,Citation27 using anti-rpL3, anti-NPM, anti-Sp1 antibodies or normal mouse IgG. Aliquots of DNA-proteins immunocomplexes were resolved by 12% SDS-gel electrophoresis and transferred into nitrocellulose filters analyzed by western blotting. The chromatin was extracted from the remaining aliquots of DNA-proteins immunocomplexes, the protein-DNA crosslink was reversed, and the proteins were digested with proteinase K. DNA was then purified and analyzed by real-time PCR analysis with the primers reported in (p21 promoter -194/+88 and p21 3′-UTR +3455/+3771), using SYBR GreenEr qPCR SuperMix Universal (Invitrogen) following the manufacturer's instructions.
Dual luciferase assay
Luciferase assays were performed with the Dual-Glo Luciferase assay system (Promega) following manufacturer’s instructions. Samples were read with Turner Luminometer and expressed as relative luciferase, i.e., RT/RC, where RT and RC are (Firefly luciferase)/(Renilla luciferase).
Flow cytometry
Cell cycle analysis, BrdU (5′-Bromo-2'-deoxyuridine) incorporation and tetramethylrhodamine ethyl ester (TMRE) staining were performed as previously reported.Citation46,Citation47 Samples were analyzed by a CyAn ADP Flow Cytometer (DAKOCytomation) and quantified using Summit Software.
Acknowledgments
The Authors wish to thank Prof. Angelo Lupo for mediating our contact with Dr. Yoshihiro Sowa, who donated the p21 promoter plasmids and Prof. Tommaso Russo for kindly providing the plasmid expressing p21 shRNA. This work was supported by Regione Campania, L5/2002 (grant number A.10057.RUSGRC05C, 2005).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Jung YS, Qian Y, Chen X. Examination of the expanding pathways for the regulation of p21 expression and activity. Cell Signal 2010; 22:1003 - 12; http://dx.doi.org/10.1016/j.cellsig.2010.01.013; PMID: 20100570
- Chang LJ, Eastman A. Decreased translation of p21waf1 mRNA causes attenuated p53 signaling in some p53 wild-type tumors. Cell Cycle 2012; 11:1818 - 26; http://dx.doi.org/10.4161/cc.20208; PMID: 22510560
- Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 2009; 9:400 - 14; http://dx.doi.org/10.1038/nrc2657; PMID: 19440234
- Tsao YP, Huang SJ, Chang JL, Hsieh JT, Pong RC, Chen SL. Adenovirus-mediated p21((WAF1/SDII/CIP1)) gene transfer induces apoptosis of human cervical cancer cell lines. J Virol 1999; 73:4983 - 90; PMID: 10233960
- Yang HL, Pan JX, Sun L, Yeung SC. p21 Waf-1 (Cip-1) enhances apoptosis induced by manumycin and paclitaxel in anaplastic thyroid cancer cells. J Clin Endocrinol Metab 2003; 88:763 - 72; http://dx.doi.org/10.1210/jc.2002-020992; PMID: 12574211
- Kraljevic Pavelic S, Cacev T, Kralj M. A dual role of p21waf1/cip1 gene in apoptosis of HEp-2 treated with cisplatin or methotrexate. Cancer Gene Ther 2008; 15:576 - 90; http://dx.doi.org/10.1038/cgt.2008.28; PMID: 18483502
- Macleod KF, Sherry N, Hannon G, Beach D, Tokino T, Kinzler K, et al. p53-dependent and independent expression of p21 during cell growth, differentiation, and DNA damage. Genes Dev 1995; 9:935 - 44; http://dx.doi.org/10.1101/gad.9.8.935; PMID: 7774811
- Koutsodontis G, Moustakas A, Kardassis D. The role of Sp1 family members, the proximal GC-rich motifs, and the upstream enhancer region in the regulation of the human cell cycle inhibitor p21WAF-1/Cip1 gene promoter. Biochemistry 2002; 41:12771 - 84; http://dx.doi.org/10.1021/bi026141q; PMID: 12379120
- Wu J, Lingrel JB. KLF2 inhibits Jurkat T leukemia cell growth via upregulation of cyclin-dependent kinase inhibitor p21WAF1/CIP1. Oncogene 2004; 23:8088 - 96; http://dx.doi.org/10.1038/sj.onc.1207996; PMID: 15361832
- Gartel AL, Ye X, Goufman E, Shianov P, Hay N, Najmabadi F, et al. Myc represses the p21(WAF1/CIP1) promoter and interacts with Sp1/Sp3. Proc Natl Acad Sci USA 2001; 98:4510 - 5; http://dx.doi.org/10.1073/pnas.081074898; PMID: 11274368
- Koutsodontis G, Kardassis D. Inhibition of p53-mediated transcriptional responses by mithramycin A. Oncogene 2004; 23:9190 - 200; PMID: 15489892
- Chew YC, Adhikary G, Wilson GM, Xu W, Eckert RL. Sulforaphane induction of p21(Cip1) cyclin-dependent kinase inhibitor expression requires p53 and Sp1 transcription factors and is p53-dependent. J Biol Chem 2012; 287:16168 - 78; http://dx.doi.org/10.1074/jbc.M111.305292; PMID: 22427654
- Kondoh N, Shuda M, Tanaka K, Wakatsuki T, Hada A, Yamamoto M. Enhanced expression of S8, L12, L23a, L27 and L30 ribosomal protein mRNAs in human hepatocellular carcinoma. Anticancer Res 2001; 21:4A 2429 - 33; PMID: 11724303
- Mduff FK, Hook CE, Tooze RM, Huntly BJ, Pandolfi PP, Turner SD. Determining the contribution of NPM1 heterozygosity to NPM-ALK-induced lymphomagenesis. Lab Invest 2011; 91:1298 - 303; http://dx.doi.org/10.1038/labinvest.2011.96; PMID: 21709672
- Amsterdam A, Sadler KC, Lai K, Farrington S, Bronson RT, Lees JA, et al. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol 2004; 2:E139; http://dx.doi.org/10.1371/journal.pbio.0020139; PMID: 15138505
- Castro ME, Leal JF, Lleonart ME, Ramon Y Cajal S, Carnero A. Loss-of-function genetic screening identifies a cluster of ribosomal proteins regulating p53 function. Carcinogenesis 2008; 29:1343 - 50; http://dx.doi.org/10.1093/carcin/bgm302; PMID: 18515283
- Deisenroth C, Zhang Y. Ribosome biogenesis surveillance: probing the ribosomal protein-Mdm2-p53 pathway. Oncogene 2010; 29:4253 - 60; http://dx.doi.org/10.1038/onc.2010.189; PMID: 20498634
- Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol Cell 2008; 32:180 - 9; http://dx.doi.org/10.1016/j.molcel.2008.08.031; PMID: 18951086
- Morgado-Palacin L, Llanos S, Serrano M. Ribosomal stress induces L11- and p53-dependent apoptosis in mouse pluripotent stem cells. Cell Cycle 2012; 11:503 - 10; http://dx.doi.org/10.4161/cc.11.3.19002; PMID: 22262176
- Colombo E, Marine JC, Danovi D, Falini B, Pelicci PG. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat Cell Biol 2002; 4:529 - 33; http://dx.doi.org/10.1038/ncb814; PMID: 12080348
- Lo D, Lu H. Nucleostemin: Another nucleolar “Twister” of the p53-MDM2 loop. Cell Cycle 2010; 9:3227 - 32; http://dx.doi.org/10.4161/cc.9.16.12605; PMID: 20703089
- Wanzel M, Russ AC, Kleine-Kohlbrecher D, Colombo E, Pelicci PG, Eilers M. A ribosomal protein L23-nucleophosmin circuit coordinates Mizl function with cell growth. Nat Cell Biol 2008; 10:1051 - 61; http://dx.doi.org/10.1038/ncb1764; PMID: 19160485
- Cuccurese M, Russo G, Russo A, Pietropaolo C. Alternative splicing and nonsense-mediated mRNA decay regulate mammalian ribosomal gene expression. Nucleic Acids Res 2005; 33:5965 - 77; http://dx.doi.org/10.1093/nar/gki905; PMID: 16254077
- Russo A, Catillo M, Esposito D, Briata P, Pietropaolo C, Russo G. Autoregulatory circuit of human rpL3 expression requires hnRNP H1, NPM and KHSRP. Nucleic Acids Res 2011; 39:7576 - 85; http://dx.doi.org/10.1093/nar/gkr461; PMID: 21705779
- Russo A, Siciliano G, Catillo M, Giangrande C, Amoresano A, Pucci P, et al. hnRNP H1 and intronic G runs in the splicing control of the human rpL3 gene. Biochim Biophys Acta 2010; 1799:419 - 28; http://dx.doi.org/10.1016/j.bbagrm.2010.01.008; PMID: 20100605
- Russo G, Cuccurese M, Monti G, Russo A, Amoresano A, Pucci P, et al. Ribosomal protein L7a binds RNA through two distinct RNA-binding domains. Biochem J 2005; 385:289 - 99; http://dx.doi.org/10.1042/BJ20040371; PMID: 15361074
- Wu S, Cetinkaya C, Munoz-Alonso MJ, von der Lehr N, Bahram F, Beuger V, et al. Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter. Oncogene 2003; 22:351 - 60; http://dx.doi.org/10.1038/sj.onc.1206145; PMID: 12545156
- Cen B, Deguchi A, Weinstein IB. Activation of protein kinase G Increases the expression of p21CIP1, p27KIP1, and histidine triad protein 1 through Sp1. Cancer Res 2008; 68:5355 - 62; http://dx.doi.org/10.1158/0008-5472.CAN-07-6869; PMID: 18593937
- Huang L, Sowa Y, Sakai T, Pardee AB. Activation of the p21WAF1/CIP1 promoter independent of p53 by the histone deacetylase inhibitor suberoylanilide hydroxamic acid (SAHA) through the Sp1 sites. Oncogene 2000; 19:5712 - 9; http://dx.doi.org/10.1038/sj.onc.1203963; PMID: 11126357
- Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol 2007; 35:495 - 516; http://dx.doi.org/10.1080/01926230701320337; PMID: 17562483
- Oliver FJ, de la Rubia G, Rolli V, Ruiz-Ruiz MC, de Murcia G, Murcia JM. Importance of poly(ADP-ribose) polymerase and its cleavage in apoptosis. Lesson from an uncleavable mutant. J Biol Chem 1998; 273:33533 - 9; http://dx.doi.org/10.1074/jbc.273.50.33533; PMID: 9837934
- Caldarola S, De Stefano MC, Amaldi F, Loreni F. Synthesis and function of ribosomal proteins--fading models and new perspectives. FEBS J 2009; 276:3199 - 210; http://dx.doi.org/10.1111/j.1742-4658.2009.07036.x; PMID: 19438715
- Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene 2003; 313:17 - 42; http://dx.doi.org/10.1016/S0378-1119(03)00629-2; PMID: 12957375
- Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim Biophys Acta 2010; 1803:673 - 83; http://dx.doi.org/10.1016/j.bbamcr.2009.10.009; PMID: 19879902
- Lempiäinen H, Shore D. Growth control and ribosome biogenesis. Curr Opin Cell Biol 2009; 21:855 - 63; http://dx.doi.org/10.1016/j.ceb.2009.09.002; PMID: 19796927
- Miliani de Marval PL, Zhang Y. The RP-Mdm2-p53 pathway and tumorigenesis. Oncotarget 2011; 2:234 - 8; PMID: 21406728
- Hölzel M, Burger K, Mühl B, Orban M, Kellner M, Eick D. The tumor suppressor p53 connects ribosome biogenesis to cell cycle control: a double-edged sword. Oncotarget 2010; 1:43 - 7; PMID: 21293052
- Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell 2009; 16:369 - 77; http://dx.doi.org/10.1016/j.ccr.2009.09.024; PMID: 19878869
- Donati G, Montanaro L, Derenzini M. Ribosome biogenesis and control of cell proliferation: p53 is not alone. Cancer Res 2012; 72:1602 - 7; http://dx.doi.org/10.1158/0008-5472.CAN-11-3992; PMID: 22282659
- Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat Rev Cancer 2006; 6:663 - 73; http://dx.doi.org/10.1038/nrc1954; PMID: 16915296
- Koutsodontis G, Tentes I, Papakosta P, Moustakas A, Kardassis D. Sp1 plays a critical role in the transcriptional activation of the human cyclin-dependent kinase inhibitor p21(WAF1/Cip1) gene by the p53 tumor suppressor protein. J Biol Chem 2001; 276:29116 - 25; http://dx.doi.org/10.1074/jbc.M104130200; PMID: 11384995
- Gartel AL. The conflicting roles of the cdk inhibitor p21(CIP1/WAF1) in apoptosis. Leuk Res 2005; 29:1237 - 8; http://dx.doi.org/10.1016/j.leukres.2005.04.023; PMID: 15946739
- Chopin V, Toillon RA, Jouy N, Le Bourhis X. P21(WAF1/CIP1) is dispensable for G1 arrest, but indispensable for apoptosis induced by sodium butyrate in MCF-7 breast cancer cells. Oncogene 2004; 23:21 - 9; http://dx.doi.org/10.1038/sj.onc.1207020; PMID: 14712207
- Hsu SL, Chen MC, Chou YH, Hwang GY, Yin SC. Induction of p21(CIP1/Waf1) and activation of p34(cdc2) involved in retinoic acid-induced apoptosis in human hepatoma Hep3B cells. Exp Cell Res 1999; 248:87 - 96; http://dx.doi.org/10.1006/excr.1999.4397; PMID: 10094816
- Wu Q, Kirschmeier P, Hockenberry T, Yang TY, Brassard DL, Wang L, et al. Transcriptional regulation during p21WAF1/CIP1-induced apoptosis in human ovarian cancer cells. J Biol Chem 2002; 277:36329 - 37; http://dx.doi.org/10.1074/jbc.M204962200; PMID: 12138103
- Pacifico F, Crescenzi E, Mellone S, Iannetti A, Porrino N, Liguoro D, et al. Nuclear factor-kappaB contributes to anaplastic thyroid carcinomas through up-regulation of miR-146a. J Clin Endocrinol Metab 2010; 95:1421 - 30; http://dx.doi.org/10.1210/jc.2009-1128; PMID: 20061417
- Iannetti A, Pacifico F, Acquaviva R, Lavorgna A, Crescenzi E, Vascotto C, et al. The neutrophil gelatinase-associated lipocalin (NGAL), a NF-kappaB-regulated gene, is a survival factor for thyroid neoplastic cells. Proc Natl Acad Sci USA 2008; 105:14058 - 63; http://dx.doi.org/10.1073/pnas.0710846105; PMID: 18768801